Abstract
5-Hydroxymethylcytosine (5hmC) is an epigenetic marker that has recently been shown to promote homologous recombination (HR). In this study, we determine the effects of 5hmC on the structure, thermodynamics, and conformational dynamics of the Holliday junction (the four-stranded DNA intermediate associated with HR) in its native stacked-X form. The hydroxymethyl and the control methyl substituents are placed in the context of an amphimorphic GxCC trinucleotide core sequence (where xC is C, 5hmC, or the methylated 5mC), which is part of a sequence also recognized by endonuclease G to promote HR. The hydroxymethyl group of the 5hmC junction adopts two distinct rotational conformations, with an in-base-plane form being dominant over the competing out-of-plane rotamer that has typically been seen in duplex structures. The in-plane rotamer is seen to be stabilized by a more stable intramolecular hydrogen bond to the junction backbone. Stabilizing hydrogen bonds (H-bonds) formed by the hydroxyl-substituent in the 5hmC or from a bridging water in the 5mC structure provide approximately 1.5 to 2 kcal/mol per interaction of stability to the junction, which is mostly offset by entropy compensation, thereby leaving the overall stability of the G5hmCC constructs similar to the GCC core. Thus, both methyl and hydroxymethyl modifications are accommodated without disrupting the structure or stability of the Holliday junction. Both 5hmC and 5mC are shown to open up the structure to make the junction core more accessible. The overall consequences of incorporating 5hmC into a DNA junction are thus discussed in the context of the specificity in protein recognition of the hydroxymethyl substituent through direct and indirect readout mechanisms.
TOC Graphic
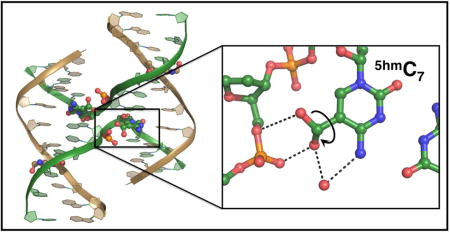
1. INTRODUCTION
Epigenetic modifications to DNA are now recognized as a complementary mechanism to expand and regulate genomic information. For example, 5-methylcytosine (5mC) serves as a mark to target gene silencing in eukaryotes—misregulation of specific gene silencing events can be hugely detrimental, even fatal, to an organism’s development1. A complex system of proteins help to regulate 5mC levels, including modifying the DNA de novo in response to external stimulus2 and maintenance inheritance of the methylation fingerprint from a previous generation of cells3. Although other DNA modifications, adenine methylation4 and N4-methylcytosine5, are found in genomes6, most of the epigenetic research on the mammalian genome has been focused on determining the effects and regulatory mechanisms of 5mC. We show here that 5-hydroxymethylcytosine (5hmC), an epigenetic marker recently shown to promote recombination7, affects the structure and stability of the DNA Holliday junction.
5-Hydroxymethylcytosine (5hmC) is a modified base that was first reported in animal cells in 19728, but it has recently seen renewed interest when Heintz observed its presence in purkinje neurons9. The ten-eleven translocation (Tet) family of dioxygenases generates 5hmC in the cell by oxidating 5-methylcytosine (5mC)10, which can further convert 5hmC to increasingly oxidized formyl- and carboxyl-cytosines11. Standard bisulphite sequencing analysis cannot distinguish between 5hmC from 5mC12. New methods, including Tet-assisted bisulphite sequencing and others, have allowed 5hmC to be mapped in genomic and physiological contexts13–17, which has resulted in a new surge of interest in the effects of 5hmC on biological processes (~94% of all 5hmC-related papers have been published since 2009, according to the Web of Science18). The initial mapping of 5hmC onto specific genomic regions, tissue types, and development stages in both normal and cancerous cells19–24 have implicated 5hmC’s involvement in gene regulation25–27, brain development19,28,29, regulation of 5mC levels30, embryonic development10,21,26, and potentially in regulating homologous recombination (HR) events7,26.
The evidence of its role in HR came initially from the observation that 5hmC’s were enriched in GC-rich regions26, which are associated with recombination hotspots15,31. This theory was further strengthened recently by the studies of Robertson et al.7, which demonstrated that 5hmC promotes homologous recombination in a sequence dependent manner. This effect was seen to be mediated by endonuclease G (Endo G), specifically through recognition and binding of the sequence 5′-GGGG5hmCCAG-3′/5′-CTGGCCCC- 3′ to induce double strand breaks that then trigger the actions of the cell’s recombination machinery. The question we raise here is whether and how 5hmC affects the structure and stability of the Holliday junction, the four-stranded DNA structure that is the intermediate formed during homologous recombination events32.
The formation of Holliday junctions has been shown to be sequence-dependent in crystals33 and in solution34. Junctions exist in two functional forms: the open-X and the stacked-X structures35. The open-X form takes a classical “cruciform DNA” shape, and allows the junction to isoenergetically migrate along stretches of DNA sequence during HR. This form of the junction is seen under low salt conditions in DNA only constructs, or in complex with proteins that require migration of the junction in order to locate a specific recombination site (as in the RuvABC DNA repair system36).
The stacked-X junction is essentially two continuous duplexes interrupted by the crossovers that connect the adjacent duplexes. The stacked-X form is observed in DNAs under high salt conditions and, since it is topologically locked and cannot migrate, is seen in complexes with sequence independent resolvases (such as the T7 bacteriophage endonuclease I37 or the T4 bacteriophage endonuclease VII38). The crystal structures of DNA only constructs have revealed that the stacked-X junction is stabilized by a trinucleotide core, a three nucleotide sequence that defines the cross-over point between adjacent duplexes of the junction. The sequence preference within this trinucleotide core is A > G > C at the first position, a C > T at the second, and C required at the third33. The specificity at the second and third positions are attributed to a unique set of hydrogen bonds (H-bonds) that form between the amino group of the cytosine bases and oxygens from the adjacent residue’s backbone phosphate group, helping to mitigate the interphosphate electrostatic repulsion along the DNA backbone as it makes the tight U-turn that connects the two adjoining duplexes of the junction.
