Summary
Maize ARGOS8 is a negative regulator of ethylene responses. A previous study has shown that transgenic plants constitutively overexpressing ARGOS8 have reduced ethylene sensitivity and improved grain yield under drought stress conditions. To explore the targeted use of ARGOS8 native expression variation in drought‐tolerant breeding, a diverse set of over 400 maize inbreds was examined for ARGOS8 mRNA expression, but the expression levels in all lines were less than that created in the original ARGOS8 transgenic events. We then employed a CRISPR‐Cas‐enabled advanced breeding technology to generate novel variants of ARGOS8. The native maize GOS2 promoter, which confers a moderate level of constitutive expression, was inserted into the 5′‐untranslated region of the native ARGOS8 gene or was used to replace the native promoter of ARGOS8. Precise genomic DNA modification at the ARGOS8 locus was verified by PCR and sequencing. The ARGOS8 variants had elevated levels of ARGOS8 transcripts relative to the native allele and these transcripts were detectable in all the tissues tested, which was the expected results using the GOS2 promoter. A field study showed that compared to the WT, the ARGOS8 variants increased grain yield by five bushels per acre under flowering stress conditions and had no yield loss under well‐watered conditions. These results demonstrate the utility of the CRISPR‐Cas9 system in generating novel allelic variation for breeding drought‐tolerant crops.
Keywords: maize, ARGOS, CRISPR‐Cas9, genome editing, drought tolerance, grain yield
Introduction
Developing more drought‐tolerant crops in a sustainable manner is one means to meet the demand of an increasing human population that will require more food, feed and fuel. Improvement in drought tolerance of crops is ultimately measured by an increase in grain yield under water‐limiting conditions. The physiological processes and metabolic networks underlying drought tolerance are complicated and often difficult to delineate. Nevertheless, the phytohormone ethylene is known to play an important role in regulating plant response to abiotic stress, including water deficits and high temperature (Hays et al., 2007; Kawakami et al., 2010, 2013). Field studies have shown that reducing ethylene biosynthesis by silencing 1‐aminocyclopropane‐1‐carboxylic acid synthase6 in transgenic maize plants improves grain yield under drought stress conditions (Habben et al., 2014). A higher yield also can be achieved by decreasing the sensitivity of maize to ethylene (Shi et al., 2015). ARGOS genes are negative regulators of the ethylene response and modulate ethylene signal transduction, enhancing drought tolerance when overexpressed in transgenic maize plants (Guo et al., 2014; Shi et al., 2015).
In addition to a transgenic approach, natural genetic variation for traits that impact drought tolerance has also been used in maize breeding programmes to improve grain yield. By applying precision phenotyping and molecular markers as well as understanding the genetic architecture of quantitative traits, maize breeders developed hybrids (AQUAmax®) with increased grain yield under drought stress conditions (Cooper et al., 2014; Gaffney et al., 2015). The drought tolerance in these hybrids is governed by multiple genes which individually have small effects. Potentially, some of these key genes could be identified and altered to generate new alleles to produce a larger effect, thus enhancing the breeding process. However, until recently, generating such allelic variation with physically or chemically induced mutagenesis was a random process, which made it difficult to produce intended DNA sequence changes at a target locus. In the past few years, efficient genome editing technologies have emerged, enabling rapid and precise manipulation of DNA sequences, and setting the stage for developing drought‐tolerant germplasm by editing major genes in their natural chromosomal context.
Four genome editing tools, meganucleases, zinc‐finger nucleases (ZFN), transcription activator‐like effector nucleases (TALEN) and the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated nuclease protein (Cas) system, have provided targeted gene modification in plants (Čermák et al., 2015; Gao et al., 2010; Li et al., 2012, 2013; Shukla et al., 2009). Among these, the CRISPR‐Cas9 system is easiest to implement and is highly efficient. The system consists of a Cas9 endonuclease derived from Streptococcus pyogenes and a chimeric single guide RNA that directs Cas9 to a target DNA sequence in the genome. CRISPR‐Cas9 genome editing is accomplished by introducing a DNA double‐strand break in the target locus via Cas9, followed by DNA repair through either the endogenous imprecise nonhomologous end‐joining (NHEJ) or the high‐fidelity homology‐directed repair (HDR) pathways. NHEJ can induce small insertions or deletions at the repair junction while HDR stimulates precise sequence alterations, including programmed sequence correction as well as DNA fragment insertion and swap, when a DNA repair template is exogenously supplied. The system has been successfully tested in staple crops, such as maize, wheat, rice and soybean (Cai et al., 2015; Du et al., 2016; Jacobs et al., 2015; Jiang et al., 2013; Li et al., 2015; Liang et al., 2014; Shan et al., 2015; Sun et al., 2016; Svitashev et al., 2015; Wang et al., 2014; Zhang et al., 2014; Zhou et al., 2014, 2015).
In maize, endogenous ARGOS8 mRNA expression is relatively low and spatially nonuniform. Previous field testing showed that constitutive overexpression of ARGOS8 in transgenic plants increases grain yield under drought stress conditions without yield penalty in nonstress environments (Shi et al., 2015). Aiming at creating novel ARGOS8 variants which would confer beneficial traits for maize breeding, the genomic sequence of ARGOS8 was edited using CRISPR‐Cas‐enabled advanced breeding technology to produce ubiquitous and elevated expression across multiple tissues and at different developmental stages. Here, we report the generation of maize lines carrying ARGOS8 genome‐edited variants and their hybrid yield performance in a field study. Our results demonstrate that modifying single native genes to change expression patterns can increase maize grain yield under drought stress conditions.
Results
Natural allelic variation of maize ARGOS8 and expression patterns
In wild‐type (WT) inbreds PH184C (proprietary) and B73 (public), ARGOS8 mRNA expression is very low in all the tissues tested, ranging from 3 to 25 transcripts per ten million (TPTM), as measured with RNA sequencing (Figure S1). The only exception is in kernels where expression was approximately 260 TPTM. For comparison, the transcripts of the ubiquitously and moderately expressed GOS2 gene, the maize homolog of rice GOS2 (de Pater et al., 1992), are about 6000 TPTM in most tissues with the highest expression occurring in internodes (13 700 TPTM) and the lowest occurring in tassels (2500 TPTM). A survey of a diverse set of 419 proprietary and public inbred lines showed that the ARGOS8 expression in leaves of 3‐week‐old seedlings only ranged from 0 to 20 TPTM (Figure S2), suggesting that the natural variation in expression levels among these inbreds was also low.
