Abstract
Proteins that lack tertiary stability under normal conditions, known as intrinsically disordered, exhibit a wide range of biological activities. Molecular descriptions for the biology of intrinsically disordered proteins (IDPs) consequently rely on disordered structural models, which in turn require experiments that assess the origins to structural features observed. For example, while hydrodynamic size is mostly insensitive to sequence composition in chemically denatured proteins, IDPs show strong sequence-specific effects in the hydrodynamic radius (Rh) when measured under normal conditions. To investigate sequence-modulation of IDP Rh, disordered ensembles generated by a Hard Sphere Collision model modified with a structure-based parameterization of the solution energetics were used to parse the contributions of net charge, main chain dihedral angle bias, and excluded volume on hydrodynamic size. Ensembles for polypeptides 10 to 35 residues in length were then used to establish power-law scaling relationships for comparison to experimental Rh from 26 IDPs. Results showed the expected outcomes of increased hydrodynamic size from increases in excluded volume and net charge, and compaction from chain-solvent interactions. Chain bias representing intrinsic preferences for α helix and polyproline II (PPII), however, modulated Rh with intricate dependence on the simulated propensities. PPII propensities at levels expected in IDPs correlated with heightened Rh sensitivity to even weak α helix propensities, indicating bias for common (φ, ψ) are important determinants of hydrodynamic size. Moreover, data show that IDP Rh can be predicted from sequence with good accuracy from a small set of physicochemical properties, namely intrinsic conformational propensities and net charge.
Keywords: dynamic light scattering, size exclusion chromatography, polyproline type II, net charge, simulation
INTRODUCTION
Structural characterizations of intrinsically disordered proteins (IDPs) provide key molecular details from which to understand the broad range of biological tasks that have been associated with this protein class.1-4 Identified by a persistent lack of tertiary stability, intrinsic disorder is common in eukaryotic proteins and protein domains.1 Functionally, IDPs have roles regulating gene expression,5,6 cellular communication,7-9 and subcellular spatial organization,10-12 as well as mechanical duties like maintaining curvature in the cell bilayer.13 Molecular descriptions of IDP biology and its associated human disorders6,9,14 thus depend on an accurate accounting of the features and properties of disordered protein structures.
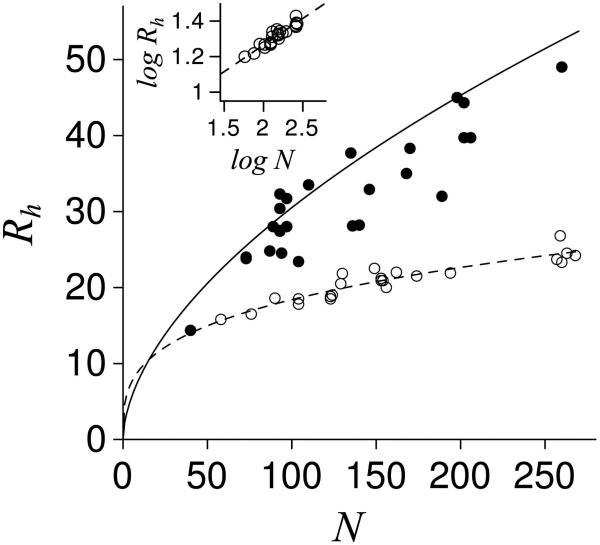
Hydrodynamic size, as represented by the radius of gyration, Rg, or the hydrodynamic radius, Rh, has been widely used for quantifying protein structures15-17 and other polymers.18-20 Rh measured under normal conditions and from published reports for IDPs21-39 and folded proteins16,40,41 are provided in Figure 1. When comparing sets of Rh (or Rg), logarithmic plots of Rh and N, the number of subunits in a polymer (e.g., the number of amino acids in a protein), are useful since the y-axis intercept and slope in the dataset trend provide a pre-factor (Ro) and exponent (v) for a power-law scaling relationship that can be used to numerically approximate Rh from N; Rh ~ Ro·Nv. More importantly, v, sometimes referred to as the Flory exponent, provides information on the nature of the molecular interactions governing the observed Rh.42,43 For v less than 0.5, polymers are considered to be in ‘poor’ solvents where chain-chain intramolecular interactions dominate relative to chain-solvent intermolecular interactions. For example, a log-log plot of Rh and N for the folded proteins in Figure 1 (see inset) yields v = 0.3, indicating that the hydrodynamic dimensions of folded proteins are established mostly from intramolecular chain-chain contacts (e.g., hydrogen bonds, ionic pairs, packing interactions) that form owing to folding and the accompanying burial of atomic surfaces. If folded proteins are transferred to solutions with high concentrations of urea or guanidine hydrochloride, causing denaturation of native folds, v increases to ~ 0.6.15,16 Polymers are considered to be in ‘good’ solvents where chain-solvent interactions dominate relative to chain-chain interactions when v is greater than 0.5.42,43 The observed v for chemically-denatured proteins thus predicts well-solvated structures with few stable tertiary contacts and minimal burial of atomic surfaces.
Figure 1.
Rh trends with N in proteins. Solid and open circles are IDPs21-39 and folded proteins,16,40,41 respectively. The trend for folded proteins, Rh = 4.65·N0.30, was determined from the inset plot and is shown by the dashed line. The trend for chemically denatured proteins, Rh = 2.21·N0.57, from Wilkins16 is shown by the solid line. Rh is reported in Å. Inset: Plot of log Rh and log N for the folded proteins, which gave a linear slope and y-intercept of 0.298 (v ~ 0.30) and 0.667 (Ro = antilog (0.667) ~ 4.65), respectively.
When comparing Rh from IDPs under normal conditions to the expected hydrodynamic size of a protein in a poor solvent, given by the power-law scaling relationship for folded proteins (see Figure 1), it is not surprising that IDP Rh are noticeably larger relative to folded proteins of similar N. IDP structures are mostly lacking in stable, tertiary contacts1-4 that are hallmarks of folded and globular proteins and, accordingly, should exhibit larger v and concomitant larger Rh. Likewise, since normal aqueous solutions are poor solvents for polypeptides relative to concentrated urea or guanidine hydrochloride solutions, as demonstrated with solubility studies44-46 and fluorescence correlation spectroscopy experiments,47 it may not be surprising that IDPs exhibit Rh that are usually, but not always, smaller than the trend for chemically-denatured proteins. From the reference of folded and chemically-denatured proteins, Rh that have been observed for IDPs are reasonable.
In contrast to chemically denatured proteins, which yield Rh and Rg that are mostly insensitive to details of amino acid sequence,15,16 IDPs with similar N often show large differences in Rh. For example, Rh ranges from ~ 24-32 Å for IDPs with N ~ 90 in the dataset of Figure 1. The molecular origins for these differences in Rh could be owing to coulombic interactions between charged side chains, since the spacing48 and sequential patterns49 of charged groups in disordered structures are known to influence Rh. Biases in main chain dihedral angles, such as propensities for (φ, ψ) associated with type II polyproline helix (PPII), are also thought to modulate Rh in disordered structures.21,48 Overall, these data indicate that certain sequence details are capable of modulating the structural features of disordered proteins under normal conditions. Identifying the influential physical properties and determining their relative strengths (i.e., energetics) and additivities are thus prerequisites for a quantitative description of IDP structures.
Here, we investigate the effects of net charge, main chain dihedral angle bias, and excluded volume on the hydrodynamic dimensions of IDPs as reported by Rh. The term ‘excluded volume’ is used to represent the exclusion of chain conformations from steric restrictions associated with atomic volumes. Computer simulation of disordered structures using a Hard Sphere Collision (HSC) model developed by Richards50 and modified to approximate chain-solvent interactions using the solvent accessible surface area (SASA) parameterization of Hilser and Freire51 provided a theoretical construct from which to parse and combine these structural effectors. Simulations were used to generated large conformational ensembles of disordered polypeptides ranging in size from 10 to 35 residues that were then used to establish power-law scaling relationships for comparison to the set of IDP Rh in Figure 1. Since prior experiments and calculations have shown that the hydrodynamic sizes of disordered ensembles are indeed sensitive to sequence details associated with net charge and intrinsic PPII propensities,21,48,49,52-54 experimental Rh from IDPs with net charge and chain bias for PPII that are statistically elevated relative to dataset norms were verified using dynamic light scattering (DLS) and analytical size exclusion chromatography (SEC) techniques. These verifications of experimental Rh were performed to control for data that could be strongly influential in our comparative study.
The results indicated that excluded volume effects owing to sequence differences were generally minor for establishing hydrodynamic size differences among IDPs. Net charge effects on Rh were mostly consistent across the IDP dataset, whereby increases in net charge correlated with increased Rh, showing agreement with previous reports.52-54 The IDP with the highest net charge, prothymosin-α, however, trended separately from the other IDPs in the dataset when assessing apparent Rh sensitivity to net charge. An important structural parameter for establishing Rh in disordered ensembles seemed to be main chain dihedral angle bias. Rh sensitivity to chain bias for α helix was detected in both a sequence analysis of the IDP dataset and computer generated ensembles simulated with the HSC model. Of note, the effects on Rh from α helix propensities, which caused compaction, were stronger when combined with PPII propensities. Overall, the results indicate a key role provided by intrinsic chain bias for determining the hydrodynamic size and show that, at least under normal conditions, IDP Rh can be described accurately from sequence-based estimates of intrinsic conformational propensities and net charge.