It was interesting to us that the Endo G recognition sequence identified by Robertson, et al7 contained the sequence motif G5hmCC, a 5hmC-modified version of the GCC trinucleotide core shown previously to stabilize junctions33. This hydroxymethyl group is potentially positioned to displace the H-bond that stabilizes the stacked-X junction structure39. The question we posed is whether the a hydroxymethyl group introduced at this position would sterically interfere with this important interaction or, since it is an H-bond donor itself, supplant this interaction, and how these perturbations would affect the conformation and stability of the junction as a whole. As a control, we compare the effects of 5hmC with the methylated variant (5mC), which would have similar steric effects, but cannot form an H-bond.
2. METHODS
Oligonucleotides
DNA were designed as self-complementary decanucleotide sequences in the motif 5′-CCGGCGXCGG-3′ (X is C, 5mC, or 5hmC), previously shown to form junctions in the presence of monovalent and divalent cations33.
Oligonucleotides were purchased from Midland Certified Reagent Company with the 5′-dimethoxytrityl (DMT) protecting group intact and remaining attached to the CPG solid support bead to facilitate purification. The CPG was removed by suspension in ammonium hydroxide and the full-length products were isolated by reverse phase HPLC on a C18 column, taking advantage of the additional hydrophobicity of the 5′ DMT. The DMT group was cleaved by resuspending the oligos in 3% acetic acid, and the final product desalted by size exclusion chromatography off a Sephadex G-25 column.
Crystallography and Structure Analysis
Crystals were grown in sitting drop trays, with 8–10 μL sample volumes containing 0.78 mM DNA (not annealed), 25 mM sodium cacodylate pH 7.0, calcium chloride (ranging from 1–15 mM) and spermine (ranging from 0.1–2.0 mM), and equilibrated against reservoir solution of 25% 2-methyl-2,4-pentanediol (MPD). These crystallization drop conditions were chosen for screening because of their propensity to yield both duplex and junction DNA crystals33.
Data were collected using a Rigaku Compact Home Lab equipped with a PILATUS detector; HLK300040 was used to index, integrate, and scale the data. The structures were solved by molecular replacement (using the GCC core junction as the starting search model, PDB 1P4Y33) and subsequently refined using Phenix41. Standard Phenix occupancy refinement routines were used to determine the occupancy of each rotamer for the 5hmC hydroxyl group. DNA structure measurements (rise, twist, slide, etc.) were performed with CURVES+ DNA structure analysis program42, and junction structure parameters (Jroll, and Jtwist) were calculated according to the methods described by Watson et al.43.
Melting Profiles by Differential Scanning Calorimetry (DSC)
DSC samples were prepared by annealing 25 μM DNA in 15 mM calcium chloride and 50 mM sodium cacodylate (pH 7.0) at 90°C for 20 min, and allowed to slowly cool over 2 hours. The DNA melting data were collected using a TA Instruments Nano DSC with 900 sec of equilibration, and scanning from 5–105°C at a rate of 1°C/min at a constant pressure of 3.0 ATM. Melting temperatures (Tm) and enthalpies of melting (ΔHm) were determined by fitting the data with TA Nano Analyze software using a two-component (junction and duplex), two-state scaled model. Each construct was measured through at least 18 replicates. Melting energies were extrapolated to a standard 25°C temperature, and the duplex melting energies were subtracted from the junction to determine the stabilization energy of the junction core44,45.
Quantum Mechanical (QM) Calculations
QM calculations were performed using Gaussian0946 at Møller-Plesset 2 (MP2) level, using the 6–31++G** basis set. Cyclohexane (ε=2) was chosen as the solvent in order to mimic the semi-sequestered and hydrophobic environment of the junction core, and a counterpoise (BSSE) correction was applied from a gas phase calculation. Geometry scanning calculations (5° increments) were first performed on the in-context dinucleotide (G6-5hmC7) to determine the minimum-energy orientation of the hydrogen from the 5hmC’s hydroxyl group for each isomer resolved in the crystal structure. To determine relative rotamer stability of the crystallographic structures, energy calculations were performed on the isolated 5hmC bases, including the optimized hydrogen positions for the in-context crystal structure (Figure 3). Dimethylphosphate was chosen to mimic the DNA backbone in the calculation to determine the Phos6—5hmC7 hydrogen bond (H-bond) energy for each 5hmC rotamer.
Figure 3.
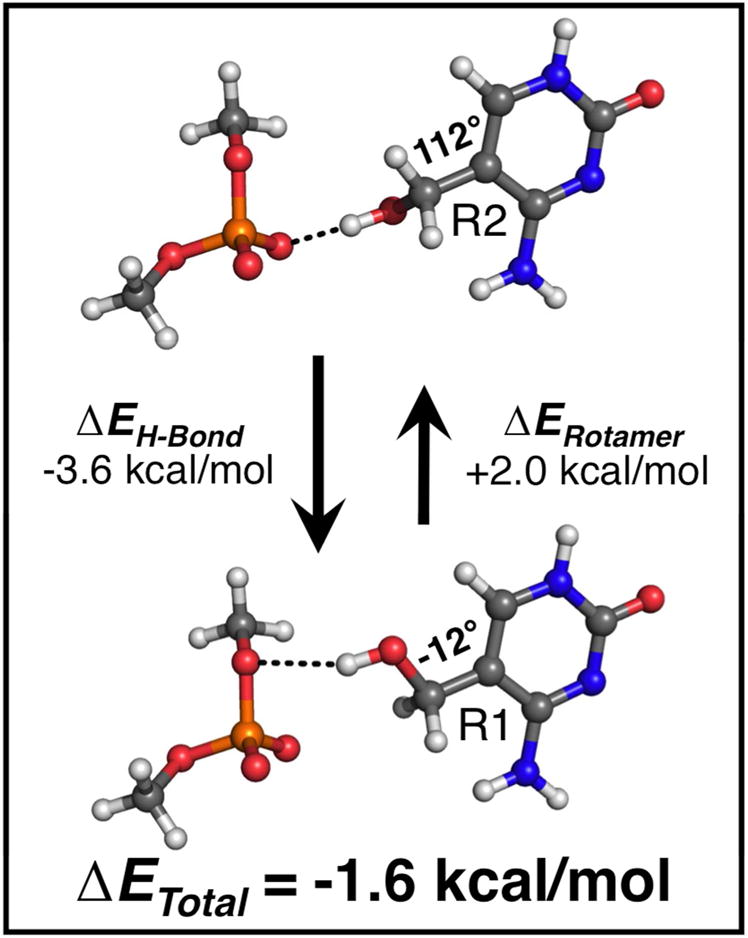
Comparison of H-bond energies (ΔEH-Bond) and rotamer energies (ΔERotamer) between the major (R1, bottom) and minor (R2, top) conformations of the hydroxymethyl substituent in the 5hmC structure. Quantum mechanical (QM) energies were calculated on small molecule models of the junction core (5hmC and dimethylphosphate), constructed from atomic coordinates taken from the crystal structure. The isolated 5hmC base has a 2.0 kcal/mol energy preference towards the R2 rotamer (112°) in the bond rotation energy. However, H-bonding interaction energy was calculated to favor the R1 rotamer (−12°) by −3.6 kcal/mol (signs of the energy terms are defined as the difference ER1 – ER2). In summation, the dominant R1 rotamer is favored by an overall energy (ΔETotal = ΔERotamer + ΔEH-Bond) of −1.6 kcal/mol.
