Abstract
The mammalian Hedgehog (Hh) signaling pathway is required for development and for maintenance of adult stem cells, and overactivation of the pathway can cause tumorigenesis. All responses to Hh family ligands in mammals require the primary cilium, an ancient microtubule-based organelle that extends from the cell surface. Genetic studies in mice and humans have defined specific functions for cilia-associated microtubule motor proteins: they act in the construction and disassembly of the primary cilium, they control cilia length and stability, and some have direct roles in mammalian Hh signal transduction. These studies highlight how integrated genetic and cell biological studies can define the molecular mechanisms that underlie cilia-associated health and disease.
Keywords: Primary cilia, Microtubules, Kinesin, Dynein, Ciliopathies, Hedgehog pathway
Vertebrate Hedgehog signaling is coupled to primary cilia
Cilia and flagella are microtubule-based organelles present on the surface of many eukaryotic cells. While motile cilia are associated with movement and fluid flow, it has been long recognized that sensory signaling by non-motile cilia is required for chemosensation in organisms as diverse as the nematode C. elegans and the alga Chlamydomonas [1]. The primary cilium serves as a specialized subcellular compartment that concentrates membrane-bound receptors and signaling complexes, thereby allowing detection of sensory inputs and ligands from the extracellular environment and relay of that information to the cell interior.
One of the most striking revelations in recent cilia biology is that vertebrate cells require a single non-motile primary cilium to respond to Hedgehog (Hh) family ligands [2,3]. The Hh signaling pathway (and therefore primary cilia) is responsible for many aspects of vertebrate embryonic development and stem cell maintenance, and it is disrupted in a spectrum of human tumors [3–5]. Specific mutations in cilia genes can block, increase, or change the spatial pattern of Hh target gene expression. In humans, perturbation of ciliary proteins causes a broad range of genetic disorders, collectively called ciliopathies [6–8] and many important aspects of ciliopathy phenotypes can be explained by misregulation of Hh signaling.
The construction and maintenance of cilia depends microtubule-based motor proteins to transport cargo into and out of the ciliary compartment. Ciliary trafficking depends on the evolutionarily conserved process of intraflagellar transport (IFT). Large protein complexes organized into IFT trains, large structures that can be visualized in the light microscope, move between the plasma membrane and the axonemal microtubules and carry cargo between the base and the tip of the cilium [9,10]. IFT trains are built from two large protein complexes, the IFT-B complex, which includes 16 subunits and is essential for anterograde IFT, and the IFT-A complex, which includes 6 subunits and is thought to have a particularly important role in retrograde IFT [11]. The anterograde motor Kinesin-II is required to move IFT trains from the cilia base to the tip (Figure 1) (that is, towards the plus-ends of axonemal microtubules) in organisms from Chlamydomonas to humans. Cytoplasmic dynein-2 is the evolutionarily conserved motor that is dedicated to retrograde IFT from the cilia tip to the base (Figure 1). The two motors move on different regions of the microtubule doublets, with anterograde trains moving on the B-tubules and retrograde trains moving on the A-tubules of the axonemal doublets (Stepanek and Pigino, 2016). For recent reviews of IFT, please see references [11,13].
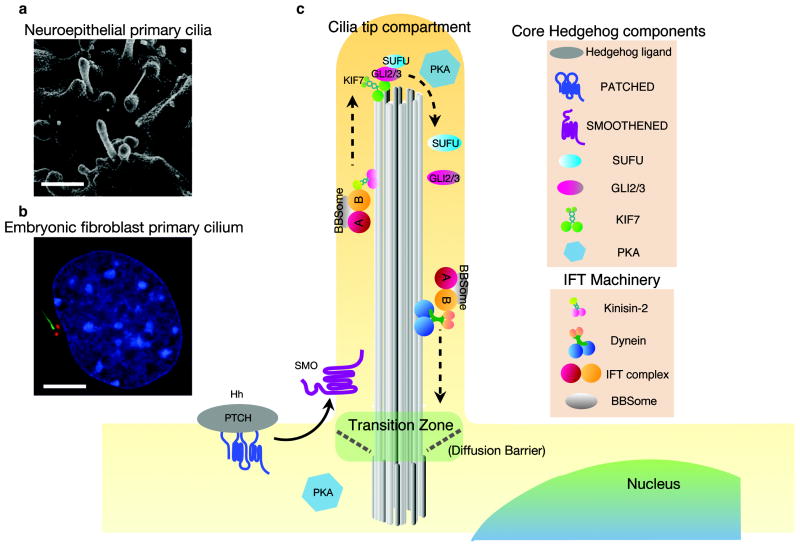
Figure 1. Primary cilia are required for mammalian Sonic hedgehog signaling.
a. Primary cilia on progenitor cells the mouse neuroepithelium at embryonic day 10.5 (E10.5) visualized by scanning electron microscopy.
b. Primary cilium of cultured mouse embryonic fibroblast formed during Go/G1 of the cell cycle. Staining of acetylated α-tubulin (green) labels the axonemal microtubule, and γ-tubulin staining (red) labels the basal body. Nucleus is labeled by DAPI in blue. Scale bars = 5 μm.
c. Core Hedgehog components localize to the primary cilium. Hh ligand inhibits Patched localization to the cilium and promotes Smoothened accumulation in the ciliary membrane. GLI transcription factors, Suppressor of Fused (SUFU) and KIF7 are enriched in the cilia tip compartment upon Hh stimulation [3]. Dissociation of the GLI-SUFU complex is required for maximal activation of the Hh pathway [145,146]. Protein Kinase A (PKA) is required for phosphorylation and proteolytic processing to GLI to form transcriptional repressors [147,148]. PKA resides at the base of cilia in neuroepithelial cells [148], but is found within the ciliary compartment in mIMCD3 cells and cholangiocytes [149,150]. The activity of PKA may be coupled to an orphan G-protein coupled receptor GPR161, which can increase intracellular cAMP when activated. Like PTCH1, GPR161 localization to the ciliary membrane is negatively controlled by Hh ligand [88]. Correct localizations of membrane-bound cilia proteins depend on both active transport through IFT trains/BBSome and diffusion.
The Hh signaling pathway was discovered and first characterized for its roles in Drosophila segmentation and patterning. However, in contrast to the broad distribution of primary cilia in vertebrates (e.g. references [14], and [15]), primary cilia are found only on sensory neurons in Drosophila and not on the cells that respond to the Hedgehog signals during Drosophila development. Although Hh pathway proteins are present in the cilia of Drosophila olfactory sensory neurons [16], mutations that inactivate IFT complex proteins or the IFT motors do not affect Drosophila Hedgehog signaling, segmentation or patterning [17]. As a result, Drosophila genetic studies have not provided information about how primary cilia are used in Hh signaling. Instead, mouse and human genetic studies have provided the foundation for the current understanding of how primary cilia are exploited in mammalian Hh signaling.
Analysis of mouse cilia mutants has focused on patterning of the neural tube and the limb, which provide reliable in vivo readouts of Hh pathway activity. These studies have revealed that genetic inactivation of Kinesin-II and most of the IFT-B proteins, including IFT172, IFT88, IFT52, IFT57 and IFT46, cause midgestation lethality around embryonic day 10.5 and produce identical phenotypes: absence of cilia, lack of all responses to Hedgehog ligand and loss of Hh-dependent ventral neural cell types [3,18]. Inactivation of mouse IFT-A proteins also causes embryonic lethality and altered Hh signaling, although the phenotypes are more complex than those of IFT-B mutations [3]. Many mutations in both the subunits of the major IFT motors and other cilia-associated motor proteins have been identified in human ciliopathies.
Cell biological experiments have demonstrated that the core components required for vertebrate Hh signal transduction are highly enriched in cilia and that they change their localization within the cilium in response to ligand [3]. The cell biological analysis of trafficking of Hh pathway proteins in wild-type and mutant mice and humans provides the foundation for our current understanding of how transport within the primary cilium promotes normal Hh signaling.