The protein encoded by the ARGOS8 gene varies among inbred lines. The ARGOS8 protein in B73 has 118 amino acids (long version) while the protein from PH184C consists of 94 amino acids (short version). The difference in protein sequence is the presence of an N‐terminal extension of 24 amino acids in the long version. The extra coding sequence in the B73 allele is a result of a 7‐bp duplication in the 5′‐untranslated region (5′‐UTR) which produces an in‐frame ATG codon upstream of the original translation start codon. The 7‐bp duplication may be a footprint left behind by a transposon excision event (Scott et al., 1996). Like the long version from B73 (Shi et al., 2015, 2016), the short version of ARGOS8 also reduces ethylene responses when overexpressed, as demonstrated in Arabidopsis transgenic plants. In the ethylene triple response assay (Bleecker et al., 1988), hypocotyls and roots of the etiolated 35S:ARGOS8 Arabidopsis seedlings were longer than that in WT controls in the presence of the ethylene precursor aminocyclopropane‐1‐carboxylic acid (Figure 1). Although the short version of the ARGOS8 protein accumulated to a higher level than the B73 version in plants with a similar level of transcripts (data not shown), the native allelic variant still produces a very low level of the ARGOS8 protein in WT plants and it was not detectable by immunoblot analysis. This short allele, as well as the long B73 allele, was not able to confer drought‐tolerant phenotypes without ectopic overexpression. Consequently, this observed functional native diversity in the ARGOS8 gene is not enough for targeted drought breeding.
Figure 1.
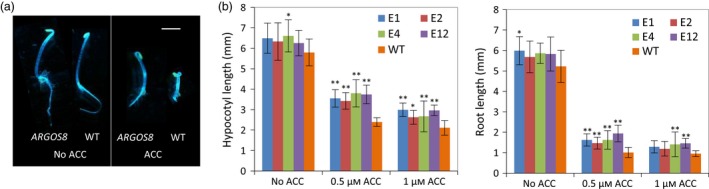
Maize ARGOS8 reduces plant responses to ethylene when overexpressed in transgenic Arabidopsis plants. (a) Ethylene triple response of Arabidopsis ARGOS8 transgenic plants (ARGOS8) and wild‐type (WT) controls to 0.5 μm of the ethylene precursor aminocyclopropane‐1‐carboxylic acid (ACC). A short version of ARGOS8 was overexpressed under control of the cauliflower mosaic virus 35S promoter (35S). Composite image of representative 3‐day‐old etiolated seedlings. Bar = 2 mm. (b) Hypocotyl and root lengths of etiolated Arabidopsis seedlings overexpressing the short version of ARGOS8. Four transgenic lines (E1, E2, E4 and E12) and wild‐type (WT) controls were grown in the dark in the presence of indicated ACC concentrations for 3 days. Data are means ± SD, n = 15. Significant differences of the transgenic plants from the WT are denoted by asterisks (*P < 0.05, **P < 0.01, ANOVA, Tukey's HSD).
Novel ARGOS8 variants generated by the CRISPR‐Cas9 system
To achieve a moderate level of constitutive expression of ARGOS8, the maize GOS2 promoter and the 5′‐UTR with an intron, hereafter collectively referred to as GOS2 PRO, were either used to replace the native promoter of the ARGOS8 gene or were inserted into the 5′‐UTR of ARGOS8 in the inbred PH184C. Because both the promoter swap and insertion require precise manipulation of genomic DNA, we employed an RNA‐guided Cas9 endonuclease to generate DNA double‐strand breaks in a site‐specific manner, integrating the GOS2 PRO into the upstream region of ARGOS8 via homology‐directed DNA repair (Figure 2a). A DNA repair template and genome editing reagents were delivered into immature embryos by particle bombardment and plantlets were regenerated from embryogenic calli. The reagents include an S. pyogenes Cas9 gene and a single guide RNA (sgRNA) gene, CRISPR RNA1 (CR1), in the GOS2 PRO insertion or two sgRNAs (CR2 and CR3) in the GOS2 PRO swap (Figure S3), as well as phosphomannose isomerase (PMI), ovule development protein2 (ODP2) and WUSCHEL (WUS) for stimulation of transformation and seedling regeneration (Svitashev et al., 2015). The DNA repair template consisted of the GOS2 PRO flanked by two DNA fragments of approximately 400‐bp homologous to genomic sequences immediately adjacent to the Cas9 cleavage sites in the ARGOS8 locus (Figure 2b). Of approximately 1000 immature embryos particle‐bombarded for the promoter insertion and swap experiments, 194 and 334 shoots, respectively, were regenerated on the selection medium (Table S1). To eliminate the shoots whose CRISPR RNA target sites (CTS) were not altered, a rapid screening was performed using a quantitative PCR (qPCR) assay (Table S2), which estimates the copy number of CTS. Shoots with no modification at CTS contained two copies of the wild‐type CTS, shoots with CTS modification in one of the two sister chromosomes had one intact copy, while modification in both chromosomes would reduce the copy number to zero. With this screening, 190 and 172 regenerated shoots from the insertion and swap experiments, respectively, were selected for genotyping with a junction PCR assay.
Figure 2.
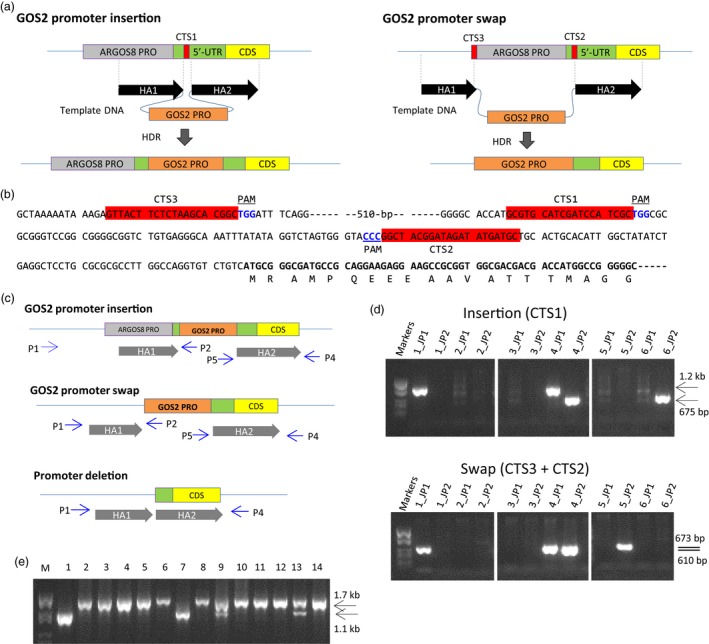
Editing the ARGOS8 genomic sequence using the CRISPR/Cas9 system to generate variants with constitutive expression. (a) Schematic drawing illustrating the insertion of GOS2 PRO into the 5′‐UTR of ARGOS8 and the promoter swap. CTS, CRISPR‐RNA target site; HA, homology arm; HDR, homology‐directed repair; GOS2 PRO, maize GOS2 promoter and the 5′‐UTR with an intron. (b) Genomic sequence of the ARGOS8 5′‐UTR and the upstream region. The CRISPR‐RNA target sites (CTS) are highlighted in red, and the protospacer adjacent motifs (PAM) are shown in blue font. The ARGOS8 coding region is shown in bold font. (c) Diagram showing primers used in junction PCR for genotyping regenerated shoots and long PCR for amplifying and sequencing the entire modification region in homozygous plants. The relative position and direction of PCR primers (P) are indicated by arrows. P1 and P2 for the HR1 junction; P5 and P4 for the HR2 junction; P1 and P4 for the long PCR. (d) Junction PCR analysis of regenerated shoots. Agarose gel images are shown for representative regenerated shoots positive for one junction or two junctions and shoots negative in the junction PCR assay. JP1, HR1 junction PCR with the primer P1 and P2; JP2, HR2 junction PCR with P5 and P4. (e) PCR screening regenerated shoots for deletion in the ARGOS8 locus. An agarose gel image is shown for PCR products amplified with the primer P1 and P4 in representative shoots (Lanes 1‐14) generated from the CRISPR RNA‐3 and RNA‐1 transformation. M, DNA molecular weight markers.