3. RESULTS & DISCUSSION
The initial premise of the current study is that the hydroxyl group of 5hmC could form an H-bond to supplant or supplement an interaction that had previously been shown to stabilize and, thus, infer sequence specificity to the four-stranded Holliday junction. We have determined the structural effects of this epigenetic modification by determining the crystallographic structure of 5hmC in the self-complementary sequence d(CCGGCG5hmCCGG). We designed this sequence motif around a GCC trinucleotide core (in bold italics), which had previously been shown to be amphimorphic (capable of forming either B-DNA duplexes or four-stranded HJs, depending on cations)33; thus, this sequence, as opposed to a strictly junction forming ACC core, would be very sensitive to any destabilizing effects of the substituents on the junction. In addition, we apply differential scanning calorimetry to correlate the structural effects on the overall stability of the junction, and interpret these energies in terms of contributions of the molecular interactions on the enthalpic and entropic effects locally and globally. A parallel study on the methylated sequence d(CCGGCG5mCCGG) allowed us to distinguish between contributions from steric and hydrogen bonding interactions.
5hmC and 5mC Modifications Are Structurally Accommodated in the Holliday Junction Core
The first observation from the crystal structures of d(CCGGCG5hmCCGG) and d(CCGGCG5mCCGG) is that both the bulky methyl and hydroxymethyl substituent groups are accommodated at the key stabilization trinucleotide of the stacked-X form of the HJ (Supplementary Table 1, Figure 1). Both sequences conform to the overall conformation of the stacked-X junction as seen in previous crystal structures33, with the DNA forming two sets of near continuous double-helices, interrupted by the crossing of the phosphodiester bond at nucleotide 6, which connects the helices to form the four-stranded junction (Figure 1). The assembly of these self-complementary sequences results in junctions in which the methyl or hydroxymethyl modifications sit at two unique nucleotide positions and, thus, experience two unique structural environments. In each structure, either a 5mC or 5hmC base sits on a continuous strand of an uninterrupted B-type duplex region, while the second similarly modified base sits at the crossing strand that joins the two duplexes of a junction. The two positions allow us to compare and contrast the effects of each modification in the conformation of the HJ relative to that of a standard B-DNA duplex within the same structure.
Figure 1.
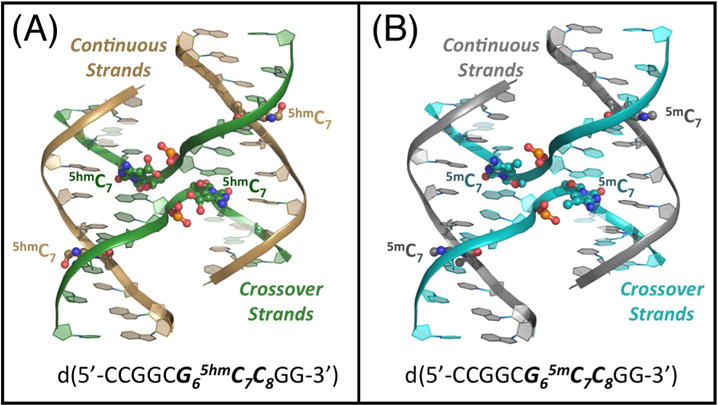
Comparison of the structures of 5-hydroxymethylcytosine (5hmC) and 5-methyl cytosine (5mC) in DNA Holliday junctions. (A) The crystal structure of the hydroxylmethylated sequence d(CCGGCG65hmC7C8GG) is shown with the DNA backbones traced as ribbons (colored gold for the outside continuous strands and green for the junction crossing strands). The 5hmC bases, along with the phosphate groups that they are H-bonded to, are rendered as ball-and-stick models, with the carbon atoms of the nucleotides along the continuous strand colored gold and those at the junction colored green. The 5hmC bases on the crossover strands have hydroxyl groups that occupy two rotamer conformations, both shown on the image. (B) The DNA backbone of the methylated sequence d(CCGGCG65mC7C8GG) is traced as ribbons (grey along the outside continuous strands and blue along the junction crossing strands). The ball-and-stick models of the 5mC bases and their interacting the phosphate groups are colored grey on the continuous and blue on the junction crossing strands.
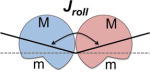
The global structures of HJ’s are described by the geometric relationships between the two sets of interconnected double-helices (Table 1, where Jtwist and Jroll define the angular relationships of the helical axes and in the plane perpendicular to their axes, respectively)43, which reflect the accessibility of the junction-cross over to the environment. A comparison of the G5hmCC and G5mCC to the parent GCC33 sequences (referring to the core trinucleotide of each sequence) shows progressively larger Jtwist and Jroll values as the size of the substituent group (H to CH3 to CH2OH) increases, resulting in a more open and potentially more accessible overall structure of the junction.
Table 1.