In addition to the anterograde and retrograde motors, a set of other motor proteins has dedicated functions in cilia, in the regulation of the control of cilia initiation, cilia length, cilia stability and cilia disassembly, although the functions of these motors have not been studied extensively. This review summarizes the general roles of cilia motor proteins in cilia function and the vertebrate Hh signaling, with a focus on observations made in mouse and human studies.
Kinesin-2: the primary anterograde motor for cilia assembly in vertebrates is essential for Hh signaling
The evolutionarily conserved kinesin responsible for anterograde trafficking of the IFT trains is the plus end-directed motor kinesin-2, a heterotrimeric complex that belongs to the Kinesin-2 family [19,20] There are two distinct forms of the Kinesin-II motor complexes in vertebrates, KIF3A/KIF3B/KIFAP3 and KIF3A/KIF3C/KIFAP3. KIF3A and KIF3B/C form the motor heads responsible for the ATP-dependent power stroke. KIFAP3 (formerly KAP3) is thought to be a regulatory subunit that is important for motility and/or cargo recognition [21] (Figure 2). Based on the similarity of the mouse phenotypes caused by the loss of KIF3A and KIF3B [22,23,24], it is likely that KIF3A/KIF3B/KIFAP3 is the major form of kinesin-2 in mammals.
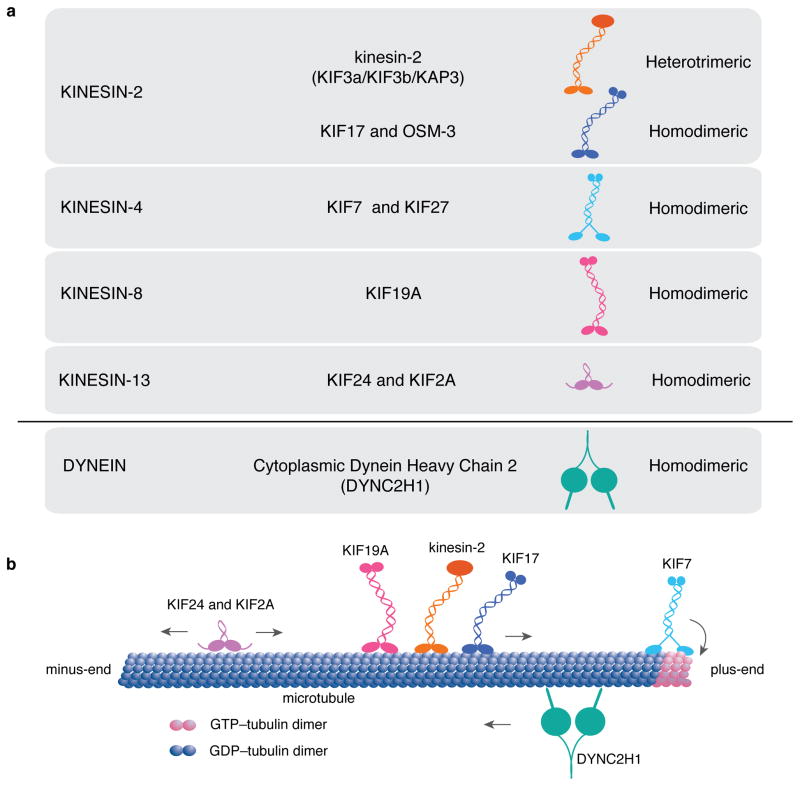
Figure 2. Vertebrate cilia-associated kinesins and IFT dynein.
a. Four families of kinesins and one dynein are known to be associated with the structure and IFT of mammalian cilia. Heterotrimeric kinesin-2 and homodimeric KIF17 belong to the Kinesin-2 family. Homodimeric KIF7 and KIF27 belong to the Kinesin-4 family. Homodimeric kinesin KIF19 belongs to the Kinesin-8 family. The motor domains of those kinesins are located at the N-terminus of the protein. KIF24 and KIF2A belong to the kinesin-13 family, with motor domains in the central region of the protein. Cytoplasmic dynein-2 is the only dynein required for intraflagellar transport. b. Both kinesin-2 and KIF17 are anterograde kinesins that move along the axoneme from the cilia base, the minus end, to the cilia tip, the plus end [56]. KIF7 lacks motility along the microtubule lattice, but can bind preferentially to GTP-bound tubulin at the growing ends of microtubule and promote catastrophe. Other members of the Kinesin-4 family, including KIF4 and KIF21A, exhibit plus-end directed motility to reach microtubule ends, where these kinesins regulate microtubule dynamics [151,152]. KIF19 is a motile kinesin and exhibits microtubule depolymerization activity towards the plus-ends of microtubules and appears to act specifically in motile cilia [132]. KIF24 and KIF2A lack motility and show microtubule-depolymerizing activity towards both ends of microtubules. Although both Kinesin- 8 and Kinesin-13 family members can shorten microtubules, the underlying actions are different [153]. Cytoplasmic dynein-2 is required for microtubule-based cargo transport from plus ends to the minus ends of microtubules within cilia. The motor activity resides in the heavy chain, which acts as a homodimer, and a large number of smaller chains mediate cargo loading and specificity of the motor (see Figure 4c).
Null mutations in mouse Kif3a or Kif3b inactivate the kinesin-2 machinery and cause loss of primary cilia in all embryonic tissues [3,24–30]. Similar phenotypes are seen in most other ciliated species including Chlamydomonas, Tetrahymena, Drosophila, Xenopus, sea urchin and trypanosomes [19,20]. A recently identified zebrafish kif3a mutant allele with 6 base-pair inframe insertion in a highly conserved region within the motor domain causes short cilia and kidney cysts [31], a phenotype consistent with the cilia defect seen in mouse [26,32]. This phenotype is weaker than those seen in mouse Kif3a mutants, presumably because maternal gene product can rescue defects in early patterning of zebrafish embryos. Like the kif3a mutants, zebrafish Ift88 mutants have kidney cysts. For this gene, it has been shown that removal of both maternal and zygotic Ift88 has a stronger phenotype, including loss of normal Hh signaling [33,34].
KIF3A is required for cilia formation and Hh signaling in every cell type tested in mouse. Signaling by Sonic hedgehog (Shh) or Indian hedgehog (Ihh) depends on Kif3a in processes as diverse as bone formation [35,36], tooth development [37], and stem cell proliferation in the embryonic and adult brain [30,38–40]. Similar in vivo analyses have shown that KIF3A and cilia are required for tumorigenesis in Hedgehog-driven tumors such as basal cell carcinoma and medulloblastoma [28,41].
While KIF3A is essential for cilia formation in both mammals and zebrafish, the roles of other Kinesin-II subunits appear to be species-specific. Zebrafish kif3b mutants lack cilia in some tissues but not others: cilia in many tissues are affected, but cone photoreceptor cilia and some kinocilia of the inner ear (specialized primary cilia) appear normal in the mutants. Furthermore, mouse Kif3c null mutants are also viable, fertile and show normal behavior, suggesting that the activity of kinesin-2 machinery mainly depends on KIF3A and KIF3B [42]. [43]. Inactivation of zebrafish kif3c does not have an obvious effect on cilia formation. However, morpholino knockdown of zebrafish kif3c in a kif3b mutant background led to a general loss of cilia, suggesting that Kif3b and Kif3c play functionally redundant roles in zebrafish within the Kinesin-II complex [42].
In addition to its role in building cilia, kinesin-2 could, in principle, also directly transport proteins required for Hh signal transduction. Recent work showed that both KIF3A and KIFAP3 can directly interact with GLI proteins in vitro, and suggested that GLI binding to KIFAP3 restricts the function of GLI activators but does not affect GLI repressors [44]. A direct interaction with KIF3A in cilia would be surprising, as the motor is believed to interact with cargo through IFT protein intermediates [25]. The authors proposed that the interaction of KIFAP3 with GLI proteins is most consistent with a role outside cilia; for example, in the transport of GLI proteins to the nucleus [45]. Although Kif3ap3 mutations (which inactivate both KIFAP3 isoforms) cause embryonic lethality [46], it is not known whether deletion of KIFAP3 affects cilia structure or Hh signaling in the mouse. Analysis of cilia and Hedgehog-dependent patterning defects in Kifap3 mutants should help test whether kinesin-2 can act directly on GLI proteins, independent of the IFT-B complex.