MATERIALS AND METHODS
Computer Generation of Disordered Structures
Conformers for polypeptide chains restricted to the 20 normal amino acids were generated by a random search of conformational space using the HSC model.40,50 A detailed description of the computer algorithm is available elsewhere.55 Briefly, this model uses van der Waals atomic radii56,57 as the only scoring function to eliminate grossly improbable conformations. The procedure to generate a random conformer starts with a unit peptide and all other atoms for a chain are determined by the rotational matrix.58 Backbone atoms are generated from the dihedral angles φ, ψ, and ω and the standard bond angles and bond lengths.59 To sample conformational space efficiently, (φ, ψ) are restricted to the allowed Ramachandran regions.60 For peptide bond dihedral angles (ω), non-PRO amino acids were given 100% trans form (180°) and PRO sampled the cis form (0°) at a rate of 10% if the preceding amino acid was non-PRO, and at a rate of 6% if the preceding amino acid was PRO.61 The dihedral angle ω was given a Gaussian fluctuation of ± 5° around the value of 180° or 0°. Of the two possible positions of the Cβ atom, the one corresponding to L-amino acid residues was used throughout the studies. The positions of all other side chain atoms were determined from random sampling of rotamer libraries.62 To calculate state distributions typical of protein ensembles, a structure-based energy function parameterized to solvent-accessible surface areas was used to population-weight the generated structures.63-71
Expression and purification of recombinant protein
Genes coding for Hdm2-ABD and prothymosin-α, each including an N-terminal histidine tag and thrombin cleavage site, were cloned into plasmid expression vectors by DNA 2.0 (Menlo Park, California). Hdm2-ABD and prothymosin-α were individually expressed, isolated from bacterial lysate, and affinity tag removed using the expression and purification protocols described elsewhere for recombinant human p53(1-93).72 The same expression and purification protocols were used for recombinant wild type human p53(1-93) and the PRO− and ALA−PRO− variants.21 Recombinant PGR was provided as a gift from Andrew Herr (Cincinnati Children’s Hospital, Cincinnati, Ohio, USA). Rh for each IDP was measured by techniques based on dynamic light scattering (DLS) and size exclusion chromatography (SEC), described below. SEC methods require a linear relationship between KD (distribution coefficient) and Rh for the tested range, which was demonstrated using the folded proteins Staphylococcal nuclease, bovine erythrocyte carbonic anhydrase, chicken egg albumin, and horse heart myoglobin. Nuclease was expressed and isolated using the protocols described elsewhere.73 Carbonic anhydrase, albumin, and myoglobin were purchased from Sigma-Aldrich (St. Louis, MO) and further processed by ion exchange chromatography and extensive dialysis to remove residual contaminants.
DLS measured Rh
Dynamic light scattering readings used noninvasive backscatter optics and were measured using a Zetasizer Nano ZS with Peltier temperature control from Malvern Instruments (Worcestershire, UK). All measurements were performed at 25 °C and used 1-cm path-length quartz cuvettes, as described elsewhere.21,40 Samples contained 600 μL of ~ 0.5 mg/mL protein and were filtered immediately prior to use by 0.2-μm PVDF syringe-driven filters. Rh reported for each protein was the average of at least 5 measurements.
SEC measured Rh
Size exclusion chromatography (SEC) experiments used Sephadex G-75 (GE Healthcare, Piscataway, NJ) equilibrated in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7, following previously described protocols.21,40 Elution volumes (Ve) for each protein were determined from chromatograms measured using a Bio-Rad BioLogic LP System equipped with a UV absorbance monitor (Hercules, CA). Each sample was 100 µL and contained 0.5–1 mg/mL protein in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7 with 3 mg/mL blue dextran and 0.3 mg/mL 2,4-dinitrophenyl-L-aspartate added as indicator dyes for determining the void (Vo) and total (Vt) column volumes, respectively. For PGR and prothymosin-α, which lack PHE residues and have low absorbance at 280 nm, 250 µL samples containing 3-4 mg/mL of protein were used. KD for each protein was determined as KD = (Ve - Vo)/(Vt - Vo). KD reported for each protein was the average of at least 3 room-temperature (~ 22 °C) measurements.
RESULTS
Composition of experimental Rh dataset
The hydrodynamic dimensions of disordered proteins under normal conditions are known to depend on sequence, specifically proline54 and alanine content,21 chain length,54 chain bias for PPII,48 net charge,52-54 and charge distributions where mixing of positive and negative charged residues show compaction relative to linear stretches of like charges.48,49 IDPs in the experimental dataset, which numbered 26, were selected to provide sequence-based variations associated with these known modulators of Rh. N ranged from 40 to 260 and net charge from 1 to 43, where low net charge was from mixing positive and negative groups (e.g., SNAP25 and securin) or from relatively few charged residues (e.g., ShB-C). The dataset includes IDPs known to aggregate under certain conditions (e.g., CFTR-R74 and α-synuclein75) or form dimers (e.g., sml139), as well as IDPs that have high monomeric solubility (e.g., p53 TAD22 and prothymosin-α24). Sequence content among dataset IDPs was generally diverse (see Fig. S1 in Supporting Information) with, for example, the fractional number of PRO residues (i.e., (# PRO residues)/N) ranging from 0 to 0.29, ALA from 0 to 0.24, SER from 0.02 to 0.20, and GLU from 0.03 to 0.31. The identity, sequence, and experimentally determined Rh for each IDP is provided in Table S1 of the Supporting Information. Rh measured for proteins containing histidine affinity tags were avoided since these affinity tags are known to compact Rh.54 Folded proteins destabilized by mutation (e.g., CTL9-I98A76) were not included to limit the investigation to IDP sequences.
As mentioned above, numerous studies have demonstrated that increases in net charge correlate with increases in Rh for IDPs.52-54 For each IDP in Table S1, net charge was estimated from sequence as the absolute value in the number of GLU and ASP residues minus the number of LYS and ARG residues. This sequence-estimated net charge was normalized to IDP size,
| (1) |
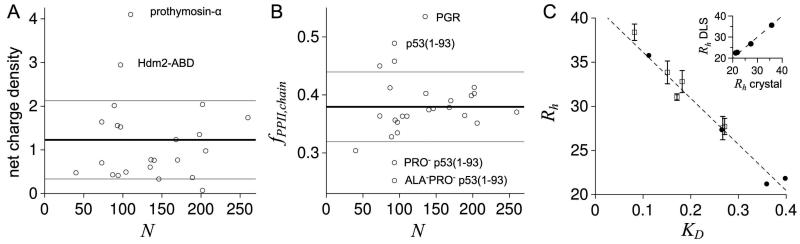
using N and the Flory exponent for IDPs54 to calculate a net charge density. Figure 2A shows that net charge density calculated for 23 of 26 IDPs were within 1 standard deviation of the dataset average, 1.2 ± 0.9. Outlier IDPs are indicated in the figure and, of the 3, sequences from Hdm2-ABD and prothymosin-α gave net charge density values that were significantly above the dataset norm. Based on these data, charge effects on structure may be pronounced in Hdm2-ABD and prothymosin-α relative to the cumulative effects of charge on structure in the other dataset IDPs. Accordingly, experimental Rh from Hdm2-ABD and prothymosin-α may hold greater influence for assessing the effects on Rh owing to net charge. To verify Rh for these two IDPs, recombinant Hdm2-ABD and prothymosin-α were expressed, isolated from bacterial lysate, and Rh measured using DLS and SEC techniques. Results from these experiments are shown in Figure 2C and demonstrate that Rh for Hdm2-ABD and prothymosin-α were found to be 31.7 Å and 33.6 Å, respectively. Our measurements for prothymosin-α gave Rh that were practically identical to the literature reported value of 33.7 Å.24 The literature reported value for Hdm2-ABD was 25.7 Å,78 which was lower than our measurements. Changing solution conditions to match the prior report (10 mM sodium phosphate, 10 mM sodium chloride, pH 6) did not correct the discrepancy, yielding 34.2 Å for Rh for Hdm2-ABD when determined from SEC-measured KD. We assume the increase in Rh for our second measurement was from decreased solution ionic strength, which could increase net electrostatic repulsion in Hdm2-ABD from its high net charge.
Figure 2.
A) Net charge density calculated from sequence for each dataset IDP. B) Chain-averaged PPII propensity calculated from sequence for each dataset IDP using experimental propensities.79 In A and B, bold lines are dataset averages, whereas grey lines are averages ± the standard deviation. C) Rh (in Å) and KD for the IDPs identified in panels A and B and the folded proteins chicken egg albumin, bovine erythrocyte carbonic anhydrase, Staphylococcal nuclease, and horse heart myoglobin. Open squares are DLS-measured Rh, from largest to smallest, for the IDPs PGR (38.4 ± 0.9Å), prothymosin-α (33.8 ± 1.3Å), p53(1-93) (32.8 ± 1.2Å), Hdm2-ABD (31.0 ± 0.5Å), ALA−PRO− p53(1-93) (27.7 ± 0.9Å), and PRO− p53(1-93) (27.5 ± 1.3Å). Filled circles are Rh estimated as one-half the maximum Cα-Cα distance in the crystallographic structures of albumin,81 carbonic anhydrase,82 nuclease,83 and myoglobin.84 KD is the distribution coefficient determined by SEC. Standard deviations from measuring KD were < 0.007. The dashed line is a linear fit of Rh to KD applied to the filled circles (folded proteins), which was used to estimate Rh from KD for the IDPs. Table S1 lists the averages of the DLS-measured and KD-estimated Rh for each of the 6 IDPs. Inset: Filled circles are Rh measured by DLS for the folded proteins compared to Rh estimated from crystal structures as one-half the maximum Cα-Cα distance, showing good agreement. DLS-measured Rh for each folded protein was: albumin (35.6 ± 0.5Å), carbonic anhydrase (26.8 ± 0.8Å), myoglobin (22.7 ± 0.8Å), and nuclease (22.4 ± 0.4Å).