Structural parameters of GCC33, G5mCC, and G5hmCC Holliday junctions. Parameters that describe the helical structure42 around the modified C7 cytosine of the crossover GCC trinucleotide core are listed. The standard values for these parameters in B-DNA are shown in parentheses33. The overall conformation of the junction are reflected in the parameters Jroll and Jtwist (schematics for these two are shown at the bottom, adapted from Watson et. al.43).
| DNA Core: | GCC | G5mCC | G5hmCC |
|---|---|---|---|
|
| |||
| Rotational Parameters | |||
|
| |||
| Helical Twist (34.7) | 37.3° | 34.5° | 34.4° |
| Propeller Twist (−12.0) | −20.1° | −17.6° | −14.2° |
| Tilt (−0.62) | −0.7° | 4.9° | 5.0° |
| Roll (1.74) | −2.7° | 0.9° | 3.6° |
| Buckle (−0.23) | 4.8° | −0.8° | 5.3° |
| Opening | 1.6° | 1.6° | 0.2° |
|
| |||
| Translational Parameters | |||
|
| |||
| Rise (3.30) | 3.52 Å | 3.49 Å | 3.41 Å |
| Slide (0.66) | 0.84 Å | 0.38 Å | 0.51 Å |
| Shear | 0.14 Å | −0.10 Å | −0.03 Å |
| Stretch | 0.08 Å | −0.08 Å | −0.07 Å |
| Stagger | 0.24 Å | 0.23 Å | 0.42 Å |
| Shift | −0.43 Å | 0.75 Å | 0.82 Å |
|
| |||
| Junction Parameters | |||
|
| |||
| Jroll | 135.31° | 143.04° | 150.46° |
| Jtwist | 39.95° | 40.81° | 41.10° |
|
| |||
|
|
||
A more detailed analysis of the crystal structures (Table 1) shows that the methylated and hydroxymethylated bases at the N7 position conform adopt more “ideal” geometries associated with B-DNA double-helices than the unmodified junction. In particular, the 5hmC·G shows reduced shear, propeller twist, and opening of the base pair (Table 1), and both the methylated and hydroxymethylated base pairs show helical twists that are typical of a ~10.4 bp/turn repeat as compared to the overwound 9.7 bp/turn for the unmodified structure. These analyses suggest that the direct H-bonding from C7 to the phosphate of G6 that stabilizes the unmodified GCC structure33 (Figure 2A) induces distortions to the natural geometric tendencies of stacked B-DNA base pairs, and that methylation or hydroxymethylation at this base helps to relieve some of the local conformational stress by breaking the direct H-bonding interaction of the amine.
Figure 2.
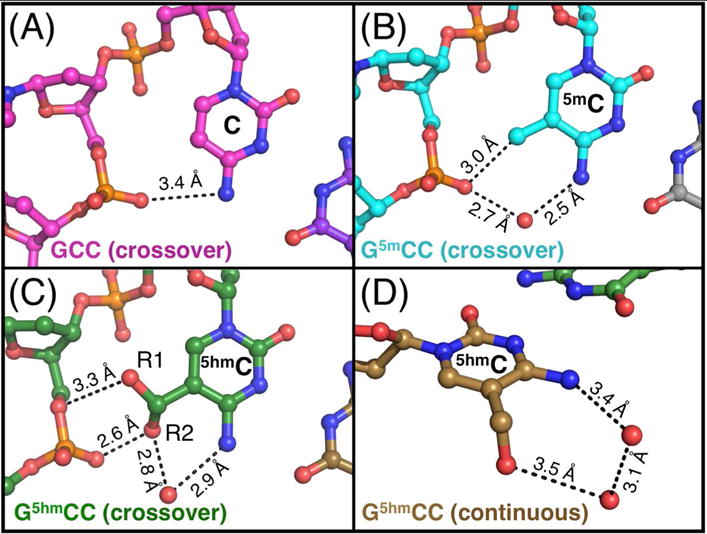
Structures of GCC (A), G5mCC (B), and G5hmCC (C, D) trinucleotide cores of DNA junctions. (A) GCC core structure (PDB entry 1P4Y2) is stabilized by an H-bond from the N4-amine of the C7 base to the neighboring G6 phosphate. No waters are observed within H-bonding distance to the base. (B) The methyl of the 5mC7 sterically interferes with the amine’s direct H-bond, which is replaced by an H-bonded water that bridges between the amine and the phosphate. The methyl group is within H-bonding distance to the phosphate, likely indicating a weak attractive force. (C) 5hmC7 stabilizes the junction core by displacing the amine to allow the hydroxyl group to H-bond with the G6 phosphate. The hydroxyl group is observed in two orientations, with the dominant rotamer in plane of the base and the minor rotamer 112° out of plane. The rotamers interact with two different oxygens on the phosphate. A water (red sphere) is held in place by H-bonds to the hydroxyl and amine in the minor form. (D) 5hmC on the continuous (not junction-stabilizing) strand adopts an out-of-plane hydroxyl position, similar to those seen in previous B-DNA structures. Two waters (red spheres) are within H-bonding distance to the base.
In the 5mC structure, the direct N4-amino to phosphate oxygen H-bond is now displaced by the methyl group, and is replaced by a water mediated interaction (Figure 2B). In the G5hmCC structure, the hydroxymethyl substituent was seen to occupy two distinct rotamer conformations (Figure 2C). The major rotamer form (R1, representing ~2/3 of the structure) sits in the plane of the cytosine base and is H-bonded to the O5′-oxygen of nucleotide G6. The minor rotamer (R2, which accounts for 1/3 of the structure) is rotated 112° out-of-plane, in a position similar to the conformation of the 5hmC on the outside continuous strand (Figure 2D) and to previous rotamers seen in B-DNA duplexes47,48. In the R2 rotamer, the OH forms an H-bond to non-linkage oxygen of the G6 phosphate, and is bridged to the N4 amino group of the cytosine base by a water. Similar waters are seen coordinated to the N4 amine and hydroxyl on the continuous strand 5hmC residue. Thus, both rotamer forms of the 5hmC place the OH in position to form an H-bond that replaces standard interaction of the N4 amine. The question is, what factors determine which conformation is dominant?
Hydroxymethyl rotamers in the 5hmC junction
It is clear from previous structures of 5hmCs in duplex DNAs47,48 that there is a rotational bias to position the hydroxyl substituent in an out-of-plane geometry. From quantum mechanical studies47, the perpendicular out-of-plane rotamer is the global energy minimum, and is ~2 kcal/mol more stable than an in-plane form, which sits at a local minimum (Supplemental Figure S1). This torsional preference explains why the 5hmC along the continuous strand of the junction is in the out-of-plane geometry, just as it has been seen in the structures of the DNA duplexes (Table 2). Spingler et al.47 suggested that the bridging waters add very little to the preference of this torsionally preferred rotamer.