A pair of junction PCR assays was designed to detect GOS2 PRO inserts or swaps at CTS due to homologous recombination (Figure 2c). In the insertion experiments, five of the 190 shoots from the initial screening were found positive for one of the two junctions, and two shoots were positive for both junctions (Figure 2d and Table S1). These shoots were transferred to rooting media, and three plantlets were regenerated. Genotyping the T0 plants with the junction PCR assays revealed that one plant contained the GOS2 PRO insert in the ARGOS8 locus. The junction PCR products were sequenced, and expected sequences were confirmed (Figure S4). This line is referred to as ARGOS8‐variant1 (ARGOS8‐v1). For the GOS2 promoter swap, 23 of the 172 shoots obtained from the initial screening were positive for at least one junction. Among them, three were positive for both junctions (Figure 2d and Table S1). From these shoots, eight plantlets were regenerated. Of these T0 plants, two produced expected junction PCR products for the promoter swap in the ARGOS8 locus. Sequencing the PCR products confirmed correct sequences from both junctions (Figure S4). One of the lines is referred to as ARGOS8‐variant2 (ARGOS8‐v2). Genotyping also revealed that the ARGOS8‐v1 and ARGOS‐v2 were heterozygous.
F1 seeds of ARGOS8‐v1 and ARGOS8‐v2 were produced by crossing the T0 plants with WT PH184C plants. F1 plants were genotyped by PCR to select those that carry the ARGOS8‐v1 or ARGOS8‐v2, but were nulls for the genome editing reagents Cas9, sgRNA, PMI, ODP2 and WUS. To eliminate the plants containing random insertions of the DNA repair template, qPCR was performed to assess the copy number of the GOS2 PRO and ARGOS8. Selected clean F1 plants were backcrossed to produce BC1 seeds, or self‐pollinated to obtain F2 seeds. Among the F2 segregants, homozygous plants were used to determine the sequence integrity of the newly created ARGOS8 variants. The entire genomic region was amplified using long PCR with primers P1 and P4, which were derived from genomic sequences further upstream and downstream of the homology arms used in the DNA repair templates (Figure 2c). Sequencing the long‐PCR products confirmed that the ARGOS8‐v1 and ARGOS8‐v2 possess the expected DNA sequences (Figure 3a). The ARGOS8‐v1 and ARGOS8‐v2 segregated in a Mendelian fashion in BC1 and F2 populations (data not shown). Quantitative reverse‐transcription PCR (qRT‐PCR) analysis showed that the abundance of ARGOS8 transcripts in leaves of homozygous plants is approximately twice as much as in the heterozygotes for both lines (Figure 3b).
Figure 3.
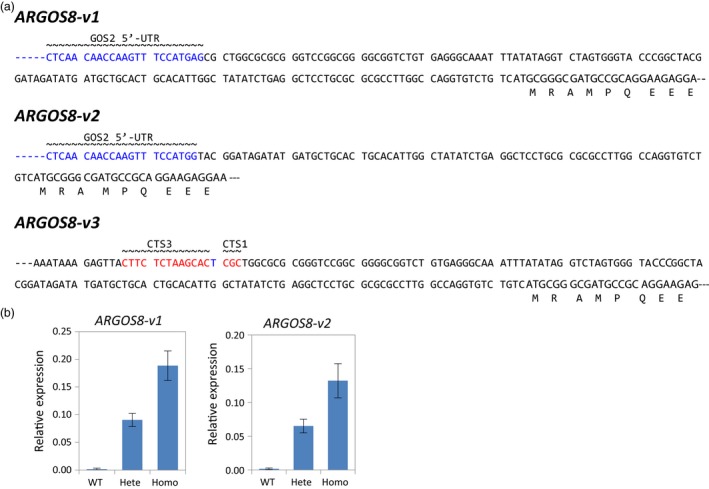
Maize genome‐edited ARGOS8 variants. (a) Genomic sequence upstream of the ARGOS8 coding region in three genome‐edited variants. The entire modification region in homozygous F2 plants was amplified using long PCR, and the PCR products were sequenced. Part of the GOS2 5′‐UTR sequence (blue font) and the remaining 5′‐UTR of ARGOS8 as well as the 5′‐terminus of ARGOS8 coding sequence are shown. In the promoter deletion variant ARGOS8‐v3, the remnant CTS3 and CTS1 sequences are highlighted. (b) Relative expression levels of ARGOS8 in leaves as measured by qRT‐PCR. Means ± SD are shown for F2 plants of 14‐day‐old ARGOS8‐v1 and 18‐day‐old ARGOS8‐v2; n = 10–24. WT, wild‐type; Hete, Heterozygote; Homo, homozygote.