IDP Rh have also been observed to trend with intrinsic propensities for PPII helix.21,48 To estimate chain bias for (φ, ψ) representative of PPII structure for each dataset IDP, experimental propensities for the 20 common amino acids from Elam and colleagues79 were used. Figure 2B shows the chain averaged propensity, fPPII,chain, determined for each IDP sequence by summing the experimental propensities across a sequence and dividing by N. For the dataset, the average fPPII,chain was 0.38 ± 0.06. The 135-residue intrinsically disordered Proline/Glycine-rich Region (PGR) of the cell surface protein Aap from Staphylococcus epidermidis80 was included in the IDP dataset specifically because its sequence yields a value for fPPII,chain that is much higher than average. Figure 2C shows that Rh measured for recombinant PGR using DLS and SEC methods gave an average of 37.7 Å. Other IDPs with calculated fPPII,chain that were substantially outside the dataset norm were p53(1-93) and the PRO− and ALA−PRO− variants of p53(1-93). Since experimental Rh from these IDPs could hold greater influence for testing the effects on Rh owing to chain bias for PPII, experimental Rh were verified using DLS and SEC. Results from these experiments are given in Figure 2C and demonstrate that Rh for wild type p53(1-93), PRO− p53(1-93), and ALA−PRO− p53(1-93) were 32.3 Å, 27.5 Å, and 27.4 Å, respectively, each of which were practically identical to the literature reported values.21
Rh calculated from simulated ensembles
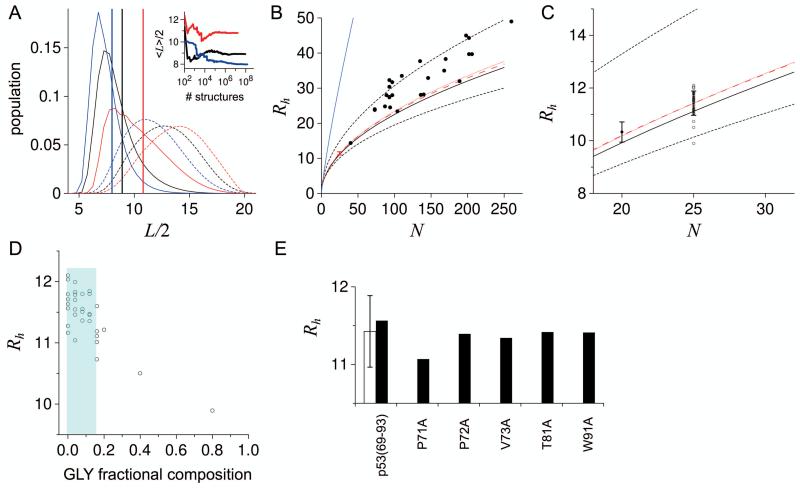
Conformers for polypeptide sequences were generated using the HSC model,40,50 which builds structures from the standard bond angles and bond lengths,59 a random sampling of backbone dihedral angles and side chain rotamer libraries,62 and discards those containing contact violations based on van der Waals atomic radii.56,57 Each structure generated for a conformational ensemble is independent of previously generated structures. Figure 3A demonstrates typical HSC simulation output, using 15-residue poly-GLY, poly-ALA, and poly-PRO sequences as examples. Although any number of structural metrics could be presented, L/2 is used to represent an approximate Rh. L was calculated as the maximum Cα-Cα distance in a generated structure and <L> is the population-weighted average for an ensemble. The inset to Figure 2C shows that L/2 is a reasonable estimate for experimental Rh when the structure of a protein is known.
Figure 3.
Rh calculated from simulated conformational ensembles. A) L/2 distribution in ensembles of poly-GLY (blue), poly-ALA (black), and poly-PRO (red) with N = 15. Stippled curves were determined by giving each structure equal statistical weight; solid curves are probability-weighted distributions using equation 2. Vertical lines show <L>/2 calculated for each ensemble by equation 3. Inset: <L>/2 calculated for ensembles with increasingly larger numbers of simulated structures. B) Filled circles are experimental IDP Rh. Lower-stippled, solid, and upper-stippled black lines show the trend in Rh with N from ensembles simulated for poly-GLY, poly-ALA, and poly-PRO, respectively. Red dot with error bar at N = 25 is the average ± standard deviation in Rh calculated from ensembles simulated for IDP fragments. Red lines extrapolate the fragment average to larger N by using the poly-ALA scaling relationship (Rh = 2.16·N0.509) with pre-factor or exponent modified for agreement with the fragment average at N = 25 (dashed line is Ro changed to 2.22 and v kept at 0.509; stippled line is Ro kept at 2.16 and v changed to 0.518). Blue line is the trend for poly-PRO when simulated with PPII propensity of 0.95 at each residue position. C) Open circles show Rh from ensembles simulated for each 25-residue IDP fragment, with the average ± the standard deviation given by the error bar (11.4 ± 0.5 Å). Rh from ensembles simulated for IDP fragments with N = 20 give an average ± standard deviation of 10.3 ± 0.4 Å, shown by the filled circle with error bar. Lines match their panel B representations. D) Open circles are Rh from ensembles simulated for the 25-residue IDP fragments and the corresponding GLY compositions determined from sequence. Shading is the range in fractional GLY composition for the natural IDP sequences in the dataset. E) Open column with error bar is the average Rh (± standard deviation) from ensembles simulated for the 33 IDP fragments with N = 25. Black columns show Rh calculated from ensembles simulated for 6 different sequences based on the C-terminal 25-residue fragment from p53(1-93) wild type. Shown are p53(69-93) (left-most black column) and p53(69-93) with ALA point substitutions applied at the PRO-71, PRO-72, VAL-73, THR-81, and TRP-91 positions as labeled. Rh is in Å.
If the generated structures are given equal statistical weight, symmetric distributions are obtained for L/2 in ensemble populations. Relatively few conformers for an ensemble exhibit highly extended or highly compacted structures and most are structurally in-between these two extremes. To approximate the effects of chain-solvent interactions on an ensemble, a structure-based energy function that has been parameterized to SASA51 and tested extensively63-71 was used to estimate the solution energetics of each generated conformer. This SASA-based energy function does not include terms accounting for the energetics of common intramolecular forces such as intra-chain hydrogen bonds and charge-charge interactions, which could be added separately.48 The energy function calculates a Gibbs free energy for each structure (ΔGi) and sets probabilities by the Boltzmann distribution,
| (2) |
where Pi is the probability of structure i, Ki is the statistical weight from the relative Gibbs free energy (Ki = e−ΔGi/RT, where R is the gas constant and T is absolute temperature), and the summation is over all structures in an ensemble. Ensemble populations for poly-GLY, poly-ALA, and poly-PRO favor compacted structures with application of the SASA-based energy function (see Fig. 3A), phenomenologically similar to the hydrophobic collapse expected of polypeptides in water.47 Energy-weighted distributions tend to show a positive skew, with the population-weighted value,
| (3) |
slightly higher than the distribution mode. To calculate Rh from a simulated ensemble for polypeptide sequences using this model, conformers were generated until <L> converged to a statistically stable value (Fig. 3A inset). <L> was considered stable if its value changed by less than 1% over a 10-fold increase in the number of conformers generated. For the computational results reported here, Rh was calculated from simulated ensembles as <L>/2 using equation 3.
Estimating excluded volume effects on Rh
Computer simulations using the HSC model to compute <L> were intractable for poly-ALA with N > 75,40 and prohibit direct simulation of full sequences from each IDP in the dataset. Polypeptide sequences containing large, bulky side chains (e.g., PHE, TRP, PRO) also exhibit less efficient conformer sampling from generating steric conflicts at higher rates. To manage these computational limitations associated with polypeptide length, Rh (i.e., <L>/2) calculated from ensembles for short sequences were extrapolated to larger N. For example, simulation results for poly-ALA, poly-GLY, and poly-PRO are shown in Figure 3B and compared to IDP Rh from the experimental dataset. Rh for poly-GLY and poly-PRO were calculated from ensembles using N = 10, 15, 20, 25, and 35. Log-log plots of Rh and N were constructed to obtain the pre-factor (Ro) and exponent (v) for the poly-GLY and poly-PRO curves shown in the figure. For poly-ALA, the trend of Rh with N determined previously40 was used.
Comparing Rh from simulated ensembles for poly-ALA, poly-PRO, and poly-GLY demonstrates the limits of sequence-based excluded volume effects on IDP Rh in this model. Removing a heavy atom (Cβ) attached to the main chain at each residue position results in decreases in Rh at all N, as shown by poly-GLY relative to poly-ALA. In contrast, attaching an additional heavy atom (Cδ) directly to the main chain at each residue position increases Rh, as shown by poly-PRO relative to poly-ALA.