Table 2.
Torsion angles relating atoms C6–C5–C5A–O5 of the 5hmC bases in cross-over and continuous strands in the current junction structure, and in B-DNA duplexes from the literature. A torsion angle of 0° indicates the hydroxyl is in plane with the base and pointed towards the glycosidic bond, while a 180° angle points the hydroxyl towards the N4 amine group. Positive angles place the hydroxyl above the plane of the base in the 5′ direction and negative angles are in the 3′ direction.
| Junction DNA (PDB 5DSB) | |
|
| |
| Crossover Strand (R1) | −12.0° |
| Crossover Strand (R2) | 111.4° |
| Continuous Strand | 92.3° |
|
| |
| Published out of plane rotamers | |
|
| |
| 4HLIa | 132.8° |
| 4GLCa | 126.9° |
| 4GLCa | 96.1° |
| 4GLHa | 115.7° |
| 4GLHa | 109.8° |
| 4I9Vb | 111.0° |
| 4I9Vb | 112.1° |
| Average (Standard Deviation) | 114.9° (12.0°) |
|
| |
| Published rotamer outliers | |
|
| |
| 4HLIa | 72.6° |
| 4I9Vb | 24.7° |
Renciuk, D. et al. Nucleic Acids Res. 2013
Szulik, M.W. et al., Biochemistry 2015
The relatively small difference in occupancy between R1 and R2 at the junction’s crossover in the current structure suggests that additional interactions, in this case H-bonds to the phosphate group, could readily shift the rotamer preference. We therefore applied an MP2 calculation on the two rotamer forms of 5hmC at the H-bonding geometries seen in the crystal structures, using a dimethylphosphate as a model for the H-bond acceptor of the junction backbone (Figure 3), to compare their H-bonding energies (ΔEH-Bond). From this calculation, we estimate ~3.5 kcal/mol difference in ΔEH-Bond that favors the R1 over R2 rotamer. The two contributing energies (intrinsic torsional energy versus H-bonding) oppose each other, resulting in an overall preference for R1 by ~1.5 kcal/mol, which would explain the approximate two-fold preference for this rotamer in the crystal structure.
Energetic effects of hydroxymethyl and methyl substituents in solution
With the atomic details elucidated, we then asked whether and how the various interactions observed in the crystal structures (the direct hydroxyl H-bonds in G5hmCC and the water-mediated H-bond seen in the G5mCC structures) confer stability to the HJ in solution. We had previously shown that the sequence-dependent formation of HJs identified in crystals translates well to the stability of junctions in solution34. In the current study, we can directly apply differential scanning calorimetry (DSC) to determine the effects of these molecular interactions on the melting energies and, thus tease out their effects on the stabilization of the four-stranded junction45,49.
In order to determine the effects of hydroxymethyl or methyl modifications on the DSC energies, we take advantage of the concentration dependence for the formation of four-stranded junctions by self-complementary decanucleotides, in which DNAs, at lower concentrations show melting parameters of duplexes, while higher concentrations reflect those of junctions50. We chose a DNA concentration for our DSC studies that showed both duplex and junctions in solution, thereby allowing us to measure the energies of the DNA species simultaneously (supplemental figure 2). Since the stacked-X junction is essentially composed of two duplexes and the interruption of the crossover region, the difference between junction and duplex DSC energies (scaled per two strands of DNA) isolates the stabilization energy associated with just the interactions at the junction core. In this way, we were able to determine the energetic contributions (ΔH, ΔS, and ΔG) of the core trinucleotides to junction stabilization. Furthermore, by subtracting the thermodynamic values for GCC from either those of G5hmCC or of G5mCC, we can specifically determine the effect of each substituent at the C7 nucleobase on the stability of the junction.
DSC melting profiles for each construct were best fit using a two-component analysis, indicating the presence of both duplex and junction DNA in each sample. An analysis of the melting temperatures shows that cytosine methylation has an overall effect of stabilizing the duplex (increased Tm, Table 3) relative to the unmodified DNA, while hydroxymethylation slightly destabilizes the duplex. We see very similar effects of the substituents on the Tms for the junction, where the methyl group is associated with the highest Tm, and the hydroxymethyl with the lowest. However, when we subtract the Tms of each duplexes from those of the junctions, we see that the methylcytosine results in a smaller difference in ΔTm compared to the native GCC sequence, suggesting that methylation has a destabilizing effect on the junction. In contrast, this analysis of ΔTm suggests that hydroxymethylation would have a slightly stabilizing effect on the Holliday junction relative to its duplex. This is consistent with the hydroxyl groups forming additional stabilizing H-bonds to the junction core. The magnitude of the ΔHm difference between the junction and duplex forms of the hydroxymethylated G5hmCC construct (ΔΔHm = 30.6 kcal/mol) is indeed larger than that of the parent GCC (27.8 kcal/mol).
Table 3.
The melting temperature (Tm) and melting enthalpy (ΔHm) measured by DSC of GCC, G5mCC, and G5hmCC core DNA constructs in solution. The entropy of melting (ΔSm) for each construct is calculated from the Tm and ΔHm by ΔSm = ΔHm/Tm, with the assumption that at the melting temperature, the concentrations of folded and denatured DNA are equal and, thus, ΔGm = 051.
| Duplex | Junction | J–D | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| DNA Core: | Tm (°C) | ΔHm (kcal/mol) | ΔSm (cal/mol K) | Tm (°C) | ΔHm (kcal/mol) | ΔSm (cal/mol K) | ΔTm (°C) |
| GCC | 68.01 ± 0.11 |
81.7 ± 0.7 |
239 ± 2 |
73.50 ± 0.08 |
109.5 ± 1.4 |
316 ± 4 |
5.49 ± 0.14 |
| G5mCC | 70.06 ± 0.10 |
68.1 ± 1.0 |
198 ± 2 |
75.20 ± 0.06 |
98.7 ± 1.0 |
284 ± 3 |
5.14 ± 0.12 |
| G5hmCC | 65.03 ± 0.09 |
66.7 ± 0.5 |
196.9 ± 1.6 |
70.60 ± 0.05 |
97.3 ± 0.7 |
283 ± 2 |
5.57 ± 0.10 |
We find that the G5mCC DNA constructs are the most thermally stable (highest Tm) of the species studies. At the Tm, GCC stabilization was the most enthalpically driven, in contrast to larger entropic stabilization of the G5hmCC. The duplex and junction constructs follow similar trends with respect to their relative melting parameters (Table 3), suggesting the energetic effects of the modified bases are similar in both duplex and junction.