A role for kinesin-2-mediated trafficking of membrane proteins is also controversial. It has been reported that KIF3A acts with β-arrestin1/2 to promote SMO localization to cilia [45], but in mouse embryonic fibroblasts that lack both β-arrestin-1 and -2, cilia localization of SMO appears normal [47]. Although it is possible that KIF3A could directly transport SMO within cilia, a series of elegant pulse-chase experiments and single-molecule imaging studies demonstrated that SMO translocation to the cilia depends on lateral diffusion rather than by kinesin-mediated IFT [48,49]. However, given the presence of a size-dependent physical barrier at the base of cilia [50,51], SMO could require kinesin-2 to get through the diffusion barrier.
Are there additional anterograde motors in vertebrate primary cilia?
In C. elegans, two different kinesins, kinesin-2 and OSM-3, act as anterograde motors for formation of cilia. Cilia on amphid/phasmid channel sensory neurons have two regions, a “middle” segment, which includes the transition zone and the proximal part of the axoneme that is made of doublet microtubules, and a distal segment, which consists of singlet microtubules. OSM-3, a homodimeric kinesin of the Kinesin-2 family, can compensate for the loss of Kinesin-2 activity in these specialized cilia. The function redundancy model suggested that kinesin-2, a slow motor, and OSM-3, a fast motor, may be mechanically coupled and travel as a dual-motor IFT complex in the middle segment of sensory cilia [52]. Recent work using high-resolution fluorescence microscopy in live C. elegans reveals a functional differentiation between the two motors [53]. At single-molecule sensitivity, the live imaging data show that kinesin-2 localizes to the middle segment and is required to import IFT trains and cargo across doublet microtubules (the transition zone and proximal axoneme), whereas OSM-3 resides at the distal segment and acts as a long-haul motor along the singlet microtubule to deliver IFT trains to the cilia tip. The differences in motor distribution and motor activity may account for efficient and dynamic IFT transport observed in the specialized sensory cilia.
The vertebrate homologue of OSM-3 is KIF17. In zebrafish, both kinesin-2 and KIF17 associate with microtubule doublets of photoreceptor connecting cilia (Figure 3). Deletion of zebrafish kif17 does not have an apparent effect in early embryogenesis, but does lead to severe disruption in the morphogenesis of the photoreceptor outer segment [54,55]. Sensory cilia of mammalian olfactory neurons also exhibit a compartmentalized axonemal structure similar to the cilia of nematode amphid/phasmid channel sensory neurons [56]. Fluorescently tagged KIF17 and Kinesin-II are present along the full length of these cilia, and show bidirectional processive movement along the axoneme. Nevertheless, a null mutation of Kif17 in mouse does not affect photoreceptor morphology or function and is dispensable for cilia assembly and Hh signaling during embryonic development [57]. It is possible that in the absence of KIF17 motility, KIF17 associated vesicles are transported via kinesin-II or by diffusion-mediated mechanisms [56].
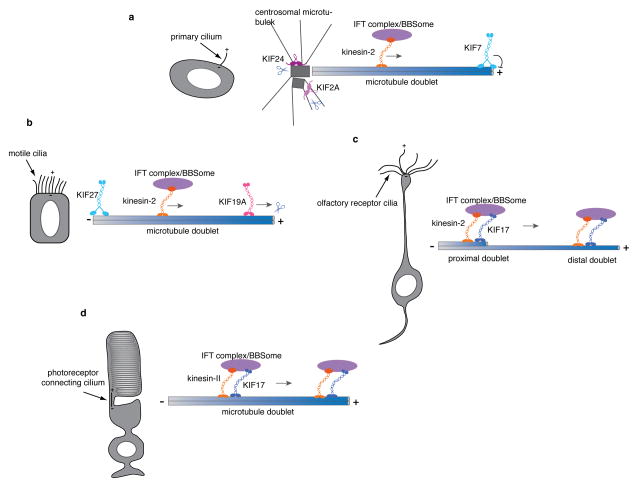
Figure 3. Cell-type dependent functions of ciliary kinesins.
a. In primary cilia, kinesin-2 is required for the anterograde transport of IFT complexes, BBSome and ciliary cargo along the axoneme, which is composed of nine microtubule doublets. Kinesin-2 dependent transport is essential for cilia formation in all kinds of cilia (a–d). KIF7, which can autonomously track the plus-ends of growing microtubules, localizes to the tip of cilia, where axonemal microtubule plus ends are located. KIF7 controls the structure and length of primary cilia. KIF24 and KIF2A exhibit microtubule-depolymerizing activities and are thought to act on centrosomal microtubules to provide temporal control of cilia formation during the cell cycle.
b. While kinesin-2 is required for motile cilia formation, the length of motile ciliary axonemes is controlled by KIF19. KIF19 relies on its plus-end directed motility to reach the axonemal plus ends at the motile cilia tip, where the kinesin plays a role in shortening the length of the axoneme. KIF27 localizes to the base of motile cilia and is required for motility.
c. Olfactory receptor cilia are much longer than motile cilia and primary cilia. The proximal segment of the axoneme consists of doublet microtubules, whereas the distal segment has only singlet microtubules. KIF17 is present along the axoneme, but it is unclear whether the motor activity of KIF17 is required for its targeting to the distal end or if KIF17 is necessary for IFT.
d. The connecting cilium of the photoreceptor does not directly project into the extracellular environment but bridges the inner segment and outer segment of the photoreceptor cells, such as rod and cone cells. Kinesin-2, IFT proteins and KIF17 appear to form a complex that moves along the connecting ciliary axoneme. KIF17 is required for the morphogenesis of the outer segment during an early stage of photoreceptor development in zebrafish.
It is striking that no mutations in KIF3A, KIF3B, KIF3C, KIFAP3 or KIF17 have been identified in human ciliopathy patients, particularly in contrast to the large number of ciliopathy-associated mutations in the subunits of the retrograde motor and in KIF7 (Table 1; see below). This might suggest some functional redundancy between anterograde motors. An alternative possibility is raised by the recent publication of large-scale human exome (protein-coding) sequence, which shows that KIF3A is intolerant to loss-of-function mutations, which suggests that heterozygotes have a disadvantage in survival or reproduction [58].
Table 1.