To obtain controls for analysing ARGOS8 gene expression in the ARGOS8 variants, the shoots regenerated from particle‐bombarded immature embryos were screened for ARGOS8 promoter deletions in the promoter swap experiments using CR3 and CR1. Of 185 shoots screened, 30 produced a PCR product shorter than that expected for WT plants (Figure 2c and e), indicating a deletion between CTS3 and CTS1. Sequencing the PCR products from two T0 plants showed that both had an extra base pair (one line having T and the other A) at the junction of the nonhomologous end‐joining (Figures 3a and S4). Similarly, approximately 13% (23 of 176) of the regenerated shoots were found contain deletion at the target sites in the swap experiments using CR3 and CR2 (data not shown). The deletion of the 550‐bp genomic DNA fragment between CTS3 and CTS1 removed part of the ARGOS8 5′‐UTR and the upstream promoter sequence (Figures 2b and 3a). One of the lines was referred to as ARGOS8‐variant3 (ARGOS8‐v3). The ARGOS8 transcripts and proteins were undetectable in ARGOS8‐v3 (Figure 4a and b). The line had normal growth and development, and no phenotypic defects were observed under normal growing conditions, indicating that the ARGOS8 gene is likely dispensable.
Figure 4.
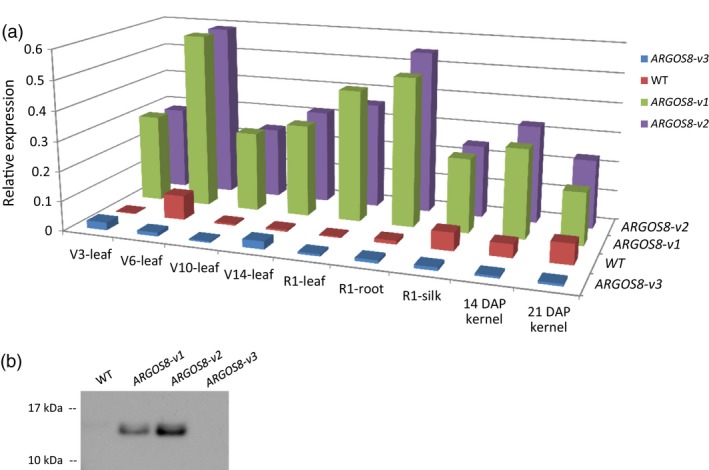
Comparison of the ARGOS8 expression in genome‐edited variants and wild‐type maize plants. (a) Relative expression of ARGOS8 in a selection of maize tissues and stages. mRNA was quantified with qRT‐PCR. Six individual plants were analysed for the genome‐edited variants and two plants for WT controls. DAP, days after pollination. (b) ARGOS8 protein expression in developing kernels. Immature kernels (21 DAP) were analysed by immunoblotting using a monoclonal anti‐ARGOS8 antibody.
Expression patterns of ARGOS8 in genome‐edited variants
The mRNA expression of ARGOS8 in the genome‐edited variants was analysed in leaves, roots, silks and kernels using qRT‐PCR. In the uppermost collared leaves of plants at the developmental stages V3, V6, V10 and V14, the ARGOS8 transcript level in ARGOS8‐v1 and ARGOS8‐v2 was significantly higher than that in WT plants with the highest expression found at V6 (Figure 4a). At the developmental stage R1, silks, roots and leaves all had higher levels of the ARGOS8 mRNA in the genome‐edited plants relative to WT controls. In developing kernels 14 and 21 days after pollination (DAP), the ARGOS8 mRNA was also more abundant in the ARGOS8‐v1 and ARGOS8‐v2 than the WT. The ARGOS8 protein was detectable by immunoblot analysis in the developing kernels of the genome‐edited variants, but not in the WT (Figure 4b). The two variants had a similar level of ARGOS8 mRNA expression in all the tissues tested (Figure 4b).
Improved grain yield under drought stress environments
The two genome‐edited variants ARGOS8‐v1 and ARGOS8‐v2 were crossed with an inbred tester to create a hybrid for field evaluation. These variants were compared to a wild‐type hybrid that had not undergone genome editing. Entries were evaluated across multiple environments at eight locations throughout the United States. At the end of the growing season, locations were grouped into three environmental types based on the occurrence of drought stress. Four locations had yields near or above 200 bushel per acre; these were classified as optimal locations (OPT) where water deficits were not a constraint. The remaining locations were grouped as either flowering stress (FS) or grain‐filling stress (GFS), based on the EnClass location classification system (Loffler et al., 2005).
Significant differences among entries were observed for grain yield in the FS location group, with the ARGOS8‐v1 and ARGOS8‐v2 entries yielding approximately five bushel per acre more than the control (Table 1). In contrast, there was no significant difference in grain yield between the variants and WT in the GFS or OPT locations (Table 1). The GFS locations were characterized by limited soil moisture availability due to soil texture, and drought stress developed very quickly. This may have resulted in early cessation of grain filling in the ARGOS8‐v1 and ARGOS8‐v2 entries; grain moisture was significantly less than in the control for ARGOS8‐v1 (Table S3). Plant height and ear height increased by a small (2.6 and 3.2 cm, respectively) but significant amount in the ARGOS8‐v2 (Table S3) in the OPT locations. No differences were observed in thermal time to silk or to shed.
Table 1.
Grain yield of ARGOS8 genome‐edited variants and wild type under flowering stress, grain‐filling stress and optimal (well‐watered) conditions
| Flowering | Grain‐filling | Optimal | |
|---|---|---|---|
| Stress | Stress | ||
| ton ha−1 (bushel acre−1) | |||
| ARGOS8‐v1 | 8.67 (138.0)a | 7.47 (119.0) | 13.13 (209.0) |
| ARGOS8‐v2 | 8.67 (138.0)a | 7.54 (120.0) | 13.19 (210.0) |
| WT | 8.34 (132.8) | 7.72 (122.9) | 13.01 (207.1) |
Data are from two individual genome‐edited variants (ARGOS8‐v1, ARGOS8‐v2) and wild type tested as one hybrid at eight locations in 2015. Predicted difference for each variant is compared with the wild type. All analyses were implemented using ASReml with output of the model presented as best linear unbiased predictions (see Experimental procedures).
Predicted difference significant at P < 0.1.
Discussion
Constitutive overexpression of ARGOS8 using a transgenic approach increases grain yield in maize under drought stress conditions (Shi et al., 2015). To explore the feasibility of recapitulating the ARGOS8 transgene effect using a conventional breeding approach, we determined the native expression levels of ARGOS8 in a set of public and proprietary maize inbred lines. None of the inbreds we examined had mRNA expression levels great enough to match those in the transgenic events. In addition, a naturally occurring variant of ARGOS8 encoding a shorter protein also was not able to confer desired phenotypes without overexpression. Therefore, conventional breeding with this gene was not deemed worthwhile, and we elected to use a CRISPR‐Cas enabled advanced breeding technology to generate new ARGOS8 variants by changing the DNA sequence at the native ARGOS8 locus. Replacement of the ARGOS8 promoter with a maize GOS2 promoter (GOS2 PRO), or insertion of a GOS2 PRO into the 5′‐UTR of the ARGOS8 gene, led to a change in the ARGOS8 expression pattern from tissue preferred to ubiquitous, and from relatively low mRNA expression levels to significantly increased ARGOS8 expression levels. Precise modification of the nucleotide sequence of ARGOS8 at its native location in the genome was achieved, as determined by PCR assays of the entire region followed by sequencing. The ARGOS8 variants were found to be stably inherited via analysis of over four generations. Field testing showed that the novel ARGOS8 variants increased grain yield under drought stress conditions. These yield results are similar to previous results obtained from transgenic plants overexpressing ARGOS8 (Shi et al., 2015). These results demonstrate the utility of genome editing in creating novel allelic variation for enhancing crop drought tolerance.