A better measure for the effect of each substituent on the energetics of the junction is to determine the ΔΔG° relative to the duplex at a standard temperature (25°C). In order to determine the interaction energies of each substituent group in the DNA junction45,49, we first extrapolate the DSC energies to a common reference temperature (25°C) using standard relationship (eqs. 1 and 2). Following those extrapolations, the duplex energies were subtracted from those of the junctions, leaving only the junction core stabilization energy. Finally the GCC core energy was subtracted from the modified cores (G5mCC and G5hmCC), reported as ΔΔH25°C, ΔΔS25°C, and ΔΔG25°C, to narrow the analysis to the specific interaction energies associated with each modification (methylation or hydroxymethylation, Table 4).
| eq. 1 |
| eq. 2 |
Table 4.
Thermodynamic stabilization of G5mCC and G5hmCC junction cores relative to the GCC junction core. The enthalpic, entropic, and overall free energies (at 25°C) for each modified construct are listed with the values from the parent construct subtracted (ΔΔH25°C, ΔΔS25°C, and ΔΔG25°C, respectively). Values reflect stabilization per interaction at each crossover strand; therefore, each complete junction structure is stabilized by twice the tabulated energies.
| DNA Core | ΔΔH25°C (kcal/mol) | ΔΔS25°C(cal/mol·K) | ΔΔG25°C (kcal/mol) |
|---|---|---|---|
| G5mCC - GCC | −2.1 ± 1.0 | −6 ± 3 | −0.22 ± 0.13 |
| G5hmCC - GCC | −1.5 ± 0.9 | −5 ± 3 | −0.01 ± 0.12 |
The most immediate observation is that methylation or hydroxymethylation has little effect on the overall free energies (ΔΔG25°C ≈ 0), indicating that the modified bases cause minimal disruption to the stability of the Holliday junction. However, we observed compensatory enthalpic and entropic effects, which contribute to these very small ΔΔG25°C values. The G5hmCC and G5mCC gain 1.5 and 2.0 kcal/mol of enthalpic energy respectively (calculated per interaction, meaning twice this energy is stabilizing the whole junction), which suggests either stronger core H-bond stabilization, or reduction in conformational strain on the residue 7 base pair (Table 1).
The stabilizing enthalpies are compensated by unfavorable energy from entropic terms (−5 or −6 cal/molK, equivalent to ~1.5 kcal/mol of unfavorable energy at 25°C) in the modified constructs. We had seen this type of enthalpy-entropy compensation previously in a DNA junction that is stabilized through halogen bonds49. In this latter case, we attributed the loss in entropic stabilization to reduced dynamics, as reflected in the smaller B-factors associated with the nucleotide bases and phosphates that were involved in the stronger molecular interaction. A similar B-factor analysis on these structures, however, showed that restriction of the conformational dynamics from stronger molecular interactions is not the rationale for the loss in entropy in the G5hmCC and G5mCC structures. A comparison of B-factors indicates that the modified constructs are more locally dynamic at the junction core than the unmodified junction (Figure 4). The GCC construct shows the pattern that is typical of H-bond stabilized junctions, where the B-factors for nucleotides 6 to 8 (where the stabilizing H-bonding interactions occur) are lower than the overall junction. This same pattern is also seen with the G5hmCC and G5mCC structures; however, the modifications result in less constrained atoms at the junction core compared to the GCC structure, particularly at base of C7 and the phosphate of position 6, the specific positions involved in the 5hmC or 5mC interactions.
Figure 4.
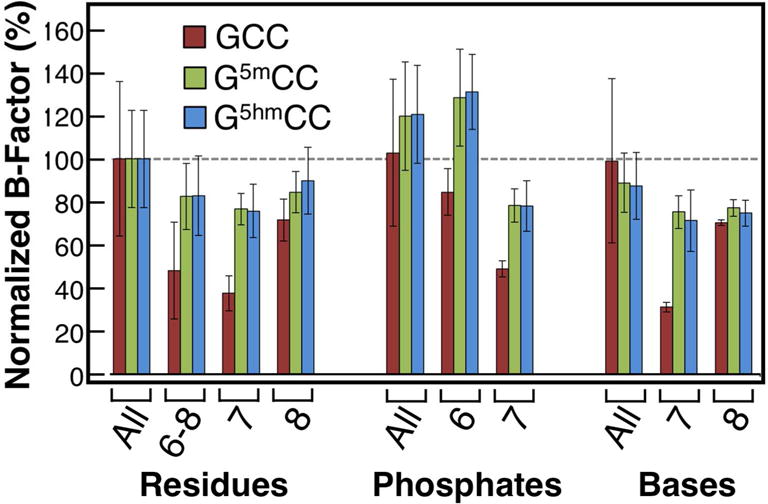
Normalized temperature factors of DNA junctions for the unmodified (GCC), methylated (G5mCC), and hydroxymethylated (G5hmCC) structures. Temperature factors (B-factors) were normalized to the average value for the non-solvent atoms in each structure (100%=average), and each structure was normalized on its own scale independently of the others. Error bars represent the standard deviation of B-factors for the atoms in the selected group.
The H-bond associated with the C8 amino to the C7 phosphate that is essential for the stabilizing the junction continues to constrain the dynamics of these interacting groups relative to the overall junction. The methyl and hydroxymethyl modifications, however, do appear to also increase the dynamics of the C8 nucleotide and the C7 phosphate, indicating that these substituents do affect the overall conformational dynamics of the entire junction core. It is clear, therefore, that the entropic compensation for the stabilizing enthalpy of folding does not come explicitly from loss in conformational dynamics of the nucleotides involved in the H-bonding interactions.