Cilia motor proteins associated with mammalian Hh signaling
| Gene | Mouse mutations | Human ciliopathy |
|---|---|---|
| Kif3a | Targeted null [23,24]; ENU-induced null allele [133]. Homozygous mutants of both targeted knockout and ENU induced-null die at midgestation; lethal with loss of cilia and loss of Hh response. | None described. |
| Kif3b | Targeted null [22]. Homsozygous mutants die at midgestation; lethal with loss of cilia; loss of Hh response was not examined [134]. | None described. |
| Kif3c | Targeted null is viable [43]. No phenotype observed in homozygous null retina photoreceptor cells. | None described. |
| Kifap3 | Targeted null associated with midgestation lethality [46]; cilia and Hh defects not described. | None described. |
| Dync2h1 | Gene trap and ENU- induced null and hypomorphic alleles [65–67,102,135]; midgestation lethal with short, bloated cilia and loss of Hh signaling response; polydactyly; congenital heart disease. | Short rib polydactyly syndrome and asphyxiating thoracic dystrophy (with or without polydactyly) [69,136–140, 80,141]. |
| Dync2li1 | Targeted null [81]; midgestation lethal with short bloated cilia and loss of Hh signaling response | Short rib polydactyly syndrome and skeletal ciliopathy [76,142] |
| Wdr34 | No mutants described | Short rib polydactyly syndrome and severe asphyxiating thoracic dysplasia [79,143] |
| Wdr60 | No mutants described | Short rib polydactyly and Jeune syndromes [79,80]. |
| Tctex1d2 | No mutants described | Jeune asphyxiating thoracic dystrophy [144] |
| Dynll1 | Gene trap allele [86]; midgestation lethal with short, bloated cilia and loss of Hh signaling response; polydactyly; congenital heart disease; abnormal lung development. | None described. |
| Kif7 | Targeted null and ENU-induced alleles [102–104,120,135]; perinatal lethal with mild expansion of ventral neural cell types; polydactyly, skeletal defects; congenital diaphragmatic hernia; primary cilia of variable length and abnormal tip structure. | Joubert and acrocallosal syndrome [100,112–117,119]. Patients exhibit global developmental delay, polydactyly, brain abnormalities (particularly corpus callosum agenesis and classical molar tooth sign), facial dysmorphism, cleft palate; |
Cytoplasmic Dynein-2: the retrograde ciliary motor
Seven of the nine major mammalian dynein complexes [59,60] are localized within motile cilia (or flagella) and are responsible for cilia motility. Two additional dynein complexes are dedicated to cytoplasmic trafficking and intraflagellar transport, respectively: cytoplasmic dynein-1 drives minus-end directed transport in the cytoplasm, whereas cytoplasmic dynein-2, which is conserved from the alga Chlamydomonas to mammals, functions exclusively in cilia and flagella to mediate retrograde IFT [61,62]. The cytoplasmic dynein-2 heavy chain includes six C-terminal AAA+ domains required for ATP hydrolysis and motor activity, a linker domain for microtubule attachment, and an N-terminal region that mediates homodimerization and subsequent interaction with dynein regulatory subunits [63] (Figure 4). The seven accessory subunits are the dynein light chains, intermediate chains, and light intermediate chain [64].
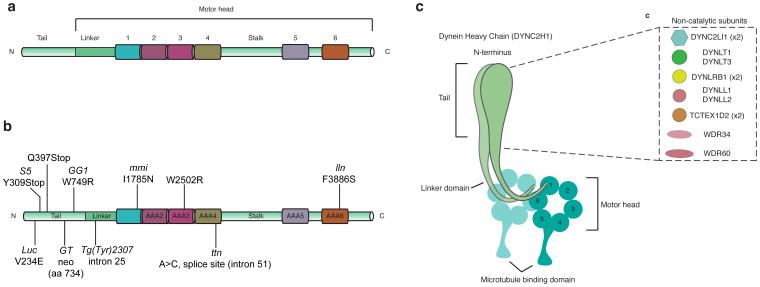
Figure 4. Mutations that disrupt function of the mouse cytoplasmic dynein-2.
a. Schematic of the domains of cytoplasmic dynein heavy chain 2 (DYNC2H1). The N-terminal regulatory domain is responsible for dynein oligomerization, cargo and non-catalytic subunits binding, and is not part of the catalytic motor domain that generates motility. The motor domain consists of the linker, six non-identical AAA+ modules (1–6), the coiled-coil stalk and strut/buttress (not depicted), as well as the C-terminal region. b. All mutations found in mouse Dync2h1 cause disruption in cilia structure and Hh signaling. For mutations in the human DYNC2H1 that cause ciliopathies, see [77]. c. Subunits of mammalian cytoplasmic dynein-2 complex. Mutations in the human genes encoding these subunits, including TCTEX1D2, DYNC2LI1, WDR34, WDR60 and DYNC2LI1, cause ciliopathies and presumably disrupt retrograde ciliary trafficking and Hh signaling.
Unlike kinesin-2, cilia can form in the absence of cytoplasmic dynein-2 heavy chain and its regulatory subunits. Mutations in mammalian cytoplasmic dynein-2 heavy chain (DYNC2H1) cause short, bulbous cilia that accumulate IFT particles and Shh components [65–67]. In all mouse Dync2h1 alleles analyzed, Shh signaling is reduced in the neural tube and mutants lack the floor plate and V3 progenitors, which are specified by the highest level of Shh activity [65,67]. The Hh signaling defects in dynein-2 mutants are similar to, but slightly milder than, loss of Kif3a or IFT-B mutants, suggesting that the loss of the retrograde motor disrupts cilia structure almost as completely as the loss of anterograde trafficking. Consistent with defects caused by mutations, chemical antagonists specific to cytoplasmic dyneins (the ciliobrevins) can inhibit dynein activity in vitro and in cells, and reduce both retrograde and trafficking and lead to the accumulation of IFT88 at the tips of cilia [68]. Mutations in human DYNC2H1 are implicated in two specific ciliopathies characterized by a constricted thoracic cage, short ribs, and shortened long bones: Jeune asphyxiating thoracic dystrophy (JATD) and short rib polydactyly syndrome (SRPS) (See table 1). Cilia from patients with DYNC2H1 mutations, like the Chlamydomonas and C. elegans mutants, accumulate IFT proteins and Shh components in cilia [69].
Biochemical studies indicate that ATP hydrolysis by the AAA1 domain is essential for movement along microtubules [70–72]. Structural and biochemical analysis of human cytoplasmic dynein-2 has indicated that the AAA2-AAA4 domains form a rigid structure that is required to modulate dynein-microtubule affinity upon ATP hydrolysis, and the AAA5-6 domains stabilize the conformational switch of the motor [72]. These data parallel the phenotypes of mouse Dync2h1 mutations: Dync2h1lln, the allele with the mildest effect on cilia morphology, affects the sixth AAA+ domain, whereas Dync2h1mmi, a more severe allele, affects the first AAA+ domain (Figure 4) [73].
Cytoplasmic dyneins share a set of light chain subunits (DYNLRB1, DYNLRB2, LC8-1 (DYNLL1), LC8-2 (DYNLL2), TCTEX-1 (DYNLT1), and TCTEX-3 (DYNLT3)). In addition, each of the cytoplasmic dynein complexes has a set of subunits required for cargo loading [74]. Cytoplasmic dynein-1 includes the dynactin complex, which enables cytoplasmic dynein-1 to carry vesicular cargo, whereas the cytoplasmic dynein-2 complex includes two intermediate chains (WDR34, WDR60), one light intermediate chain (DYNC2LI1, also called D2LIC) and another light chain (TCTEX1D2) [75]. Mutations in the subunits of cytoplasmic dynein-2 that are not shared with cytoplasmic dynein-1 can cause ciliopathies: human mutations TCTEX1D2, WDR34 and WDR60 are associated with the same types of ciliopathies caused by mutations in the heavy chain, JATD and SRPS [76–80] It is likely that these components are also important for mammalian Hh signaling. Indeed, mutations in human DYNC2LI1 cause SRPS, and fibroblasts derived from patients with missense and nonsense mutations in DYNC2LI1 confirmed that alleles of the gene cause abnormal cilia morphology [76]. Inactivation of mouse Dync2Ii1 and Dynll1 cause midgestation lethality and Hh pathway phenotypes like those seen in Dync2h1 mutants [81, 86] A role for Dynlt1 in ciliary length control has also been described in cell culture and in zebrafish [74,82].
Based on defects in retrograde trafficking in mutants, proteins of the IFT-A complex are postulated to cooperate with cytoplasmic dynein-2 in retrograde transport. However, the phenotypes of mouse embryos that lack cytoplasmic dynein-2 differ from those of IFT-A mutants: dynein mutants show loss of Hh activity, whereas IFT-A mutants show ectopic Hedgehog pathway activation, including dorsal expansion of Shh-dependent ventral progenitor domains [83,84]. The contrasting Hh-dependent phenotypes may reflect differential requirements of cytoplasmic dynein-2 and IFT-A in retrograde IFT. Scanning electron microscopy and immuofluorescent staining analysis show that most IFT-A mutants have bulges and accumulate IFT-B complex at the tip of primary cilia [83–85]. In contrast, mutants associated with partial loss of cytoplasmic dynein-2 function exhibit bulges and abnormal accumulation of IFT particles near the base of cilia [67,85,86], suggesting that, in addition to retrograde transport, cytoplasmic dynein-2 may be required to export cilia cargo through the transition zone diffusion barrier. Analyses in mouse mutants show that in the absence of IFT-A, several membrane proteins, including Smoothened, and soluble cargo such as TULP3 and GPR161, two negative regulators of the Hh pathway, failed to localize to mutant cilia, indicating that IFT-A plays a role in anterograde transport as well as retrograde transport [85,87,88]. In contrast, inactivation of cytoplasmic dynein-2 activity results in abnormal accumulation of both membrane proteins and soluble cargo in the cilia [67,73].