The mutation rate at CRISPR‐RNA target sites in the regenerated shoots ranged from 60% to 98%, similar to that reported in maize gene modification studies using stably transformed lines (Svitashev et al., 2015). In the promoter swap experiments using two guide RNAs, we observed a frequency of approximately 16% (30 of 185) for DNA fragment deletion due to the nonhomologous end‐joining. The homology‐directed DNA swap at the ARGOS8 locus occurred in approximately 1% (3 of 334) of the regenerated shoots. A comparable frequency (2 of 194) was found for insertion when one guide RNA was used. In a previous study, the insertion frequency at the maize liguleless1 locus was 2.5%–4.1% (Svitashev et al., 2015). CRISPR‐RNA target sites, the surrounding genomic DNA sequences, insert sequences and the genotype of host plants as well as other factors may contribute to the difference in insertion frequencies. We also observed that nearly 60% (19 of 30; Table S1) of the regenerated shoots failed to produce T0 plants. Of the five two‐junction‐positive events identified in the shoot stages, only three T0 plants were recovered, indicating more genome‐edited lines can be obtained by improving maize inbred transformation.
The maize ARGOS8 gene is a negative regulator of ethylene responses. ARGOS8 proteins physically interact with the ethylene receptor signalling complex, modulating ethylene perception and the early stages of the ethylene signal transduction (Shi et al., 2016). Transgenic maize and Arabidopsis plants overexpressing ARGOS genes exhibit reduced sensitivity to ethylene (Rai et al., 2015; Shi et al., 2015), enhanced cell elongation and/or division resulting in taller plants, larger leaves, and longer ears in maize (Guo et al., 2014; Shi et al., 2015), as well as creating larger organs in other plant species (Feng et al., 2011; Hu et al., 2003; Kuluev et al., 2011). Drought stress often reduces plant growth and can adversely affect development, leading to grain yield loss in crops. These stress‐induced changes at the whole plant level are largely due to reduced cell number and/or size. Constitutively overexpressed ARGOS likely counteracts the effect of water deficiency by promoting cell expansion and/or division, mitigating the yield loss by enhancing plant growth under drought stress. Here, we show that maize variants of the ARGOS8 gene generated by altering its regulatory elements can deliver a significant increase in grain yield under a flowering stress condition with no yield loss under an optimal condition, similar to that of ARGOS8 transgenic plants (Shi et al., 2015). However, this was not the case when variants were exposed to a grain‐filling stress (Table 1). This result was not surprising given that much of the yield increase resulting from ectopic expression of ARGOS8 under abiotic stress comes from an increase in kernel set (Shi et al., 2015), which is primarily determined at flowering time.
Unlike transgenic ARGOS8 plants, the maize inbred lines carrying the genome‐edited variants contain no transformation selection markers or any nonmaize DNA. All the reagents (i.e. helper genes) used in maize DNA sequence editing, including Cas9, sgRNA, PMI, ODP2 and WUS as well as plasmid backbones, were not required for the function of newly generated ARGOS8 variants and were removed by backcrossing in the early stages of breeding. Instead of Agrobacterium‐mediated transformation, particle bombardment was employed to deliver the genome editing reagents; thus, no plant pathogen was involved in the generation of these variants. The DNA repair template (GOS2 promoter flanked by homology arms) originated from maize genomic DNA; only the maize GOS2 promoter is site‐specifically integrated into the ARGOS8 locus via homologous recombination, leading to the designed modification of ARGOS8 expression.
The ARGOS8 editing process can be summarized as a two‐step procedure: duplication of the GOS2 promoter and translocation to the ARGOS8 locus. Both duplication and translocation of DNA fragments occur naturally in maize (Wang et al., 2015; Zhang et al., 2000). During its evolutionary history, the maize genome has undergone several rounds of whole‐genome duplication. In addition, segmental duplication, which involves DNA fragments of different sizes ranging from a few base pairs up to many megabases which may or may not contain intact, functional genes, also play an important role in shaping the maize genome and increasing genetic diversity (Lai et al., 2004; Schnable et al., 2009). Similarly, this process occurs naturally in other plants and animals, including humans (Zhang et al., 2005). Comparative genomic studies have shown that although segmental duplication drives the formation of clusters of closely related genes, duplicated sequences can translocate to different chromosomal locations, resulting in dispersal of paralogs throughout the genome (Freeling et al., 2008; Lai et al., 2004; Mendivil Ramos and Ferrier, 2012). For example, the plant disease resistance NBS‐LRR genes are particularly prone to being transposed (Ameline‐Torregrosa et al., 2008; Baumgarten et al., 2003; Leister et al., 1998; Richly et al., 2002), vividly attesting to segmental duplication and translocation occurring in plants. This natural rearrangement of DNA fragments occurs spontaneously at a low frequency in maize and has been exploited by breeders over the decades for maize improvement. Similarly, the CRISPR‐Cas‐enabled advanced breeding technology allows precise integration of duplicated genetic elements into a target locus and can enhance the grain yield of maize.
Since the adoption and widespread use of hybrid maize, natural allelic variations in a large number of genes, each with small effects, have improved drought tolerance, even though it has been suggested that the stress‐tolerant alleles are present at relatively low frequencies in most elite breeding populations (Blum, 1988). With increasing knowledge on plant response to drought stress and molecular understanding of gene networks underlying the physiological processes that impact drought tolerance, monogenic drought tolerance in maize has become possible by a transgenic approach. Indeed, there are multiple examples of the validation of the efficacy of transgenes in elite hybrids under field conditions (Castiglioni et al., 2008; Guo et al., 2014; Habben et al., 2014; Leibman et al., 2014; Nuccio et al., 2015; Shi et al., 2015). With the advent of CRISPR‐Cas enabled advanced technology, a new technique is now available to provide new sources of genetic variation for plant breeding. This genome editing study of ARGOS8 demonstrates that single endogenous genes can be modified to create novel variants that have a significantly positive effect on a complex trait such as drought tolerance. In short, the generation and use of genome‐edited variants is a seminal addition to the precision breeding toolbox that can enhance changes to the plant genome in a predictable manner.