To estimate excluded volume effects on Rh owing to sequence differences among IDPs, 25-residue polypeptide fragments were selected from the IDP dataset using the following protocol. First, IDPs with the two highest and two lowest net charge density values (Fig. 2A) were selected. These were Hdm2-ABD, prothymosin-α, ShB-C, and securin. Next, IDPs with the two highest and two lowest fPPII,chain values (Fig. 2B) were selected. These were PGR, p53(1-93), PRO− p53(1-93), and ALA−PRO− p53(1-93). Finally, the two IDPs with fPPII,chain closest to the dataset average (Fos-AD and HIF1-α-530) and the two IDPs with net charge density closest to the dataset average (Fos-AD and tau-K45) were selected. The goal from the selection process was to obtain IDP sequences at the extremes and at the averages for physical properties known to modulate Rh. From each of these 11 different IDPs, 25 residue fragments from the N-terminus, C-terminus, and center of each sequence were extracted, producing 33 25-residue sequences that were simulated using the HSC model to compute Rh. A table is provided in Supporting Information listing the fragment sequences and Rh that were computed from simulation (Table S2). This table also provides net charge density and fPPII,chain determined for each fragment for comparison to the parent sequences. Since the SASA-based energy function does not include terms for main chain dihedral angle bias or charge effects on structure, the variance in Rh for these fragments should provide a rough estimate of the extent of Rh differences among typical IDPs owing to sequence-based excluded volume effects on hydrodynamic size.
Figure 3C shows that Rh for the 25-residue fragments ranged from 12.1 Å to 9.9 Å in the simulations, and averaged 11.4 ± 0.5 Å. For comparison, Rh for poly-ALA and N = 25 should be ~ 11.1 Å, estimated from HSC simulations performed using identical methods.40 Rh trended with GLY content (Fig. 3D) and fragments that reported the smallest and second smallest Rh had 80% and 40% GLY composition, respectively. Fragments producing the largest Rh in these simulations were those containing sequentially adjacent PRO residues and with higher compositions of branched (LEU, ILE, VAL) and aromatic (PHE, TRP, TYR) side chains (see Fig. S2). Each of the 4 fragments containing sequentially adjacent PRO residues were among the fragments with largest Rh, although no obvious correlation was observed when comparing Rh to fractional PRO composition (Fig. S2). Using the 25-residue fragment from the C-terminal end of p53(1-93) to showcase these effects, point substitution to ALA at PRO positions adjacent to and preceding another PRO residue generally produced larger reductions in Rh when compared to ALA substitutions at positions containing branched or aromatic residues (Fig. 3E). Also, no obvious correlation was observed when comparing Rh in these simulations to net charge or number of charged groups in a sequence (Fig. S2); a result that should be expected considering the energy function that was used. Overall, these data seemed to indicate that Rh generally follows the hydrodynamic dimensions of poly-ALA when only excluded volume effects on the chain are considered, with the exception of high fractional GLY composition (Fig. 3D) or the occurrence of sequentially adjacent PRO residues (Fig. S2). To demonstrate this, if the pre-factor (Ro) or exponent (v) for the poly-ALA scaling relationship is changed such that the poly-ALA curve agrees with the fragment average (11.4 Å) at N = 25, the red lines in Figures 3B and 3C are obtained (dashed line = pre-factor changed; stippled line = exponent changed). These results are consistent with reports that excluded volume effects on the structural dimensions of random polypeptide chains are sensitive mostly to the addition or subtraction of heavy atoms covalently attached to the main chain, i.e., PRO and GLY substitution effects.85,86 Repeating these simulations for IDP fragments with N = 20 gave quantitatively similar results (Fig. 3C). It should be noted that there are only 14 instances of adjacent PRO residues out of 2958 total residue positions in the IDP dataset, when redundancy is removed owing to multiple variants of the N-terminal region of p53 and overlap in the Mlph(147-240) and Mlph(147-403) sequences. This indicates that adjacent PRO residues are not uncommon but also not over-represented among IDPs. Likewise, GLY content ranged from 0.01 (multiple IDPs) to 0.15 (PGR) in the natural IDP sequences of the dataset, with redundancy removed. For fractional GLY composition from 0 to 0.15, little to no correlation with Rh was apparent among the IDP fragments that were simulated (Fig. 3D).
Effects of main chain bias for PPII on Rh
An issue with HSC-based simulations, as used above, is that energetic minima associated with main chain dihedral angles are not accounted for, though conformational bias for certain (φ, ψ) have been detected in experimental studies with disordered peptides79,87-89 and surveys of protein structures.86,90-92 To demonstrate this issue, the blue line in Figure 3B represents Rh for poly-PRO when the propensity for PPII at each residue position was modeled as 95% in simulated ensembles.40 Poly-PRO peptides are known to adopt PPII under normal conditions.93 Comparison of Rh for poly-PRO from PPII-biased and non-biased ensembles shows that main chain dihedral angle preferences can affect Rh substantially in these simulations.
The effects of PPII propensities on Rh calculated from HSC-simulated ensembles have been investigated systematically, whereby propensities for PPII were modeled by applying a sampling bias to the main chain dihedral angles (φ, ψ).40,48 For example, a 20% sampling bias for PPII at residue position i was equivalent to 20% of (φ, ψ) at position i located in the region of (−75±10, 145±10) and 80% distributed randomly in the allowed Ramachandran areas outside of (−75±10, 145±10). Following the van der Waals check, the propensity, fPPII,i, is calculated as the sum of the probabilities, given by equation 2, for structures with (φ, ψ) in the region (−75±10, 145±10) at position i. Since conformers containing contact violations are discarded and ensemble populations are weighted toward compacted structures (see Fig. 3A), fPPII,i does not necessarily match the applied sampling bias.40
For poly-ALA, Rh was observed in simulations to depend on the chain-averaged PPII propensity by,
| (4) |
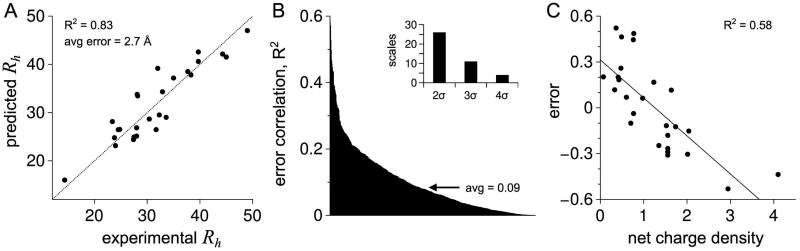
where Rh is in Å and fPPII,chain is the sum of fPPII,i over all residue positions and divided by N.48 Accordingly, fPPII,chain from a simulation is calculated in a manner analogous to its use in Figure 2B with intrinsic PPII propensities applied to an IDP sequence. The structural relationship between Rh, N, and fPPII,chain described by equation 4 was independent of position-specific patterns of PPII bias and was capable of predicting IDP Rh with good agreement to the experimental value.48 Figure 4A shows Rh predicted from sequence for each dataset IDP and compared to experimental Rh when using the intrinsic PPII propensities of Elam and colleagues.79 Good agreement (coefficient of determination, R2 = 0.83) and a small average error (± 2.7 Å) were observed. It is important to recognize that equation 4 was derived solely from conformational ensembles simulated with the HSC model and the agreement between predicted and experimental Rh represents a key test.
Figure 4.
Predicting Rh from intrinsic PPII propensities. A) Filled circles show Rh (in Å) predicted from sequence for each dataset IDP using equation 4. The stippled line is the identity line. B) Prediction error, normalized for IDP size using equation 5, was compared to 567 amino acid scales (544 from the Amino Acid Index database,94 20 representing fractional composition for the common amino acids, the net charge density, the net charge per residue, and κ). Scales from the Amino Acid Index database were summed by sequence and normalized to chain length. The correlation (R2) for each comparison is in rank order from left to right. Inset: Number of amino acids scales with R2 greater than the average plus 2, 3, and 4 times the standard deviation (σ). C) Comparison of prediction error to net charge density for each dataset IDP. The line is the observed trend; error = −0.25·(net charge density) + 0.31.
The error in predicting Rh for IDPs by equation 4 has been compared to the net charge.48 Since this error indeed correlated with net charge, as expected,52-54 a simple search for other amino acid properties that could influence IDP Rh was conducted. To do this, the prediction error was normalized for IDP size by,
| (5) |
The error determined for each dataset IDP was then compared to 544 different amino acid scales available from the Amino Acid Index database.94 To test for bias from sequence composition, the prediction error was also compared to the fractional composition of each amino acid type in each IDP. The fractional composition of each IDP sequence is provided in Figure S1, demonstrating substantial dataset variation. When comparing prediction error in the dataset to the various amino acid scales, the observed correlations were mostly weak (average R2 = 0.09 ± 0.1; Figure 4B). A few exceptions, however, gave R2 ≥ 0.5, but none exceeded the correlation observed for the net charge density (R2 = 0.58).
Additional charge-based metrics commonly used to classify IDP sequences, net charge per residue and κ,49 were also tested for correlation with the error. Since net charge per residue is determined from sequence almost identically to the net charge density, i.e., as the net charge divided by N rather than N0.5, a similar error correlation was observed (R2 = 0.57; Figure S3). For κ, which is a measure of the mixing of positive and negative charges in a sequence, correlation with the error was small and weaker than average (R2 = 0.05; Figure S3). Generally, κ is used to compare sequences with similar charge composition.49 κ ranges from 0 to 1, with values near 0 indicating that residues with opposite charge are well-mixed in a sequence. Values close to 1 are from sequences with segregated charge types, allowing for long-range attraction between oppositely charged regions of a disordered protein. The low correlation to the prediction error for κ here likely represents the diverse compositions and weak charge segregation among the IDP sequences in the dataset (dataset average = 0.211 ± 0.086 with minimum and maximum of 0.058 and 0.423, respectively). Values for net charge density, net charge per residue, and κ are provided in Table S3 for each IDP sequence.