Remarkably, when the activities of both DYNC2H1 and the IFT-B complex (or cytoplasmic dynein-2 and the IFT-A complex) are compromised, cilia structure as well as normal localization of Smoothened and GLI2 are partially rescued [67]. These data suggests that the balance between anterograde and retrograde trafficking is essential for correct localization of Hh pathway components and cilia structure.
The BBSome and IFT25/27: candidate motor adaptors for Hh proteins in primary cilia
Key steps in trafficking through the cilium are cargo loading at the base of the cilium for anterograde trafficking and the release of cargo at the cilia tip, then reloading of appropriate cargo and release at the base of the after retrograde movement. In general, it is believed that cargo associates with IFT particles through relatively low-affinity interactions that allow a wide variety of cargo molecules to be carried and released [13]. However, as discussed above, at least some of the components of the Hedgehog pathway do not traffic with IFT and therefore their movement is not directly regulated by kinesin-2 and cytoplasmic dynein-2. SMO, for example, appears to move into cilia by a lateral diffusion mechanism and by diffusion within cilia (see above). Single particle imaging has also shown that soluble proteins, such as EB1, can also move into and within cilia by diffusion and can accumulate in cilia by capture by specific binding [89].
The BBSome is an octameric complex consisting of highly conserved BBS proteins that are mutated in human Bardet-Biedl syndrome, a ciliopathy associated with cystic kidneys and obesity. The BBSome moves together with the IFT complex and is believed to tether specific cargo to the IFT machinery [3,90–92] and is particularly important in retrograde trafficking [93]. Consistent with a role for the BBSome in retrograde trafficking of Hedgehog pathway proteins, the membrane proteins SMO and PTCH1 accumulated in the cilia of MEFs from Bbs mutants even in the absence of Hh ligand stimulation [92,94].
Two small IFT proteins, IFT25 and IFT27 appear to have a distinctive role in IFT that is related to BBSome activity. Both Ift25 and Ift27 mouse mutants survive to the end of gestation, when they show clear, but mild Hh pathway phenotypes [92,95]. Ift25 and Ift27 mutant cells and embryos do not accumulate the normal amount of GLI2 at cilia tips; mutant cilia also accumulate the membrane proteins PTCH1 and SMO, suggesting a defect in retrograde trafficking. IFT27 is a member of the RAB family of small GTPases and a recent structure of the Chlamydomonas IFT25/27 subcomplex suggests that other IFT proteins may function as a GAP to activate the GTPase function of IFT27 [96]. BBS proteins accumulate in Ift27 mutants, which suggested the model that IFT25 and IFT27 link the BBS complex to the IFT machinery required for retrograde trafficking [97] and that PTCH1 and SMO retrograde trafficking is directly mediated by the BBS complex [92].
Despite the data implicating the BBSome in trafficking of Hh pathway components in mouse embryo fibroblasts, mouse Bbs mutant embryos do not have Hh-related phenotypes, indicating that the BBSome is not essential for mouse Hh signaling. It has been shown, however, that some Bbs mutations can exacerbate Hh pathway phenotypes of weak cilia mutants. For example, while homozygotes for both a weak allele of Ift88 (Ift88orpk) and Bbs7 homozygotes are viable, the double homozygotes die during gestation (~E13.5) with an open cranial neural tube, broad limbs and other Hh-related phenotypes [94]. Similarly, double mutants that lack both the transition zone protein Tectonic1 (TCTN1) and BBS1 show more extreme polydactyly and neural tube closure defects, and cilia fibroblasts from the double mutants have more pronounced defects in ciliary trafficking than Tctn1 single mutants [98]. Thus other factors appear to act in parallel with the BBSome to link Hh pathway proteins and the IFT machinery. Alternatively, the BBSome may act as a back-up for a diffusion-and-capture mechanism [89] to concentrate Hh pathway proteins in cilia.
Cilia-associated kinesins at the tip and the base of cilia
In addition to direct roles in transport within the cilium, other kinesins play specific roles in shaping microtubules at the tip and the base of the cilium through modulation of tubulin dynamics. The GLI transcription factors and their repressor SUFU are highly enriched at cilia tips. The cilia tip compartment is also crucial in trafficking, as anterograde cargo is released at the tip, retrograde is loaded and IFT complexes change their structure to accommodate the switch from the use of the kinesin-2 motor to the cytoplasmic dynein-2 motor. The base of the cilium is the site where cilium assembly is controlled and where the cargo that enters the cilium is determined. Both of these compartments are regulated by dedicated kinesin motors.
KIF7: a regulator of the cilia tip compartment that links Hh signaling to the primary cilium
KIF7, a member of the Kinesin-4 family, is the only evolutionarily conserved component of the core Hh signal transduction pathway that is required for the structure of primary cilia [99] (Figure 3a). The primary cilia of cultured fibroblasts carrying either mouse and human Kif7 mutations are longer and less stable than wild type control, which demonstrates that KIF7 plays a role in regulating cilia structure [99,100].
KIF7 localizes to the tip of the cilium throughout ciliogenesis and accumulates further at cilia tips in response to Hh activation. Mammalian KIF7 is the homolog of Drosophila Cos2, and both are required to relay signals from the transmembrane protein SMO to the Ci/GLI transcription factors [25,101]. Genetic inactivation of mammalian KIF7 causes developmental defects associated with mild ectopic activation of Hh signaling, including dorsal expansion of ventral cell types in the neural tube, preaxial polydactyly and skeletal malformations [102–104] (Table 1). Zebrafish Kif7 also localizes to primary cilia and regulates Hh signaling in the mesoderm [105,106]. Enrichment of KIF7 at cilia tips in response to Hh pathway activation may depend on PPFIA1, a subunit of the serine/threonine PP2A phosphatase complex, which resides at the base of primary cilia in MEFs [107].
Although the KIF7 motor domain has a crystal structure superimposable on that of conventional kinesin heavy chain and has the sequence hallmarks of a plus-end directed microtubule motor [108,109], the overall rates of anterograde and retrograde transport do not change in the absence of KIF7 [99]. In addition, KIF7 does not act as an anterograde motor for Hh pathway proteins, as SMO and GLI proteins localize to cilia in the absence of KIF7 [99]. Instead, single-molecule imaging showed that motor domain of KIF7 binds preferentially and autonomously to the plus-ends of growing microtubules, where it regulates the dynamics of microtubule growth at plus-ends. At the plus-ends of microtubules, purified KIF7 motor both decreases the rate of microtubule polymerization and induces microtubule catastrophe, causing an overall reduction in the length of microtubules. These activities of the KIF7 motor on microtubule dynamics can account for the long cilia phenotype seen in Kif7 mutants [99,100].
Although cilia localization of SMO and GLI proteins does not depend on KIF7, KIF7 has been shown to bind GLI proteins and SUFU [104,110], just as Drosophila Cos2 binds to the homologous Hh pathway proteins Ci and Sufu [111]. In the absence of KIF7, the GLI-SUFU complex, assessed by co-immunoprecipitation, fails to dissociate in response to Hh ligand, indicating that KIF7 helps promote GLI-SUFU dissociation and subsequent GLI activation [110]. Further structure-function analysis and in vitro reconstitution experiments will be required to define whether dissociation of the GLI-SUFU complex is directly regulated through KIF7 motor activity and require the presence of microtubule.