Experimental procedures
Plasmids and reagents used for plant transformation
Plasmids were designed to insert a DNA fragment into the 5′‐UTR of target genes using a single guide RNA (sgRNA), the Cas9 endonuclease gene derived from Streptococcus pyogenes and a DNA repair template. For the DNA fragment swap, two sgRNA were used. The sgRNA gene was adapted from Mali et al. (2013) and consists of a maize U6 polymerase III promoter (Svitashev et al., 2015), a CRISPR RNA (crRNA), a transactivating CRISPR RNA (tracrRNA) and a terminator (Figure S3). The Cas9 expression cassette contains the maize UBIQUITIN1 promoter and potato protease inhibitor II terminator. The Cas9 sequence was maize codon optimized and added the potato ST‐LS1 intron as well as the nuclear localization signals from the SV40 and the Agrobacterium tumefaciens Vir D2, as previously described (Svitashev et al., 2015), for appropriate expression and nuclear targeting in maize. The DNA repair template plasmid carries the maize GOS2 promoter (NCBI GenBank accession no. GQ184457; nucleotides 218 974–220 796 in reverse direction; Barbour et al., 2003) that was inserted between two homology arms each approximately 400 bp derived from the genomic sequence flanking the CRISPR‐RNA target site (CTS; Figure 2a). The vector also contains a multiple cloning site of 61‐bp immediately upstream of the GOS2 promoter. Constructs were assembled using chemically synthesized DNA fragments with standard DNA techniques (Sambrook et al., 1989). To facilitate delivery of the genome editing reagents into maize cells and regeneration of plants, three expression cassettes encoding transformation selection marker phosphomannose isomerase (PMI) as well as cell division and callus growth‐promoting proteins ovule development protein2 (ODP2) and WUSCHEL (WUS) were constructed as previously described (Ananiev et al., 2009; Svitashev et al., 2015).
Maize transformation
Biolistic‐mediated transformation of maize immature embryos was performed according to Svitashev et al. (2015). Briefly, gold particles, 0.6 μm in diameter, were washed with cold, 100% [v/v] ethanol and sterile distilled water. The DNA purified with QIAprep Spin Miniprep (Qiagen) was precipitated on the washed gold particles using a water‐soluble cationic lipid TransIT‐2020 (Mirus). Fifty microlitres of gold particles (water solution of 10 mg/mL) and 1 μL of TransIT‐2020 water solution were added to the premixed DNA, mixed gently and incubated on ice for 10 min. DNA‐coated gold particles were then centrifuged at 8 000 g for 1 min. The pellet was rinsed with 100 μL of 100% [v/v] ethanol and resuspended by a brief sonication. Immediately after sonication, DNA‐coated gold particles were loaded onto the centre of a macrocarrier (10 μL of each) and allowed to air dry. Immature embryos 9–11 days after pollination were bombarded using a PDS‐1000 Helium Gun (Bio‐Rad) with a rupture pressure of 425 psi. Postbombardment culture, selection and plant regeneration were carried out as previously described (Gordon‐Kamm et al., 2002). Regenerated shoots were sampled for initial qPCR screening and junction PCR. Leaf discs were taken from T0 seedlings 2 weeks post‐transplanting for genotyping. F1 seeds were produced by crossing the T0 with wild‐type plants. BC1 and F2 seeds were produced by backcrossing and self‐pollination, respectively.
Arabidopsis transformation and ethylene response assay
The 35S::ARGOS8 construct was assembled and transgenic Arabidopsis plants generated as described (Shi et al., 2015). Transgenic lines were selected based on the expression of the fluorescent marker yellow fluorescence protein in T1 seeds. The ARGOS8 transgene expression was confirmed by qRT‐PCR. To determine the activity of ARGOS8 in reducing plant response to ethylene, the ethylene triple response assay (Bleecker et al., 1988) was carried out in the presence of the ethylene precursor aminocyclopropane‐1‐carboxylic acid (ACC). Surface‐sterilized and stratified seeds were germinated in the dark for 3 days in agar that contained one‐half‐strength Murashige and Skoog salts and 1% [w/v] sucrose supplemented with 0, 0.5, or 1 μm ACC. Hypocotyl and root lengths of the etiolated seedlings were measured by photographing the seedlings with a digital camera and using image analysis software ImageJ (National Institutes of Health).
DNA extraction, PCR genotyping and sequencing
DNA was extracted from regenerated shoots or leaf discs as described in Gao et al. (2010). PCR was performed using REDEtract‐N‐Amp PCR readyMix (Sigma) or Phusion High‐Fidelity PCR Master Mix (NEB) according to the manufacturer's instructions. Quantitative PCR (qPCR) was carried out as described in Svitashev et al. (2015). The primer sequences used in PCR and qPCR are listed in Supporting Information Table S2. The PCR products were sequenced directly or cloned into the pCR2.1‐TOPO vectors (Thermo Fisher Scientific) before sequencing.
RNA extraction, qRT‐PCR and RNA sequencing
Total RNA was isolated with Qiagen RNA Isolation Kit (Qiagen). The DNaseI Enzyme Kit (Roche) was used to remove DNA from the RNA samples. Complementary DNA was synthesized from the total RNA using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). PCR amplifications were performed using the TaqMan probe‐based detection system according to the manufacturer's instructions (Applied Biosystems). Primers and probes are shown in Table S2. Relative quantification values were determined using the difference in Ct from the target genes and the reference gene, maize UBIQUITIN5.
RNA sequencing (RNA‐seq) was performed as described (Shi et al., 2015). In brief, total RNA was isolated from frozen maize tissues and used to prepare sequencing libraries using the TruSeq mRNA‐Seq Kit (Illumina), and sequenced on the Illumina HiSeq 2000 system with Illumina TruSeq SBS v3 reagents. On average, 10 million sequences were generated for each sample. The resulting sequences were trimmed based on quality scores and mapped to the maize B73 reference genome sequence V2 and normalized to reads per kilobase of transcript per ten million mapped reads. The generated data matrix was visualized and analysed in GeneData Analyst software (Genedata AG, Basel, Switzerland).
Immunoblot analysis
To detect ARGOS8 proteins, extracts were prepared from immature kernels, proteins separated by SDS‐PAGE, blotted to a nitrocellulose membrane and probed with a monoclonal anti‐ARGOS8 antibody. The primary antibodies were detected with a HRP‐conjugated goat anti‐mouse secondary antibody and the Pierce SuperSignal® West Dura Extended Duration Substrate (Thermo Fisher Scientific).