Table I lists the ten highest ranking amino acid scales95-101 in terms of R2, each 3 standard deviations or more above the average. Net charge per residue was omitted from this list because of its high similarity to the net charge density. It is interesting to note that the list is dominated by physicochemical properties associated with charge (e.g., net charge density, ASP fractional composition, amino acid net charge) and main chain conformational bias (e.g., beta-structure-coil equilibrium, frequency as first residue in a turn, positional frequency in an α helix). Showing only slightly lower correlations were partial specific volume102 and apparent partial specific volume103 with R2 of 0.36 and 0.37, respectively. These results emphasize that while the HSC model seems to perform well for generating statistical ensembles of disordered polypeptide structures, the absence of specific terms in the energy function accounting for the effects of charge, non-ALA excluded volumes (i.e., when using poly-ALA derived relationships like equation 4), and preferences for certain main chain dihedral angles are serious limitations. Since the prediction error correlated best with the net charge density, the effects of net charge on IDP Rh were investigated in more detail. Contributions from other physicochemical properties to IDP Rh are also discussed below.
Table I.
Sequence properties and amino acid scales with best correlation (R2) to equation 4 error.
| R 2 | scale |
|---|---|
| 0.58 | net charge density |
| 0.53 | beta-structure-coil equilibrium95 |
| 0.50 | ASP fractional composition |
| 0.47 | positional residue frequency at helix termini N' 96 |
| 0.46 | helix initiator at position i-197 |
| 0.45 | amino acid net charge98 |
| 0.44 | frequency as 1st residue in turn99 |
| 0.43 | amino acid isoelectric point100 |
| 0.43 | positional residue frequency at helix termini N" 96 |
| 0.39 | frequency in beta-bends101 |
Effects of net charge on Rh
The trend in the prediction error with net charge density was generally consistent across the IDP dataset (Fig. 4C). This could be considered surprising. While some studies have shown that IDP Rh are sensitive to net charge,52-54 results from other studies indicate that describing charge effects on Rh via the net charge could prove inaccurate since changes in the spacing between48 and the sequential patterns of charged groups49 are capable of producing large changes in Rh in the absence of net charge changes.
To compare Rh sensitivity to net charge among the dataset IDPs, Rh predicted by equation 4 was corrected for apparent net charge effects using the observed linear error trend. From equations 1, 4, and 5, the trend-corrected Rh is,
| (6) |
where 0.25 and 0.31 are from the slope and intercept, respectively, of the trend in Figure 4C, Q is the net charge from sequence (Table S1), and fPPII,chain is the sequence sum, divided by N, of the experimental PPII propensities79 (Fig. 2B and Table S1). The goal is to identify charge patterns in the IDP dataset that influence Rh atypically by identifying IDPs that deviate from the dataset trend according to equation 6. Specifically, if Rh depends on the net charge strongly for an IDP, and if that dependence differs from the other dataset IDPs, removal of such an IDP from the training set should produce discernible changes in equation 6 that could be used to identify atypical structural behavior and/or sequence patterns.
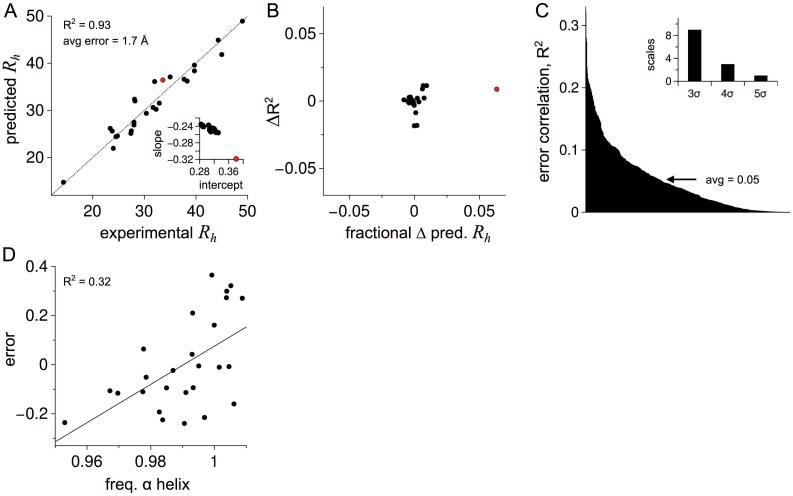
Figure 5A shows Rh predicted from sequence using equation 6 and compared to experimental Rh for each dataset IDP. Good agreement was observed, yielding an increased R2 of 0.93 and a decreased average error of ± 1.7 Å, relative to the correlation and average error obtained when predicting Rh from sequence without the net charge correction (see Fig. 4A). Next, each IDP was removed from the dataset, individually, and Rh from sequence recalculated for the remaining IDPs. First, using equation 4 to regenerate the error trend with net charge density. The trend slope and trend intercept showed small variations with the removal of individual dataset IDPs (Fig. 5A inset), with the exception of removing prothymosin-α (highlighted red in Figs. 5A and B). As such, removing an IDP from the dataset produced commensurate changes in equation 6. Rh was then recalculated from sequence by equation 6, using new slope and intercept values, for the dataset IDPs. Figure 5B shows that the correlation between predicted and experimental Rh changed only slightly (± 0.02) when removing an individual IDP from the dataset, relative to the correlation obtained with the full dataset (R2 = 0.93). Rh predicted by equation 6 also changed by 1% or less for most of the IDPs when one was removed from the training set and then equation 6, re-calculated, was applied to the missing IDP. Again, the exception was prothymosin-α that reported a 6.3% change. These results indicate that hydrodynamic size for IDPs can be well-estimated from sequence using equation 6, however, its use should be circumspect owing to apparent differences in Rh sensitivity to net charge among IDPs.
Figure 5.
Predicting Rh from intrinsic PPII propensities and net charge. A) Filled circles show Rh (in Å) predicted from sequence for each dataset IDP using equation 6. The identity line is shown by the stippled line. Inset: slope and y-intercept for the linear trend in Fig. 4C when a singular IDP is removed from the training set. B) Each filled circle represents the removal of a singular IDP from the training set. The concomitant change in correlation for predicted and experimental Rh (ΔR2) is compared to the fractional change in predicted Rh for the removed IDP. C) Prediction error, normalized for IDP size by equation 5, was compared to 567 amino acid scales (same scales as Fig. 4B). The correlation (R2) for each comparison is in rank order from left to right. Inset: Number of amino acids scales with R2 greater than the average plus 3, 4, and 5 times the standard deviation (σ). D) Comparison of prediction error to the best performing scale, normalized frequency of α helix in all α class,104 showing the trend line.
The fractional composition of charged residues in prothymosin-α is very high (fractional composition = 0.57; from 53 GLU and ASP, 10 LYS and ARG, out of 110 total residues) when compared to the other dataset IDPs (average = 0.25 ± 0.05). Considering that net charge density is at the dataset maximum for prothymosin-α (Fig. 2A), it could be argued that charge effects on structure, in sum, should be pronounced in prothymosin-α more so than typical. Thus, it may be counter-intuitive to observe that the magnitude of the slope representing Rh sensitivity to Q increased by ~ 25% when prothymosin-α was removed from the training set (Fig. 5A inset). Specifically, these data indicate that the effects of net charge on Rh were weaker in prothymosin-α, on a per-charge basis, than for the other IDPs. Since prothymosin-α was a lone outlier in this analysis, additional tests are needed. If prothymosin-α is removed from the dataset and the entire resampling analysis repeated, removing a second IDP, no obvious outliers were detected (Fig. S4). At a minimum, these results argue that net charge is a poor metric from which to model charge effects on hydrodynamic size since Figure 5B predicts that some IDP structures respond differently than others to small changes in the net charge. Of note, prothymosin-α had the largest value for κ among the sequences in the IDP dataset (κ = 0.423). Larger κ values trend in molecular dynamics simulations with compacted disordered structures,49 possibly providing an explanation for the somewhat reduced effect of net charge on Rh in prothymosin-α.
Effects of main chain bias for α helix on Rh
The HSC model has many limitations, as discussed above, and use of equation 6 for estimating IDP Rh will have errors from excluded volume and charge effects on structure, and possibly from position-specific perturbations to the intrinsic PPII propensities from neighboring residues.89 To test for additional error sources, Rh prediction error was again compared to 544 amino acid scales from the Amino Acid Index database,94 the fractional composition of each amino acid type, net charge density, net charge per residue, and κ. Prediction errors from equation 6 were normalized for IDP size using equation 5 and the results are given in Figure 5C. Correlations of prediction error to amino acid properties were weak, with an average R2 of 0.05 ± 0.05 and a maximum of 0.32. Amino acid properties that trend best with the prediction error were associated with α helix propensities, representing the 3 highest and 15 of the top 20 correlations (Table S4).96,99,101,104-115 Error correlation to net charge density, net charge per residue, and κ were 0, 0.0007, and 0.005, respectively.
The correlation of prediction error with α helix propensities could be another example of Rh sensitivity to intrinsic (φ, ψ) preferences, similar to the effects of intrinsic PPII propensities on IDP Rh.21,48 Figure 5D shows that increasing chain propensities for α helix seem to follow increasing prediction error, which was observed for each α helix propensity scale listed in Table S4 (Fig. S5). Inspection of equation 5 indicates that increasing prediction errors are associated with decreasing experimental Rh, providing a testable hypothesis. Specifically, these data predict Rh compaction with increasing α helix propensities.