4. CONCLUSIONS
The recent evidence that 5hmC promotes recombination7,26 prompted us to study the impact of this base modification on the structure and stability of the DNA Holliday junction, and consider its potential impact on HR. We modified C7 cytosine of the G6C7C8 trinucleotide core (to form G5hmCC) of the sequence d(CCGGCGCCGG), a construct that is sensitive to environmental effects to junction stability33. As a steric control, we also considered the effects of cytosine methylation at this same cytosine position on the properties of the junction. We show that there is minimal effect of either the hydroxymethyl or methyl substituent on the overall thermodynamic stability of the junction, although the general structure becomes more open, leaving the trinucleotide core more accessible.
The H-bond from the C7 base to the G6 phosphate, which helps define the sequence dependence of junction formation33,39, is seen to be disrupted in both modified constructs, with the hydroxyl group of the 5hmC providing compensatory H-bonds. In this case, the hydroxymethyl adopts two different rotamer conformations, with the prevalent interaction being associated with a less favorable rotation. We thus see that, although there is a preferred intrinsic rotamer for the 5hmC substituent, as seen here and in previous structures of B-DNA duplexes47,48, a strong intramolecular interaction can overcome the energy barrier for the hydroxymethyl to adopt a less favored rotation. In the case of the 5mC construct, the lost H-bond of the native GCC core is replaced by a water, which serves to bridge the N4 amino of the cytosine back again to the G6 phosphate. Such water mediated H-bonds have been shown to compensate well for direct H-bonds in DNA, for example, in providing stability to GT mismatches relative to standard GC Watson-Crick base pairs52. The resulting compensatory H-bonds (directly from the hydroxymethyl of 5hmC or through water mediation in the 5mC construct) resulted in a slight enthalpic stabilization of the GCC trinucleotide core in the junction.
The enthalpic stabilization in both the 5hmC and 5mC construct junctions is counter balanced by losses in entropic stabilization. We had previously seen this entropy-enthalpy compensation effect when a halogen bond was engineered to stabilize the DNA junction, with the energetically stable halogen bond resulting in a less dynamic junction core49 (as reflected in the reduced B-factors of the core). The increased conformational dynamics for the 5hmC and 5mC modified junctions, as reflected in crystallographic B-factor analysis, however was initially perplexing, as it appears to be in contrast with the decreased entropy of these constructs as measured by DSC. Clearly, the entropic penalty for folding is not associated with reduced conformational dynamics resulting from stabilization of the junction core, specifically by the methyl or hydroxymethyl groups. For the 5hmC base, some entropy loss may be attributed to constraining the hydroxyl substituent to the two specific rotamer conformations required to form the H-bonds to the junction backbone, which would impose an entropic penalty relative to the range of energetically favorable rotamers observed for the unconstrained 5hmC base (Table 2). As the C and 5mC do not have multiple rotational states available, the H-bond conformation is not a constraint. One likely explanation is a change in the solvent entropy due to constrained water molecules around the junction core. In the G5mCC crystal structure, we observe a highly structured water molecule bound near the junction core, and this water is absent in the native GCC core structure. Similarly, a highly structured water is observed in the 5hmC junction, but in this case, the water bridges between the N4 amino and the OH of the hydroxymethyl substituent and, thus, does not help to stabilize the overall junction.
The G5hmCC and G5mCC junction cores are quite different from the GCC core from a structural perspective, both in terms of direct and indirect readout implications. A hypothetical resolvase recognizing the junction core could distinguish the different cores, and hence this would be a TET-regulated control for sites of HR. In terms of indirect readout, the steric bulkiness of the 5hmC and 5mC do impact the overall junction structure by opening up the junction and relieving some strain on the contorted base pairs and backbone that kink to enable junction formation. This opening of the junction provides more space between the two duplex arms, possibly facilitating the ability of a protein to probe for specific interactions at that site.
Although the enthalpy-entropy compensation does not result in an overall more stable Holliday junction, it may affect the kinetics of junction migration, which in turn would affect the role of both the hydroxymethyl and methyl modifications on homologous recombination. Khuu et al.53 proposed a model in which the sequence specificity of junction-cleaving proteins (resolvases) results from pausing migration at sequences that help stabilize the stacked-X junction structure. The kinetics of pausing, however, may not be reflected in the overall free energy of the stacked-X junction, but in the energetic barriers. The increased H-bonding interactions in both the 5hmC and 5mC junctions, thus, would provide such barriers, which may slow the migration of the junction away from the GCC core and provide sufficient time for a resolvase to indirectly recognize these modifications. It would be interesting to determine the effects of these epigenetic markers on the kinetics of junction migration and explore the concept of sequence-dependent pausing.
To conclude, we see that the methyl substituent pushes the C·G base pair away from the junction cross-over, resulting in a more open structure, as reflected in the larger Jroll. The hydroxymethyl has an even greater effect. Given that select few sequences are capable of stabilizing a stacked-X junction33, there is great potential for direct as well as indirect readout of these base modifications, which distort the stacked-X structure without dismantling it. In the context of the Khuu model53, the H-bonds of 5hmC could kinetically pause migration, as discussed, while the more open junction provides access for a protein to directly recognize the modified base, with the alternative rotamer allowing the hydroxymethyl group in the junction to be distinguished from the standard rotamer in a B-DNA duplex.
Supplementary Material
Acknowledgments
Dr. Adam Robertson was supported in part as a member of Professor Arne Klungland’s research group at the Oslo University Hospital.
FUNDING: Research reported in this publication was supported by grants from the National Science Foundation (MCB-1515521) to PSH, a predoctoral fellowship from the National Institute of General Medical Sciences of the National Institutes of Health (F31GM113580) to CMVZ, and a start-up grant from the South-Eastern Norway Regional Health Authority (Helse Sør Øst, project number 2014017) to ABR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, the National Institutes of Health, or the South-Eastern Norway Regional Health Authority.