Maize hybrid yield testing
To evaluate the genome‐edited variants, field trials were conducted across multiple environments in small plots (approximately 4 m2) with 2–4 replications at each of eight locations. Hybrid seed for these trials was generated by crossing the genome‐edited ARGOS8 variants with an inbred tester. A wild‐type hybrid containing the native ARGOS8 allele served as the comparator. The experimental variants and control were grown in field environments at research centres in Woodland, CA; Garden City, KS; Plainview, TX; York, NE; Marion, IA; Johnston, IA; and Princeton, IN in 2015. Some environments were managed to impose various levels of drought stress while others were managed for optimum yield/nonstress conditions. Fertilizer at each location was applied to achieve maximum yields. Weeds and pests were controlled according to local practices. Grain mass and grain moisture data were collected using a small plot combine. Grain yield was adjusted to a constant 15% moisture. Additional agronomic characteristics evaluated at selected locations were plant and ear height and thermal time to shed and silk.
The field experimental design was set up as a randomized complete block arrangement. Data analysis was by ASREML (VSN International Ltd), and the values reported are BLUPs (Best Linear Unbiased Predictions; Cullis et al., 1998; Gilmour et al., 2009). A mixed‐model framework was used to perform the analysis. The model included replicate, row, column and heterogeneous residual variance with a separable autoregressive correlation for both row and column directions (AR1*AR1) within each location to reduce the impact of spatial variation in the field. In the analysis, the main effect of location type was considered a fixed effect. The main effect of entry and its interaction with location type were considered random effects. Statistical significance is reported with a P‐value of 0.1 in a two‐tailed test.
Conflict of Interest
The authors are employees of DuPont Pioneer.
Supporting information
Figure S1 Maize ARGOS8 gene expression. The transcript abundance of ARGOS8 in various tissues of maize inbred PH184C was measured by RNA sequencing. Samples were taken from the plants at the developmental stage of V10, VT/R1 and R4. TPTM, transcript per ten million.
Figure S2 Comparison of ARGOS8 gene expression among 419 maize inbred lines. The ARGOS8 transcript levels in leaves of 3‐week‐old seedlings were measured by RNA sequencing. TPTM, transcript per ten million.
Figure S3 Single guide RNA genes used in editing the genomic sequence of ARGOS8. (a) Scheme illustrating the structure of a single guide RNA (sgRNA) gene. (b) DNA sequences of the transcription region in the sgRNA genes CRISPR RNA1 (CR1), CR2 and CR3. U6 PRO, maize U6 polymerase III promoter; crRNA, CRISPR RNA (in red font) functioning as a guide; tracrRNA, transactivating CRISPR RNA, the sgRNA scaffold.
Figure S4 DNA sequences of the junction PCR products amplified from ARGOS8 variants.
Table S1 Frequency of the GOS2 promoter insertion with one target site and the promoter swap with two target sites in T0 maize plants.
Table S2 Primers and probes used for generation and characterization of ARGOS8 genome editing variants.
Table S3 Plant height (PLTHT), ear height (EARHT) and grain moisture (MST) of ARGOS8 genome‐edited variants and wild type under flowering stress, grain‐filling stress and optimal (well‐watered) conditions.
Acknowledgements
We thank Wally Marsh, Susan Wagner, Min Zeng, Megan Schroder, Joshua Young, Bruce Drummond, Mary Trimnell, Ben Weers, Jesse Ourada, Wang‐Nan Hu, Dennis O'Neill, Andy Baumgarten, Norbert Brugiere, Gina Zastrow‐Hayes, Mary Beatty, Jennifer Hanks, Matthew Hanson, Laura Church, Keith Allen and Jacque Hockenson for excellent assistance with this study. We acknowledge our colleagues at outlying breeding stations who conducted first‐rate yield trials. We are grateful to Carl Simmons, Nathan Coles, Mark Cigan and Maria Fedorova for their critical review of this manuscript. We thank Mike Lassner and Mark Cigan for their support of this project. We also thank Tom Greene and Mark Cooper for their organizational leadership and helpful input.
References
- Ameline‐Torregrosa, C. , Wang, B.B. , O'Bleness, M.S. , Deshpande, S. , Zhu, H. , Roe, B. , Young, N.D. et al (2008) Identification and characterization of nucleotide‐binding site‐leucine‐rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 146, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V. , Wu, C. , Chamberlin, M.A. , Svitashev, S. , Schwartz, C. , Gordon‐Kamm, W. and Tingey, S. (2009) Artificial chromosome formation in maize (Zea mays L.). Chromosoma, 118, 157–177. [DOI] [PubMed] [Google Scholar]
- Barbour, E. , Meyer, T.E. and Saad, M.E. (2003) Maize GOS2 Promoters. U.S. Patent No. 6504083 B1, Pioneer Hi‐Bred International.