To test this hypothesis, propensities for α helix were modeled using the same simulation strategy that was employed for PPII propensities.40,48 Briefly, a sampling bias was applied to main chain dihedral angles (φ, ψ) in simulations that computed Rh from HSC-generated ensembles of poly-ALA peptides. Surveys of α helix structures yield an average (φ, ψ) of (−64±7, −41±7).116 Accordingly, a sampling bias for α helix at residue position i, for example a 20% sampling bias, was equivalent to 20% of (φ, ψ) at position i located in the region of (−64±10, −41±10) and 80% distributed randomly in the allowed Ramachandran areas outside of (−64±10, −41±10). Following the van der Waals check, the α helix propensity, fα,i, was calculated as the sum of the probabilities (equation 2) for structures with (φ, ψ) in the region (−64±10, −41±10) at position i. No provisions accounting for the favorable energetics of intra-chain hydrogen bonds were added to the energy function, though the authors recognize that intra-chain hydrogen bonds contribute to α helix structural stability.116
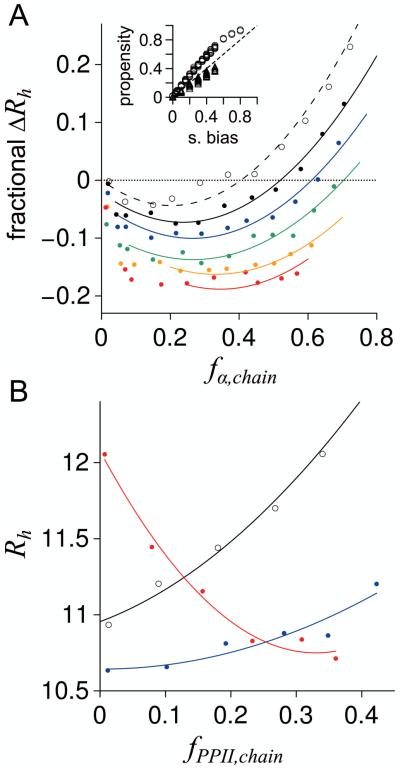
The effects of α helix propensities on Rh calculated from ensembles of poly-ALA peptides with N = 25 are shown in Figure 6. In these simulations, sampling biases were applied equally at each residue position, although the sampling bias for α helix did not necessarily match the applied sampling bias for PPII in any particular ensemble. Of note, fα,chain, calculated as the sum of fα,i for each position i and divided by N, was generally greater than the applied sampling bias for α helix (Fig. 6A inset). In contrast, fPPII,chain was smaller than the applied PPII sampling bias, consistent with previous results.40 For poly-ALA with no PPII sampling bias, fα,chain from 0.1 – 0.2 reduced Rh by ~ 5%, relative to Rh when no sampling preference for α helix was applied (Fig. 6A). For fα,chain > 0.2, Rh gradually increased with increasing fα,chain. When sampling biases for PPII were applied, the percent reduction in Rh from α helix propensities generally increased with increasing PPII bias.
Figure 6.
Rh for poly-ALA (N = 25) simulated with intrinsic propensities for α helix and PPII. A) Open circles are ensembles with no applied sampling bias for PPII. Filled circles are ensembles simulated with applied PPII sampling biases of 0.10 (black), 0.20 (blue), 0.30 (green), 0.40 (orange), and 0.50 (red). Shown is the change in Rh relative to Rh expected from fPPII,chain using equation 4. Curves in both panels (A and B) represent data fits to second order polynomials and have no physical meaning; they were provided to highlight trends. Inset: Chain propensities calculated for each ensemble, fα,chain (circles) and fPPII,chain (triangles), and compared to the applied sampling bias. The stippled line is the identity line. B) Open circles are ensembles with no applied sampling bias for α helix. Filled circles are ensembles simulated with applied α helix bias resulting in fα,chain of ~ 0.05 – 0.07 (blue) and fα,chain of ~ 0.50 – 0.60 (red).
Several observations from these simulations are noteworthy. First, Rh compaction was observed owing to α helix propensities, consistent with the motivating hypothesis from the observed dataset trend (Fig. 5D). Second, weak fα,chain values produced substantial compaction in Rh. For ensembles with fPPII,chain from 0.3 – 0.5, which should be expected for most IDPs (see Fig. 2B), chain averaged α helix propensities of just 0.05 – 0.1 resulted in 10 – 20% Rh compaction. This contrasts with simulation results for PPII bias, whereby weak PPII propensities (fPPII,chain ≤ 0.1) produced relatively small changes in Rh.40,48 Third, the effects of α helix propensities on Rh were modulated by PPII propensities. And fourth, the reciprocal relationship was also observed where the effects of PPII propensities on Rh were modulated by α helix propensities. Figure 6B shows that when no bias for α helix is applied to a poly-ALA chain, Rh increases with increasing chain propensity for PPII. For fα,chain of ~ 0.05 – 0.07, however, compaction occurs and increases in Rh from increasing PPII propensities are reduced. When the chain bias for α helix was very high (fα,chain ~ 0.50 – 0.60), the trend is reversed and increasing chain propensities for PPII yield decreasing Rh. Considering that the trend among experimental Rh is for decreasing Rh with increasing α helix propensities (Fig. 5D) and increasing Rh with increasing PPII propensities,21 these observations argue for intrinsic α helix propensities that are generally weak in IDPs, when averaged for a chain. In summary, these results predict that intrinsic bias to preferred (φ, ψ) regions in disordered polypeptides, even at levels below experimental79,87-89,117 and computational118,119 estimates for the common amino acids, are capable of establishing sequence-dependent variability in the structural dimensions of the disordered protein conformational ensemble and concomitant sequence-dependent effects on IDP Rh.
DISCUSSION
Identifying the molecular properties that regulate disordered protein structures, and to what extent, is critical for establishing molecular descriptions of IDP-mediated biology. Here, conformational ensembles of polypeptides were simulated using a model based on the HSC algorithm to assess the contributions of net charge, main chain dihedral angle bias, and excluded volume to the hydrodynamic dimensions of disordered structures. To test and evaluate simulation results, Rh calculated from simulated ensembles were compared to experimental Rh from 26 IDPs showing diverse sequence compositions, chain lengths, charge patterns, and net charge. There are benefits to using a HSC model for simulating disordered structures. First, it is reductionistic and thus useful for identifying salient phenomena by subtracting out uninteresting results. For example, the overall magnitude of Rh for a disordered ensemble may not be interesting, since its value is largely driven by excluded volume effects and chain-solvent interactions that are somewhat similar among different IDPs, at least in the simulated data (Fig. 3B). The deviations in experimental Rh from this random coil approximation accordingly could yield detail on the intra- and inter-molecular interactions that bias state distributions in the protein conformational ensemble. Second, proposed structural effectors, such as chain bias for (φ, ψ) representative of PPII40 and coulombic interactions between charged groups,48 could be added to the model and the capabilities of these changes tested. The HSC model also has limitations. As used above, and in addition to the limitations already noted, it is not capable of establishing why there is a particular (φ, ψ) bias, or net charge effect on Rh, only that the comparative analysis indicates that these specific structural effectors are present in IDP systems.
The results of this study indicate that IDP Rh can be described from simple physicochemical properties associated with the polypeptide chain. The main structural determinants, other than N, seem to be excluded volume effects that can be reasonably approximated using poly-ALA (Fig. 3B), chain-solvent interactions that cause a general compaction in the hydrodynamic dimensions (Fig. 3A), charge effects that, when averaged across a dataset of IDP structures, cause a general expansion that trends with the net charge density (Fig. 4C), and bias in the main chain dihedral angles for common (φ, ψ) that have been associated with PPII and α helix (Figs. 4A and 6). Most likely, this list of pertinent structural effectors is not complete and the identification of additional Rh sensitive properties awaits further study. For example, energetic preferences for (φ, ψ) in other areas of the allowed Ramachandran regions,90-92 position-specific perturbations to the intrinsic conformational propensities from neighboring residues,89 and a more accurate accounting of charge effects on structure than provided by the net charge density are probably needed and currently underestimated in the model (i.e., assumed to be negligible). In spite of the gross simplicity of this model, it is interesting to note that IDP Rh can be predicted accurately from sequence using just experimental PPII propensities, an estimation of the protein net charge, and equation 6 (Fig. 5A).
The agreement between the observed IDP dataset trend, showing that experimental Rh generally compact with increasing chain propensities for α helix (Fig. 5D), with the effect on simulated Rh of a sampling bias to the canonical (φ, ψ) of α helix structures (Fig. 6A) offers compelling evidence for the hypothesis that main chain biases for common (φ, ψ) are key determinants for establishing hydrodynamic size in IDPs. Previously, it was shown that experimental PPII propensities are capable of describing sequence-based differences in IDP Rh,21,48 which was predicted from HSC simulations of disordered polypeptides.40 Here, the simulated data predict that IDP Rh are modulated also by intrinsic α helix propensities. Weak α helix propensities (< 0.10) were capable of producing substantial compaction in the simulations, decreasing Rh by upwards of 20%, and the effects on Rh owing to α helix propensities were dependent on the chain propensity for PPII. Likewise, PPII effects on Rh were modulated by chain propensities for α helix. While experiments that directly test these predictions have yet to be provided, the results, overall, demonstrate how intrinsic structural propensities are capable of influencing the chain dimensions in disordered proteins.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the gift of recombinant PGR from Alexander Yarawsky and Andrew Herr (Cincinnati Children’s Hospital, Cincinnati, Ohio, USA). This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award R15GM115603 and by the Division of Materials Research of the National Science Foundation under award DMR-1205670. B.J.R was supported in part by the National Institute of General Medical Sciences under award R25GM102783 to the South Texas Doctoral Bridge program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation.