Mutations in human KIF7 have been reported in several classes of severe ciliopathies associated with perinatal lethality, including fetal hydrolethalus, acrocallosal syndrome and Joubert syndrome (Figure 5) (Table 1) [112–119]. Affected individuals show a spectrum of developmental defects including brain abnormalities, skeletal malformations and craniofacial dysmorphism, consistent with a role of KIF7 in the human Hh pathway. In addition, KIF7 has also been implicated in retinoic acid signaling, congenital diaphragmatic hernia and cell proliferation in the respiratory airway [120,121]. It will be interesting to determine whether the role of KIF7 in those contexts is mediated through Hh signaling or through other activities of cilia.
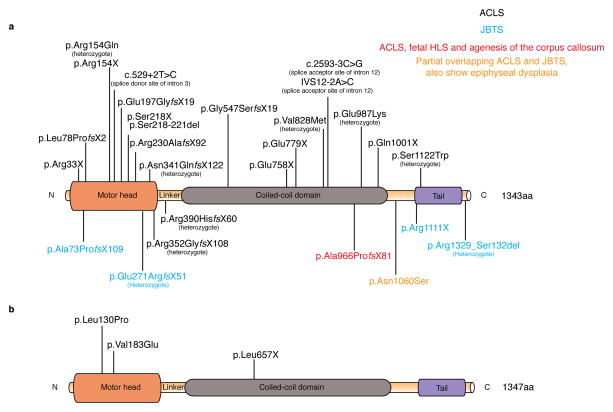
Figure 5. Kif7 mutations cause ciliopathies in humans and embryonic lethality in mice.
a. The catalytic motor head of KIF7 is located near the N-terminus and possesses microtubule-stimulated ATPase and microtubule-binding activities. Two KIF7 motor heads of a kinesin motor are coupled by the neck linker and the coiled-coil domain to form homodimers. The neck linker is important for the coordination of two motor heads as well as for the amplification of motility; the linker region of KIF7 is 100 amino acids long. This is longer than seen in most motile N-kinesins, which correlates with the lack of processive motility of the KIF7 motor. The globular domain at the C-terminus is believed to bind to cargo or adaptor proteins. Homozygous and compound heterozygous human KIF7 mutations are present in patients with Acrocallosal syndrome (ACLS), fetal hydrolethalus, agenesis of the corpus callosum, Joubert syndrome as well as mild epiphyseal dysplasia. The majority of human KIF7 mutations are found in the motor domain.
b. The mouse KIF7 protein is highly homologous to human KIF7. Two ENU induced missense mutations are present in the motor domain, while a third ENU induced nonsense mutation is found in the coiled-coil region. All characterized mouse Kif7 alleles, including two targeted null alleles of Kif7 (not depicted) in which the first 2 exons were deleted, show very similar developmental defects, associated with perinatal lethality.
A close homologue of KIF7, KIF27, is also associated with cilia but is not required for formation of primary cilia or the activity of the mammalian Hh pathway (Figure 3). Instead, KIF27 is important for the movement of motile cilia [122–124]. The ancestral planarian KIF7/KIF27 is required for cilia formation and motility [125]. It will be interesting to compare the interactions of KIF7 and KIF27 with microtubules and determine whether they both regulate microtubule dynamics by interactions with microtubule plus-ends within the cilia or whether the two proteins have distinct sets of motor activities on different types of axonemal microtubules.
Microtubule-depolymerizing kinesins at the base and tips of cilia
Microtubule-depolymerizing kinesins, M-kinesins without processive motility, have an ancient function in the regulation of the length of ciliary and flagellar axonemal microtubules, as protozoan and Chlamydomonas Kinesin-13 proteins regulate the length and motility of flagella [126]. However it is not yet clear whether depolymerizing kinesins play a role in cilia length control in animals. Human KIF24, a Kinesin-13 family member, preferentially localizes to the mother centriole in cultured cells. KIF24 has a mild microtubule destabilizing activity in vitro, much less than the mammalian MCAK KIF2C, a well characterized microtubule depolymerizing protein [127]. KIF24 recruits CP110, which caps the distal end of centrioles to prevent inappropriate ciliogenesis in cycling cultured cells (Figure 3). Thus KIF24 appears to act at the base rather than the tip of the cilium: it acts at the centriole to negatively regulate cilia initiation in concert with CP110, but is not required for either centriole or cilia length. The in vivo function of KIF24 has not been tested, while CP110 was recently shown to control centriole maturation and cilia formation in vivo [128].
Mammalian KIF2A, another member of the M-kinesin Kinesin-13 family, is also localized to the centrosome and can promote cilia disassembly from the cilia base in cell culture [129]. The microtubule-depolymerization activity of KIF2A depends on specific activation by Polo-like kinase 1 (PLK1) and is coupled to cell proliferation. Knockdown of KIF2A prevents cilia disassembly when quiescent cells re-enter the proliferative phase. Given that cilia disassembly presumably occurs from the plus-ends of microtubules at the cilia tip, it is unclear which cellular microtubules are the substrates of cilia-associated KIF2A. A role of the PLK1-KIF2A pathway in ciliogenesis has also been implicated in premature chromatid separation syndrome, which shares some features with human ciliopathies [129]. A missense mutation of KIF2A has been identified in a human patient with malformation of cortical development and microcephaly [130]. Given that the basal body may act as a trafficking hub for motor proteins, it is interesting to note that centrosome dysfunction is associated with microcephaly [131]. Cilia-associated vertebrate Kinesin-13 members may act on cytoplasmic microtubules surrounding the centrosomes or on centrosomal microtubules to control ciliogenesis and disassembly during the cell cycle.
KIF19A, a Kinesin-8 family member with a motor domain at the N-terminus, limits the length of motile cilia in vivo [132]. KIF19A uses its plus-end directed motility to reach the cilia tip, where it then depolymerizes microtubules from the plus-ends. Mouse mutants that lack Kif19A have longer motile cilia that are incapable of generating efficient fluid flow and exhibit motile cilia-associated defects, including hydrocephaly and female infertility. Kif19A mutants show normal embryonic development, suggesting that KIF19A activity is specific to motile cilia but not primary cilia and therefore does not play a role in mammalian Hh signaling [132].
Concluding remarks
The number and diversity of cilia-associated microtubule motors are remarkable. Each type of motor is responsible for distinct aspects of cilia structure and function. The IFT dynein complex is dedicated to retrograde transport, whereas kinesins play roles in anterograde transport, axonemal length control and the organization of centriolar and centrosomal microtubules. Given that mutations in Kinesin-II and IFT-dynein cause distinct ciliary structural defects but have similar effects on Hh signal transduction, it is most likely that those motor proteins act by regulating cilia structure and thereby regulating the proper localization and enzymatic activities of the Hh signaling pathway. Future investigation needs to define the mechanistic details of motor-mediated cilia trafficking and the aspects of cilia structure that are essential for Hh signaling (see Outstanding Questions).
Outstanding questions.
Do microtubules and motor protein-mediated trafficking only play a structure role in formation of the cilia, or do they actively regulate the location, composition and activity of the Hh complex?
What is the nature of Hh complex that accumulates at the tip of the cilium and how does IFT affect the dynamics of Hh complex accumulation?
Does Hh ligand or pathway activation regulate motor speed, and how do Hh proteins accumulate at the cilia tip when the pathway is activated?
How much of the Kif7 phenotype embryonic phenotype is due to cilia structure and how much to GLI/SUFU interactions?
What determines kinesin and dynein specificity to different types of cilia, and what mechanisms regulate motor proteins entering into the cilia?
Do kinesin-2 and cytoplasmic dynein-2 help carry cargo into and out of the cilium through the transition zone; if they do, are their motor mechanisms modified for these tasks?
Other motors associated with cilia have not been well-studied. Additional investigation of those motors should provide deeper understanding of the control of cilia initiation and disassembly. We anticipate that future studies will identify additional motor proteins that participate in these processes.