Footnotes
Supporting Information: Contains table of crystallographic information (PDF format), and CIF files for each structure (PDB IDs 5DSB and 5DSA)
The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic Cytosine Methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ, van der Woude M, Braaten B, Low D, LeClerc JE, Li B, Payne WL, Cebula TA, Braaten BA, Nou X, Kaltenbach LS, Low DA, Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ, Mahan MJ, Slauch JM, Mekalanos JJ, Roland KL, Martin LE, Esther CR, Spitznagel J, Vescovi EG, Soncini FC, Groisman EA, van der Woude M, Hale WB, Low DA, Hale WB, van der Woude MW, Low DA, Tavazoie S, Church GM, Bandyopadhyay R, Das J, Julio SM, Conner CP, Heithoff DM, Mahan MJ, Slauch JM, Silhavy T, Smith CL, Cantor CR, Marinus MG, Poteete A, Arraj JA. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich M, Wilson GG, Kuo KC, Gehrke CW. N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol. 1987;169:939–943. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanyushin BF. Adenine Methylation in Eukaryotic DNA. Mol Biol. 2005;39:473–481. [Google Scholar]
- 7.Robertson AB, Robertson J, Fusser M, Klungland A. Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination. Nucleic Acids Res. 2014;42:13280–13293. doi: 10.1093/nar/gku1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S-G, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson AB, Dahl JA, Vågbø CB, Tripathi P, Krokan HE, Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson AB, Dahl JA, Ougland R, Klungland A. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat Protoc. 2012;7:340–350. doi: 10.1038/nprot.2011.443. [DOI] [PubMed] [Google Scholar]
- 16.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tetassisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuters T. Web of Science [Google Scholar]
- 19.Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104:341–346. doi: 10.1016/j.ygeno.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruzov A, Tsenkina Y, Serio A, Dudnakova T, Fletcher J, Bai Y, Chebotareva T, Pells S, Hannoun Z, Sullivan G, Chandran S, Hay DC, Bradley M, Wilmut I, De Sousa P. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irier H, Street RC, Dave R, Lin L, Cai C, Davis TH, Yao B, Cheng Y, Jin P. Environmental enrichment modulates 5-hydroxymethylcytosine dynamics in hippocampus. Genomics. 2014;104:376–382. doi: 10.1016/j.ygeno.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putiri EL, Tiedemann RL, Choi JH, Robertson KD. Abstract 2319: Dynamics of TET methylcytosine dioxygenases in 5-methylcytosine and 5-hydroxymethylcytosine patterning in human cancer cells. Cancer Res. 2014;74:2319–2319. [Google Scholar]
- 24.Pfeifer GP, Xiong W, Hahn Ma, Jin SG. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 2014;356:631–641. doi: 10.1007/s00441-014-1896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson J, Robertson AB, Klungland A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochem Biophys Res Commun. 2011;411:40–43. doi: 10.1016/j.bbrc.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 26.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, Yu M, Liu X, Zhu P, Li X, Hou Y, Guo H, Wu X, He C, Li R, Tang F, Qiao J. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. 2014;15:1–17. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun W, Zang L, Shu Q, Li X. From development to diseases: The role of 5hmC in brain. Genomics. 2014;104:347–351. doi: 10.1016/j.ygeno.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, Wu H, Warren ST, Jin P, Duan R, Li X. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JU, Su Y, Zhong C, Ming G, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Osta A, Wolffe AP. DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Exp. 2000;9:63–75. doi: 10.3727/000000001783992731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 33.Hays FA, Teegarden A, Jones ZJR, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: a crystallographic map of DNA structure and conformation. Proc Natl Acad Sci U S A. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hays FA, Schirf V, Ho PS, Demeler B. Solution formation of Holliday junctions in inverted-repeat DNA sequences. Biochemistry. 2006;45:2467–2471. doi: 10.1021/bi052129x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley DMJ. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 36.Roe SM, Barlow T, Brown T, Oram M, Keeley A, Tsaneva IR, Pearl LH. Crystal Structure of an Octameric RuvA–Holliday Junction Complex. Mol Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- 37.Hadden JM, Déclais AC, Carr SB, Lilley DMJ, Phillips SEV. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007;449:621–624. doi: 10.1038/nature06158. [DOI] [PubMed] [Google Scholar]
- 38.Biertümpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 39.Eichman BF, Vargason JM, Mooers BHM, Ho PS. The Holliday junction in an inverted repeat DNA sequence: Sequence effects on the structure of four-way junctions. Proc Natl Acad Sci. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson J, Hays FA, Ho PS. Definitions and analysis of DNA Holliday junction geometry. Nucleic Acids Res. 2004;32:3017–3027. doi: 10.1093/nar/gkh631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter M, Ho PS. Assaying the Energies of Biological Halogen Bonds. Cryst Growth Des. 2011;11:5087–5095. [Google Scholar]
- 45.Vander Zanden CM, Carter M, Ho PS. Determining thermodynamic properties of molecular interactions from single crystal studies. Methods. 2013;64:12–18. doi: 10.1016/j.ymeth.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 46.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenb DJ. Gaussian 09, Revision A 02, Gaussian. Gaussian Inc; Wallingford, CT: 2009. [Google Scholar]
- 47.Renciuk D, Blacque O, Vorlickova M, Spingler B. Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine. Nucleic Acids Res. 2013;41:9891–9900. doi: 10.1093/nar/gkt738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szulik MW, Pallan PS, Nocek B, Voehler M, Banerjee S, Brooks S, Joachimiak A, Egli M, Eichman BF, Stone MP. Differential stabilities and sequence-dependent base pair opening dynamics of Watson-Crick base pairs with 5- hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine. Biochemistry. 2015;54:1294–1305. doi: 10.1021/bi501534x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter M, Voth AR, Scholfield MR, Rummel B, Sowers LC, Ho PS. Enthalpy–Entropy Compensation in Biomolecular Halogen Bonds Measured in DNA Junctions. Biochemistry. 2013;52:4891–4903. doi: 10.1021/bi400590h. [DOI] [PubMed] [Google Scholar]
- 50.Carter M, Ho PS. Assaying the Energies of Biological Halogen Bonds. Cryst Growth Des. 2011;11:5087–5095. [Google Scholar]
- 51.van Holde KE, Johnson WC, Ho PS. Principles of Physical Biochemistry. 2nd. Pearson Education Inc; 2006. [Google Scholar]
- 52.Ho P, Frederick C, Quigley G, Vandermarel G, Vanboom J, Wang A, Rich A. GT wobble base-pairing in Z-DNA at 1.0-A atomic resolution - The crystal-structure of D(CGCGTG) EMBO J. 1985;4:3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khuu PA, Voth AR, Hays FA, Ho PS. The stacked-X DNA Holliday junction and protein recognition. J Mol Recognit. 2006;19:234–242. doi: 10.1002/jmr.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