REFERENCES
- (1).van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Das RK, Ruff KM, Pappu RV. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr Opin Struct Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- (4).Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2001;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- (7).Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signaling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradović Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- (10).Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun. 2015;6:7875. doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Uversky VN, Davé V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem. Rev. 2014;114:6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- (17).Berry GC. The hydrodynamic and conformational properties of denatured proteins in dilute solutions. Protein Sci. 2010;19:94–98. doi: 10.1002/pro.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fixman M. Radius of gyration of polymer chains. J Chem Phys. 1962;36:306–310. [Google Scholar]
- (19).Flory PJ, Fisk S. Effect of volume exclusion on the dimensions of polymer chains. J Chem Phys. 1966;44:2243–2248. [Google Scholar]
- (20).Sun ST, Nishio I, Swislow G, Tanaka T. The coil–globule transition: radius of gyration of polystyrene in cyclohexane. J Chem Phys. 1980;73:5971–5975. [Google Scholar]
- (21).Perez RB, Tischer A, Auton M, Whitten ST. Alanine and proline content modulate global sensitivity to discrete perturbations in disordered proteins. Proteins. 2014;82:3373–3384. doi: 10.1002/prot.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lowry DF, Stancik A, Shrestha RM, Daughdrill GW. Modeling the accessible conformations of the intrinsically unstructured transactivation domain of p53. Proteins. 2008;71:587–598. doi: 10.1002/prot.21721. [DOI] [PubMed] [Google Scholar]
- (23).Donaldson L, Capone JP. Purification and characterization of the carboxyl-terminal transactivation domain of Vmw65 from herpes simplex virus type 1. J Biol Chem. 1992;267:1411–1414. [PubMed] [Google Scholar]
- (24).Yi S, Boys BL, Brickenden A, Konermann L, Choy WY. Effects of zinc binding on the structure and dynamics of the intrinsically disordered protein prothymosin alpha: evidence for metalation as an entropic switch. Biochemistry. 2007;46:13120–13130. doi: 10.1021/bi7014822. [DOI] [PubMed] [Google Scholar]
- (25).Sanchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1 alpha ODD domain to p53. Mol Cell. 2005;17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- (26).Campbell KM, Terrell AR, Laybourn PJ, Lumb KJ. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 2000;39:2708–2713. doi: 10.1021/bi9923555. [DOI] [PubMed] [Google Scholar]
- (27).Geething NC, Spudich JA. Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J Biol Chem. 2007;282:21518–21528. doi: 10.1074/jbc.M701932200. [DOI] [PubMed] [Google Scholar]
- (28).Soragni A, Zambelli B, Mukrasch MD, Biernat J, Jeganathan S, Griesinger C, Ciurli S, Mandelkow E, Zweckstetter M. Structural characterization of binding of Cu(II) to tau protein. Biochemistry. 2008;47:10841–10851. doi: 10.1021/bi8008856. [DOI] [PubMed] [Google Scholar]
- (29).Adkins JN, Lumb KJ. Intrinsic structural disorder and sequence features of the cell cycle inhibitor p57Kip2. Proteins. 2002;46:1–7. doi: 10.1002/prot.10018. [DOI] [PubMed] [Google Scholar]
- (30).Uversky VN, Permyakov SE, Zagranichny VE, Rodionov IL, Fink AL, Cherskaya AM, Wasserman LA, Permyakov EA. Effect of zinc and temperature on the conformation of the gamma subunit of retinal phosphodiesterase: a natively unfolded protein. J Proteome Res. 2002;1:149–159. doi: 10.1021/pr0155127. [DOI] [PubMed] [Google Scholar]
- (31).Haaning S, Radutoiu S, Hoffmann SV, Dittmer J, Giehm L, Otzen DE, Stougaard J. An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro. J Biol Chem. 2008;283:31142–31152. doi: 10.1074/jbc.M805024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Permyakov SE, Millett IS, Doniach S, Permyakov EA, Uversky VN. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- (33).Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT, Jr, Fernandez CO, Eliezer D, Zweckstetter M, Lashuel HA. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J Biol Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Baker JMR. Department of Biochemistry. University of Toronto; Toronto: 2009. Structural characterization and interactions of the CFTR regulatory region (PhD Thesis) [Google Scholar]
- (35).Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19:566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Magidovich E, Orr I, Fass D, Abdu U, Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K1 channel modulates its interaction with scaffold proteins. Proc Natl Acad Sci USA. 2007;104:13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sanchez-Puig N, Veprintsev DB, Fersht AR. Human full-length securin is a natively unfolded protein. Protein Sci. 2005;14:1410–1418. doi: 10.1110/ps.051368005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Danielsson J, Jarvet J, Damberg P, Graslund A. Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer Aβ (1-40) peptide. Determination of hydrodynamic radii of random coil peptides of varying length. Magn Reson Chem. 2002;40:S89–97. [Google Scholar]
- (39).Danielsson J, Liljedahl L, Bárány-Wallje E, Sønderby P, Kristensen LH, Martinez-Yamout MA, Dyson HJ, Wright PE, Poulsen FM, Mäler L, Gräslund A, Kragelund BB. The intrinsically disordered RNR inhibitor Sml1 is a dynamic dimer. Biochemistry. 2008;47:13428–37. doi: 10.1021/bi801040b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Langridge TD, Tarver MJ, Whitten ST. Temperature effects on the hydrodynamic radius of the intrinsically disordered N-terminal region of the p53 protein. Proteins. 2014;82:668–678. doi: 10.1002/prot.24449. [DOI] [PubMed] [Google Scholar]
- (41).Tcherkasskaya O, Davidson EA, Uversky VN. Biophysical constraints for protein structure prediction. J Proteome Res. 2003;2:37–42. doi: 10.1021/pr025552q. [DOI] [PubMed] [Google Scholar]
- (42).Flory PJ. The configuration of real polymer chains. J Chem Phys. 1949;17:303–310. [Google Scholar]
- (43).Flory PJ. Statistical mechanics of chain molecules. Vol. 432. Interscience Publishers, John Wiley & Sons; New York: 1969. [Google Scholar]
- (44).Nozaki Y, Tanford C. The solubility of amino acids and related compounds in aqueous urea solutions. J Biol Chem. 1963;238:4074–4081. [PubMed] [Google Scholar]
- (45).Nozaki Y, Tanford C. The solubility of amino acids, diglycine, and triglycine in aqueous guanidine hydrochloride solutions. J Biol Chem. 1970;245:1648–1652. [PubMed] [Google Scholar]
- (46).Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- (47).Teufel DP, Johnson CM, Lum JK, Neuweiler H. Backbone-driven collapse in unfolded protein chains. J Mol Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- (48).Tomasso ME, Tarver MJ, Devarajan D, Whitten ST. Hydrodynamic radii of intrinsically disordered proteins determined from experimental polyproline II propensities. PLoS Comput Biol. 2016;12:e1004686. doi: 10.1371/journal.pcbi.1004686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Richards FM. Areas, volumes, packing, and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- (51).Hilser VJ, Freire E. Structure-based calculation of the equilibrium folding pathway of proteins. Correlation with hydrogen exchange protection factors. J Mol Biol. 1996;262:756–772. doi: 10.1006/jmbi.1996.0550. [DOI] [PubMed] [Google Scholar]
- (52).Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Whitten ST, Yang HW, Fox RO, Hilser VJ. Exploring the impact of polyproline II (PII) conformational bias on the binding of peptides to the SEM-5 SH3 domain. Protein Sci. 2008;17:1200–1211. doi: 10.1110/ps.033647.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- (57).Iijima H, Dunbar JB, Jr, Marshall GR. Calibration of effective van der Waals atomic contact radii for proteins and peptides. Proteins. 1987;2:330–339. doi: 10.1002/prot.340020408. [DOI] [PubMed] [Google Scholar]
- (58).Jeffreys H, Jeffreys BS. Methods of mathematical physics. Cambridge University Press; New York: 1950. pp. 122–123. [Google Scholar]
- (59).Momany FA, McGuire RF, Burgess AW, Scheraga HA. Energy parameters in polypeptides. VII. Geometric parameters, partial atomic charges, nonbonded interactions, hydrogen bond interactions, and intrinsic torsional potentials for the naturally occurring amino acids. J Phys Chem. 1975;79:2361–2381. [Google Scholar]
- (60).Mandel N, Mandel G, Trus BL, Rosenberg J, Carlson G, Dickerson RE. Tuna cytochrome c at 2.0 Å resolution. III. Coordinate optimization and comparison of structures. J Biol Chem. 1977;252:4619–4636. [PubMed] [Google Scholar]
- (61).MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- (62).Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- (63).Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Murphy KP, Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- (65).Murphy KP, Bhakuni V, Xie D, Freire E. Molecular basis of cooperativity in protein folding. III. Structural identification of cooperative folding units and folding intermediates. J Mol Biol. 1992;227:293–306. doi: 10.1016/0022-2836(92)90699-k. [DOI] [PubMed] [Google Scholar]
- (66).Lee KH, Xie D, Freire E, Amzel LM. Estimation of changes in side chain configurational entropy in binding and folding: General methods and application to helix formation. Proteins. 1994;20:68–84. doi: 10.1002/prot.340200108. [DOI] [PubMed] [Google Scholar]
- (67).Xie D, Freire E. Structure based prediction of protein folding intermediates. J Mol Biol. 1994;242:62–80. doi: 10.1006/jmbi.1994.1557. [DOI] [PubMed] [Google Scholar]