Many mutations in cilia motor proteins have been linked to human ciliopathies and overlapping disease spectra; the complexity of these ciliopathy syndromes reflects the diverse functions of cilia-mediated signaling. It is remarkable that while many human mutations in KIF7 and Dynein have been identified, none has been found in Kinesin-II, raising interesting questions about redundancy or dosage sensitivity of the Kinesin-II. In vivo analysis using mouse models and in vitro biochemistry and single-molecule imaging will be powerful tools to understand basic biological questions such as how the Hh pathway is coupled to primary cilia and are likely to provide therapeutic strategies for ciliopathy patients in the future.
Trends Box.
The mammalian Hh pathway depends on primary cilia and intraflagellar trafficking. Both microtubule mediated transport and diffusion are required for cilia cargo localization.
Kinesin-II and Cytoplasmic Dynein-2 complex are evolutionarily conserved molecular motors required for cilia formation. Inactivation of Kinesin-II and Dynein complex causes distinct cilia structure defects and blocks Hh signaling in mouse embryos.
Mutations in KIF7 and Cytoplasmic Dynein-2 complex cause developmental abnormalities associated with misregulation of Hh signaling both in mouse and humans.
High resolution live imaging of fluorescently tagged cilia proteins, combined with cell permeable chemical modulators of motor proteins, can shed light on how trafficking is regulated within the cilium, and reveal how acute perturbation of motor activity can affect the outcome of Hh signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 3.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingham PW, et al. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mourão A, et al. The intraflagellar transport machinery in ciliary signaling. Curr Opin Struct Biol. 2016;41:98–108. doi: 10.1016/j.sbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 9.Kozminski KG, et al. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pigino G, et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taschner M, Lorentzen E. The Intraflagellar Transport Machinery. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 13.Lechtreck KF. IFT-Cargo Interactions and Protein Transport in Cilia. Trends Biochem Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bangs FK, et al. Lineage specificity of primary cilia in the mouse embryo. Nat Cell Biol. 2015;17:113–122. doi: 10.1038/ncb3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzhandaivel A, et al. Cilia-mediated hedgehog signaling in Drosophila. Cell Rep. 2014;7:672–680. doi: 10.1016/j.celrep.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 17.Han YG, et al. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–1686. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Lee MS, et al. IFT46 plays an essential role in cilia development. Dev Biol. 2015;400:248–257. doi: 10.1016/j.ydbio.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole DG. Kinesin-II, coming and going. J Cell Biol. 1999;147:463–466. doi: 10.1083/jcb.147.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhey KJ, et al. Kinesin motors and primary cilia. Biochem Soc Trans. 2011;39:1120–1125. doi: 10.1042/BST0391120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholey JM. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol. 2013;29:443–469. doi: 10.1146/annurev-cellbio-101512-122335. [DOI] [PubMed] [Google Scholar]
- 22.Nonaka S, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 23.Marszalek JR, et al. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda S, et al. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingham PW, McMahon AP. Hedgehog signalling: Kif7 is not that fishy after all. Curr Biol. 2009;19:R729–R731. doi: 10.1016/j.cub.2009.07.060. [DOI] [PubMed] [Google Scholar]
- 26.Lin F, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cano DA, et al. Primary cilia deletion in pancreatic epithelial cells results in cyst formation and pancreatitis. Gastroenterology. 2006;131:1856–1869. doi: 10.1053/j.gastro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 28.Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sipe CW, Lu X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 2011 doi: 10.1242/dev.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan S, et al. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development. 2013;140:4445–4451. doi: 10.1242/dev.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoder BK. Role of Primary Cilia in the Pathogenesis of Polycystic Kidney Disease. Journal of the American Society of Nephrology. 2007;18:1381–1388. doi: 10.1681/ASN.2006111215. [DOI] [PubMed] [Google Scholar]
- 33.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borovina A, Ciruna B. IFT88 plays a cilia- and PCP-independent role in controlling oriented cell divisions during vertebrate embryonic development. Cell Reports. 2013;5:37–43. doi: 10.1016/j.celrep.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 35.Haycraft CJ, et al. Intraflagellar transport is essential for endochondral bone formation. Development. 2007;134:307–316. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- 36.Koyama E, et al. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–2169. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B, et al. Primary cilia integrate hedgehog and Wnt signaling during tooth development. J Dent Res. 2014;93:475–482. doi: 10.1177/0022034514528211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 39.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. Journal of Neuroscience. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong CK, et al. Primary cilia are required in a unique subpopulation of neural progenitors. Proceedings of the National Academy of Sciences. 2014;111:12438–12443. doi: 10.1073/pnas.1321425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han YG, et al. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao C, et al. Kinesin-2 family in vertebrate ciliogenesis. Proc Natl Acad Sci USA. 2012;109:2388–2393. doi: 10.1073/pnas.1116035109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, et al. Functional analysis of mouse kinesin motor Kif3C. Mol Cell Biol. 2001;21:5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter BS, et al. The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. Journal of Cell Science. 2015;128:1034–1050. doi: 10.1242/jcs.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovacs JJ, et al. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng J, et al. The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat Cell Biol. 2005;7:474–482. doi: 10.1038/ncb1249. [DOI] [PubMed] [Google Scholar]
- 47.Pal K, et al. Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212:861–875. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milenkovic L, et al. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proceedings of the National Academy of Sciences. 2015;112:8320–8325. doi: 10.1073/pnas.1510094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye F, et al. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. Elife. 2013;2:e00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breslow DK, et al. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin YC, et al. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ou G, et al. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 53.Prevo B, et al. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 2015;17:1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- 54.Insinna C, et al. Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev Dyn. 2009;238:2211–2222. doi: 10.1002/dvdy.21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Insinna C, et al. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams CL, et al. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun. 2014;5:5813. doi: 10.1038/ncomms6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang L, et al. Kinesin family 17 osmotic avoidance abnormal-3 is dispensable for photoreceptor morphology and function. FASEB J. 2015 doi: 10.1096/fj.15-275677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts AJ, et al. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013 doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikami A, et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. Journal of Cell Science. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 62.Pazour GJ, et al. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nature Publishing Group. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou Y, Witman GB. Dynein and intraflagellar transport. Exp Cell Res. 2015 doi: 10.1016/j.yexcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 67.Ocbina PJR, et al. Complex interactions between genes controlling trafficking in primary cilia. Nat Genet. 2011;43:547–553. doi: 10.1038/ng.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.See SK, et al. Cytoplasmic Dynein Antagonists with Improved Potency and Isoform Selectivity. ACS Chem Biol. 2016;11:53–60. doi: 10.1021/acschembio.5b00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidts M, et al. Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. Journal of Medical Genetics. 2013;50:309–323. doi: 10.1136/jmedgenet-2012-101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kon T, et al. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 71.Imamula K, et al. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt H, et al. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ocbina PJR, Anderson KV. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmer KJ, et al. A role for Tctex-1 (DYNLT1) in controlling primary cilium length. Eur J Cell Biol. 2011;90:865–871. doi: 10.1016/j.ejcb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asante D, et al. Subunit composition of the human cytoplasmic dynein-2 complex. Journal of Cell Science. 2014;127:4774–4787. doi: 10.1242/jcs.159038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor SP, et al. Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat Commun. 2015;6:7092. doi: 10.1038/ncomms8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kessler K, et al. Identification of mutations in DYNC2LI1, a member of the mammalian cytoplasmic dynein 2 complex, expands the clinical spectrum of Jeune/ATD ciliopathies. Cilia. 2015 doi: 10.1186/2046-2530-4-S1-P59. [DOI] [Google Scholar]
- 78.McInerney-Leo AM, et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet. 2013;93:515–523. doi: 10.1016/j.ajhg.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huber C, et al. WDR34 mutations that cause short-rib polydactyly syndrome type III/severe asphyxiating thoracic dysplasia reveal a role for the NF-κB pathway in cilia. Am J Hum Genet. 2013;93:926–931. doi: 10.1016/j.ajhg.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cossu C, et al. New mutations in DYNC2H1 and WDR60 genes revealed by whole-exome sequencing in two unrelated Sardinian families with Jeune asphyxiating thoracic dystrophy. Clin Chim Acta. 2016;455:172–180. doi: 10.1016/j.cca.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 81.Rana AA, et al. Targeted deletion of the novel cytoplasmic dynein mD2LIC disrupts the embryonic organiser, formation of the body axes and specification of ventral cell fates. Development. 2004;131:4999–5007. doi: 10.1242/dev.01389. [DOI] [PubMed] [Google Scholar]
- 82.Schmidts M, et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat Commun. 2015;6:7074. doi: 10.1038/ncomms8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qin J, et al. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc Natl Acad Sci USA. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liem KF, et al. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goggolidou P, et al. ATMIN is a transcriptional regulator of both lung morphogenesis and ciliogenesis. Development. 2014;141:3966–3977. doi: 10.1242/dev.107755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukhopadhyay S, et al. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mukhopadhyay S, et al. The Ciliary G-Protein-Coupled Receptor Gpr161 Negatively Regulates the Sonic Hedgehog Pathway via cAMP Signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 89.Harris JA, et al. Single-particle imaging intraflagellar transport-independent transport and accumulation of EB1 in Chlamydomonas flagella. Mol Biol Cell. 2016;27:295–307. doi: 10.1091/mbc.E15-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 91.Seo S, et al. A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7:e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eguether T, et al. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;31:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lechtreck KF, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Q, et al. BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum Mol Genet. 2012;21:1945–1953. doi: 10.1093/hmg/dds004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keady BT, et al. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhogaraju S, et al. Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 2011;30:1907–1918. doi: 10.1038/emboj.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liew GM, et al. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31:265–278. doi: 10.1016/j.devcel.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yee LE, et al. Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling. PLoS Genet. 2015;11:e1005627. doi: 10.1371/journal.pgen.1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He M, et al. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Putoux A, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 102.Liem KF, et al. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheung HO-L, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 104.Endoh-Yamagami S, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 105.Maurya AK, et al. Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS Genet. 2013;9:e1003955. doi: 10.1371/journal.pgen.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tay SY, et al. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 107.Liu YC, et al. The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci Signal. 2014;7:ra117. doi: 10.1126/scisignal.2005608. [DOI] [PubMed] [Google Scholar]
- 108.Klejnot M, Kozielski F. Structural insights into human Kif7, a kinesin involved in Hedgehog signalling. Acta Crystallogr D Biol Crystallogr. 2012;68:154–159. doi: 10.1107/S0907444911053042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hirokawa N, et al. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 110.Li ZJ, et al. Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development. 2012 doi: 10.1242/dev.081190. [DOI] [PubMed] [Google Scholar]
- 111.Monnier V, et al. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC Dev Biol. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karaer K, et al. A novel KIF7 mutation in two affected siblings with acrocallosal syndrome. Clin Dysmorphol. 2015;24:61–64. doi: 10.1097/MCD.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 113.Tunovic S, et al. Novel KIF7 missense substitutions in two patients presenting with multiple malformations and features of acrocallosal syndrome. Am J Med Genet A. 2015;167:2767–2776. doi: 10.1002/ajmg.a.37249. [DOI] [PubMed] [Google Scholar]
- 114.Ibisler A, et al. Novel KIF7 Mutation in a Tunisian Boy with Acrocallosal Syndrome: Case Report and Review of the Literature. Mol Syndromol. 2015;6:173–180. doi: 10.1159/000439414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barakeh D, et al. The many faces of KIF7. Hum Genome Var. 2015;2:15006. doi: 10.1038/hgv.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walsh DM, et al. Acrocallosal syndrome: identification of a novel KIF7 mutation and evidence for oligogenic inheritance. Eur J Med Genet. 2013;56:39–42. doi: 10.1016/j.ejmg.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Ali BR, et al. Ali et al 2013. Orphanet Journal of Rare Diseases. 2012;7:1–1. doi: 10.1186/1750-1172-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Putoux A, et al. Novel KIF7 mutations extend the phenotypic spectrum of acrocallosal syndrome. Journal of Medical Genetics. 2012;49:713–720. doi: 10.1136/jmedgenet-2012-101016. [DOI] [PubMed] [Google Scholar]
- 119.Dafinger C, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121:2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Coles GL, Ackerman KG. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2013;110:E1898–905. doi: 10.1073/pnas.1222797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Coles GL, et al. KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms. PLoS Genet. 2015;11:e1005525. doi: 10.1371/journal.pgen.1005525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vogel P, et al. Congenital hydrocephalus in genetically engineered mice. Vet Pathol. 2012;49:166–181. doi: 10.1177/0300985811415708. [DOI] [PubMed] [Google Scholar]
- 123.Wilson CW, et al. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nozawa YI, et al. Fused (Stk36) is a ciliary protein required for central pair assembly and motile cilia orientation in the mammalian oviduct. Dev Dyn. 2013;242:1307–1319. doi: 10.1002/dvdy.24024. [DOI] [PubMed] [Google Scholar]
- 125.Rink JC, et al. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Piao T, et al. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kobayashi T, et al. Centriolar Kinesin Kif24 Interacts with CP110 to Remodel Microtubules and Regulate Ciliogenesis. Cell. 2011 doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 128.Yadav SP, et al. Centrosomal protein CP110 controls maturation of the mother centriole during cilia biogenesis. Development. 2016;143:1491–1501. doi: 10.1242/dev.130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miyamoto T, et al. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Poirier K, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Insolera R, et al. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Niwa S, et al. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 133.García-García MJ, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jimeno D, et al. Kinesin-2 and photoreceptor cell death: requirement of motor subunits. Exp Eye Res. 2006;82:351–353. doi: 10.1016/j.exer.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 135.Li Y, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mei L, et al. Targeted next-generation sequencing identifies novel compound heterozygous mutations of DYNC2H1 in a fetus with short rib-polydactyly syndrome, type III. Clin Chim Acta. 2015;447:47–51. doi: 10.1016/j.cca.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 137.Okamoto T, et al. Novel compound heterozygous mutations in DYNC2H1 in a patient with severe short-rib polydactyly syndrome type III phenotype. Congenit Anom (Kyoto) 2015;55:155–157. doi: 10.1111/cga.12098. [DOI] [PubMed] [Google Scholar]
- 138.Hokayem El J, et al. NEK1 and DYNC2H1 are both involved in short rib polydactyly Majewski type but not in Beemer Langer cases. Journal of Medical Genetics. 2012;49:227–233. doi: 10.1136/jmedgenet-2011-100717. [DOI] [PubMed] [Google Scholar]
- 139.Dagoneau N, et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet. 2009;84:706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Merrill AE, et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet. 2009;84:542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen LS, et al. Identification of novel DYNC2H1 mutations associated with short rib-polydactyly syndrome type III using next-generation panel sequencing. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15028134. [DOI] [PubMed] [Google Scholar]
- 142.Kessler K, et al. DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects. Sci Rep. 2015;5:11649. doi: 10.1038/srep11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schmidts M, et al. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet. 2013;93:932–944. doi: 10.1016/j.ajhg.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gholkar AA, et al. Tctex1d2 associates with short-rib polydactyly syndrome proteins and is required for ciliogenesis. Cell Cycle. 2015;14:1116–1125. doi: 10.4161/15384101.2014.985066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tukachinsky H, et al. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Humke EW, et al. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hammerschmidt M, et al. Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev. 1996;10:647–658. doi: 10.1101/gad.10.6.647. [DOI] [PubMed] [Google Scholar]
- 148.Tuson M, et al. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011 doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mick DU, et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Masyuk AI, et al. Cholangiocyte primary cilia are chemosensory organelles that detect biliary nucleotides via P2Y12 purinergic receptors. Am J Physiol Gastrointest Liver Physiol. 2008;295:G725–34. doi: 10.1152/ajpgi.90265.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bieling P, et al. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 152.van der Vaart B, et al. CFEOM1-Associated Kinesin KIF21A Is a Cortical Microtubule Growth Inhibitor. Dev Cell. 2013 doi: 10.1016/j.devcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 153.Walczak CE, et al. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]