- (68).Gómez J, Hilser VJ, Xie D, Freire E. The heat capacity of proteins. Proteins. 1995;22:404–412. doi: 10.1002/prot.340220410. [DOI] [PubMed] [Google Scholar]
- (69).D’Aquino JA, Gómez J, Hilser VJ, Lee KH, Amzel LM, Freire E. The magnitude of the backbone conformational entropy change in protein folding. Proteins. 1996;25:143–156. doi: 10.1002/(SICI)1097-0134(199606)25:2<143::AID-PROT1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- (70).Habermann SM, Murphy KP. Energetics of hydrogen bonding in proteins: A model compound study. Protein Sci. 1996;5:1229–1239. doi: 10.1002/pro.5560050702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Luque I, Mayorga OL, Freire E. Structure-based thermodynamic scale of alpha-helix propensities in amino acids. Biochemistry. 1996;35:13681–13688. doi: 10.1021/bi961319s. [DOI] [PubMed] [Google Scholar]
- (72).Schaub LJ, Campbell JC, Whitten ST. Thermal unfolding of the N-terminal region of p53 monitored by circular dichroism spectroscopy. Protein Sci. 2012;21:1682–1688. doi: 10.1002/pro.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Whitten ST, García-Moreno EB. pH dependence of stability of staphylococcal nuclease: evidence of substantial electrostatic interactions in the denatured state. Biochemistry. 2000;39:14292–14302. doi: 10.1021/bi001015c. [DOI] [PubMed] [Google Scholar]
- (74).Baȧk D, Cutting GR, Milewski M. The CFTR-derived peptides as a model of sequence-specific protein aggregation. Cell Mol Biol Lett. 2007;12:435–447. doi: 10.2478/s11658-007-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- (76).Li Y, Shan B, Raleigh DP. The cold denatured state is compact but expands at low temperatures: hydrodynamic properties of the cold denatured state of the C-terminal domain of L9. J Mol Biol. 2007;368:256–262. doi: 10.1016/j.jmb.2007.02.011. [DOI] [PubMed] [Google Scholar]
- (77).Fitzkee NC, Rose GD. Reassessing random-coil statistics in unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Sivakolundu SG, Nourse A, Moshiach S, Bothner B, Ashley C, Satumba J, Lahti J, Kriwacki RW. Intrinsically unstructured domains of Arf and Hdm2 form bimolecular oligomeric structures in vitro and in vivo. J Mol Biol. 2008;384:240–254. doi: 10.1016/j.jmb.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Elam WA, Schrank TP, Campagnolo AJ, Hilser VJ. Evolutionary conservation of the polyproline II conformation surrounding intrinsically disordered phosphorylation sites. Protein Sci. 2013;22:405–417. doi: 10.1002/pro.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Stein PE, Leslie AG, Finch JT, Turnell WG, McLaughlin PJ, Carrell RW. Crystal structure of ovalbumin as a model for the reactive centre of serpins. Nature. 1990;347:99–102. doi: 10.1038/347099a0. [DOI] [PubMed] [Google Scholar]
- (82).Saito R, Sato T, Ikai A, Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 Å resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:792–795. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- (83).Hynes TR, Fox RO. The crystal structure of Staphylococcal nuclease refined at 1.7 Å resolution. Proteins. 1991;10:92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- (84).Zahran ZN, Chooback L, Copeland DM, West AH, Richter-Addo GB. Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations. J Inorg Biochem. 2008;102:216–233. doi: 10.1016/j.jinorgbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Miller WG, Goebel CV. Dimensions of protein random coils. Biochemistry. 1968;7:3925–3935. doi: 10.1021/bi00851a021. [DOI] [PubMed] [Google Scholar]
- (86).Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- (87).Shi Z, Chen K, Liu Z, Ng A, Bracken WC, Kallenbach NR. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proc Natl Acad Sci USA. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Rucker AL, Pager CT, Campbell MN, Qualls JE, Creamer TP. Host-guest scale of left-handed polyproline II helix formation. Proteins. 2003;53:68–75. doi: 10.1002/prot.10477. [DOI] [PubMed] [Google Scholar]
- (89).Brown AM, Zondlo NJ. A propensity scale for type II polyproline helices (PPII): aromatic amino acids in proline-rich sequences strongly disfavor PPII due to proline-aromatic interactions. Biochemistry. 2012;51:5041–5051. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- (90).Serrano L. Comparison between the phi distribution of the amino acids in the protein database and NMR data indicates that amino acids have various phi propensities in the random coil conformation. J Mol Biol. 1995;254:322–333. doi: 10.1006/jmbi.1995.0619. [DOI] [PubMed] [Google Scholar]
- (91).Smith LJ, Bolin KA, Schwalbe H, MacArthur MW, Thornton JM, Dobson CM. Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J Mol Biol. 1996;255:494–506. doi: 10.1006/jmbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- (92).Anderson RJ, Weng Z, Campbell RK, Jiang X. Main-chain conformational tendencies of amino acids. Proteins. 2005;60:679–689. doi: 10.1002/prot.20530. [DOI] [PubMed] [Google Scholar]
- (93).Cowan PM, McGavin S. Structure of poly-L-proline. Nature. 1955;176:501–503. [Google Scholar]
- (94).Kawashima S, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Oobatake M, Kubota Y, Ooi T. Optimization of amino acid parameters for correspondence of sequence to tertiary structures of proteins. Bull Inst Chem Res. 1985;63:82–94. [Google Scholar]
- (96).Aurora R, Rose G. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Finkelstein AV, Badretdinov AY, Ptitsyn OB. Physical reasons for secondary structure stability: alpha-helices in short peptides. Proteins. 1991;10:287–299. doi: 10.1002/prot.340100403. [DOI] [PubMed] [Google Scholar]
- (98).Klein P, Kanehisa M, DeLisi C. Prediction of protein function from sequence properties: Discriminant analysis of a data base. Biochim Biophys Acta. 1984;787:221–226. doi: 10.1016/0167-4838(84)90312-1. [DOI] [PubMed] [Google Scholar]
- (99).Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- (100).Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- (101).Lewis PN, Momany FA, Scheraga HA. Folding of polypeptide chains in proteins: A proposed mechanism for folding. Proc Natl Acad Sci USA. 1971;68:2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Cohn EJ, Edsall JT. Density and apparent specific volume of proteins. In: Cohn EJ, Edsall JT, editors. Proteins, Amino Acids and Peptides. Van Nostrand-Reinhold; Princeton, NJ: 1943. pp. 370–381. [Google Scholar]
- (103).Bull HB, Breese K. Surface tension of amino acid solutions: A hydrophobicity scale of the amino acid residues. Arch Biochem Biophys. 1974;161:665–670. doi: 10.1016/0003-9861(74)90352-x. [DOI] [PubMed] [Google Scholar]
- (104).Palau J, Argos P, Puigdomenech P. Protein secondary structure. Studies on the limits of prediction accuracy. Int J Pept Protein Res. 1982;19:394–401. [PubMed] [Google Scholar]
- (105).Geisow MJ, Roberts RDB. Amino acid preferences for secondary structure vary with protein class. Int J Biol Macromol. 1980;2:387–389. [Google Scholar]
- (106).Qian N, Sejnowski TJ. Predicting the secondary structure of globular proteins using neural network models. J Mol Biol. 1988;202:865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- (107).Tanaka S, Scheraga HA. Statistical mechanical treatment of protein conformation. 5. A multiphasic model for specific-sequence copolymers of amino acids. Macromolecules. 1977;10:9–20. doi: 10.1021/ma60055a002. [DOI] [PubMed] [Google Scholar]
- (108).Robson B, Suzuki E. Conformational properties of amino acid residues in globular proteins. J Mol Biol. 1976;107:327–356. doi: 10.1016/s0022-2836(76)80008-3. [DOI] [PubMed] [Google Scholar]
- (109).Nagano K. Local analysis of the mechanism of protein folding. I. Prediction of helices, loops, and beta-structures from primary structure. J Mol Biol. 1973;75:401–420. doi: 10.1016/0022-2836(73)90030-2. [DOI] [PubMed] [Google Scholar]
- (110).Maxfield FR, Scheraga HA. Status of empirical methods for the prediction of protein backbone topography. Biochemistry. 1976;15:5138–5153. doi: 10.1021/bi00668a030. [DOI] [PubMed] [Google Scholar]
- (111).Burgess AW, Ponnuswamy PK, Scheraga HA. Analysis of conformations of amino acid residues and prediction of backbone topography in proteins. Isr J Chem. 1974;12:239–286. [Google Scholar]
- (112).Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- (113).Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- (114).Kanehisa MI, Tsong TY. Local hydrophobicity stabilizes secondary structures in proteins. Biopolymers. 1980;19:1617–1628. doi: 10.1002/bip.1980.360190906. [DOI] [PubMed] [Google Scholar]
- (115).Finkelstein AV, Ptitsyn OB. Theory of protein molecule self-organization. II. A comparison of calculated thermodynamic parameters of local secondary structures with experiments. Biopolymers. 1977;16:497–524. doi: 10.1002/bip.1977.360160303. [DOI] [PubMed] [Google Scholar]
- (116).Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- (117).Kashtanov S, Borcherds W, Wu H, Daughdrill GW, Ytreberg FM. Using chemical shifts to assess transient secondary structure and generate ensemble structures of intrinsically disordered proteins. Methods Mol Biol. 2012;895:139–152. doi: 10.1007/978-1-61779-927-3_11. [DOI] [PubMed] [Google Scholar]
- (118).Beck DA, Alonso DO, Inoyama D, Daggett V. The intrinsic conformational propensities of the 20 naturally occurring amino acids and reflection of these propensities in proteins. Proc Natl Acad Sci USA. 2008;105:12259–12264. doi: 10.1073/pnas.0706527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Towse CL, Vymetal J, Vondrasek J, Daggett V. Insights into unfolded proteins from the intrinsic ϕ/ψ propensities of the AAXAA host-guest series. Biophys J. 2016;110:348–361. doi: 10.1016/j.bpj.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.