- Baumgarten, A. , Cannon, S. , Spangler, R. and May, G. (2003) Genome‐level evolution of resistance genes in Arabidopsis thaliana. Genetics, 165, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A.B. , Estelle, M.A. , Somerville, C. and Kende, H. (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science, 241, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Blum, A. (1988) Plant Breeding for Stress Environments. Boca Raton, FL: CRC Press. 223p. [Google Scholar]
- Cai, Y. , Chen, L. , Liu, X. , Sun, S. , Wu, C. , Jiang, B. , Han, T. et al (2015) CRISPR/Cas9‐mediated genome editing in soybean hairy roots. PLoS ONE, 10, e0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni, P. , Warner, D. , Bensen, R.J. , Anstrom, D.C. , Harrison, J. , Stoecker, M. , Abad, M. et al (2008) Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water‐limited conditions. Plant Physiol. 147, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák, T. , Baltes, N.J. , Čegan, R. , Zhang, Y. and Voytas, D.F. (2015) High‐frequency, precise modification of the tomato genome. Genome Biol. 16, 232. doi:10.1186/s13059‐015‐0796‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, M. , Gho, C. , Leafgren, R. , Tang, T. and Messina, C. (2014) Breeding drought‐tolerant maize hybrids for the US corn‐belt: discovery to product. J. Exp. Bot. 65, 6191–6204. [DOI] [PubMed] [Google Scholar]
- Cullis, B.R. , Gogel, B.J. , Verbyla, A.P. and Thompson, R. (1998) Spatial analysis of multi‐environment early generation trials. Biometrics, 54, 1–18. [Google Scholar]
- Du, H. , Zeng, X. , Zhao, M. , Cui, X. , Wang, Q. , Yang, H. , Cheng, H. et al (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. [DOI] [PubMed] [Google Scholar]
- Feng, G. , Qin, Z. , Yan, J. , Zhang, X. and Hu, Y. (2011) Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol. 191, 635–646. [DOI] [PubMed] [Google Scholar]
- Freeling, M. , Lyons, E. , Pedersen, B. , Alam, M. , Ming, R. and Lisch, D. (2008) Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18, 1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, J. , Schussler, J. , Löffler, C. , Cai, W. , Paszkiewicz, S. , Messina, C. , Groeteke, J. et al (2015) Industry scale evaluation of maize hybrids selected for increased yield in drought stress conditions of the U.S. Corn Belt. Crop Sci. 55, 1608–1618. [Google Scholar]
- Gao, H. , Smith, J. , Yang, M. , Jones, S. , Djukanovic, V. , Nicholson, M.G. , West, A. et al (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187. [DOI] [PubMed] [Google Scholar]
- Gilmour, A.R. , Gogel, B.J. , Cullis, B.R. and Thompson, R. (2009) ASReml User Guide. Release 3.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK: [Google Scholar]
- Gordon‐Kamm, W. , Dilkes, B.P. , Lowe, K. , Hoerster, G. , Sun, X. , Ross, M. , Church, L. et al (2002) Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc. Natl Acad. Sci. USA, 99, 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Rupe, M.A. , Wei, J. , Winkler, C. , Goncalves‐Butruille, M. , Weers, B.P. , Cerwick, S.F. et al (2014) Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J. Exp. Bot. 65, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben, J.E. , Bao, X. , Bate, N.J. , DeBruin, J.L. , Dolan, D. , Hasegawa, D. , Helentjaris, T.G. et al (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought‐stress conditions. Plant Biotechnol. J. 12, 685–693. [DOI] [PubMed] [Google Scholar]
- Hays, D.B. , Do, J.H. , Mason, R.E. , Morgan, G. and Finlayson, S.A. (2007) Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci. 172, 1113–1123. [Google Scholar]
- Hu, Y. , Xie, Q. and Chua, N.H. (2003) The Arabidopsis auxin‐inducible gene ARGOS controls lateral organ size. Plant Cell, 15, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T.B. , LaFayette, P.R. , Schmitz, R.J. and Parrott, W.A. (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA‐mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, E.M. , Oosterhuis, D.M. and Snider, J.L. (2010) Physiological effects of 1‐methylcyclopropene on well‐watered and water‐stressed cotton plants. J. Plant Growth Regul. 29, 280–288. [Google Scholar]
- Kawakami, E.M. , Oosterhuis, D.M. and Snider, J. (2013) High temperature and the ethylene antagonist 1‐methylcyclopropene alter ethylene evolution patterns, antioxidant responses, and boll growth in Gossypium hirsutum. Am. J. Plant Sci. 4, 1400–1408. [Google Scholar]
- Kuluev, B.R. , Knyazev, A.V. , Iljassowa, A.A. and Chemeris, A.V. (2011) Constitutive expression of the ARGOS gene driven by dahlia mosaic virus promoter in tobacco plants. Rus. J. Plant Physiol. 58, 507–515. [Google Scholar]
- Lai, J. , Ma, J. , Swigonova, Z. , Ramakrishna, W. , Linton, E. , Llaca, V. , Tanyolac, B. et al (2004) Gene loss and movement in the maize genome. Genome Res. 14, 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibman, M. , Shryock, J.J. , Clements, M.J. , Hall, M.A. , Loida, P.J. , McClerren, A.L. , McKiness, Z.P. et al (2014) Comparative analysis of maize (Zea mays) crop performance: natural variation, incremental improvements and economic impacts. Plant Biotechnol. J. 12, 941–950. [DOI] [PubMed] [Google Scholar]
- Leister, D. , Kurth, J. , Laurie, D.A. , Yano, M. , Sasaki, T. , Devos, K. , Graner, A. et al (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl Acad. Sci. USA, 95, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al (2013) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Liu, Z.B. , Xing, A. , Moon, B.P. , Koellhoffer, J.P. , Huang, L. , Ward, R.T. et al (2015) Cas9‐guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhang, K. , Chen, K. and Gao, C. (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 41, 63–68. [DOI] [PubMed] [Google Scholar]
- Loffler, C.M. , Wei, J. , Fast, T. , Gogerty, J. , Langton, S. , Bergman, M. , Merrill, B. et al (2005) Classification of maize environments using crop simulation and geographic information systems. Crop Sci. 45, 1708–1716. [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendivil Ramos, O. and Ferrier, D.E. (2012) Mechanisms of gene duplication and translocation and progress towards understanding their relative contributions to animal genome evolution. Int. J. Evol. Biol. 2012, 846421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio, M.L. , Wu, J. , Mowers, R. , Zhou, H.P. , Meghji, M. , Primavesi, L.F. , Paul, M.J. et al (2015) Expression of trehalose‐6‐phosphate phosphatase in maize ears improves yield in well‐watered and drought conditions. Nat. Biotechnol. 33, 862–869. [DOI] [PubMed] [Google Scholar]
- de Pater, B.S. , van der Mark, F. , Rueb, S. , Katagiri, F. , Chua, N.H. , Schilperoort, R.A. and Hensgens, L.S. (1992) The promoter of the rice gene GOS2 is active in various different monocot tissues and bind rice nuclear factor ASF‐1. Plant J. 2, 837–844. [DOI] [PubMed] [Google Scholar]
- Rai, M.I. , Wang, X. , Thibault, D.M. , Kim, H.J. , Bombyk, M.M. , Binder, B.M. , Shakeel, S.N. et al (2015) The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 15, 157. doi:10.1186/s12870‐015‐0554‐x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E. , Kurth, J. and Leister, D. (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19, 76–84. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schnable, P.S. , Ware, D. , Fulton, R.S. , Stein, J.C. , Wei, F. , Pasternak, S. , Liang, C. et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science, 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Scott, L. , LaFoe, D. and Weil, C.F. (1996) Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics, 142, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Zhang, Y. , Chen, K. , Zhang, K. and Gao, C. (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Habben, J.E. , Archibald, R.L. , Drummond, B.J. , Chamberlin, M.A. , Williams, R.W. , Lafitte, H.R. et al (2015) Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 169, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Drummond, B.J. , Wang, H. , Archibald, R.L. and Habben, J.E. (2016) Maize and Arabidopsis ARGOS proteins interact with ethylene receptor signaling complex, supporting a regulatory role for ARGOS in ethylene signal transduction. Plant Physiol., 171, 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, V.K. , Doyon, Y. , Miller, J.C. , DeKelver, R.C. , Moehle, E.A. , Worden, S.E. , Mitchell, J.C. et al (2009) Precise genome modification in the crop species Zea mays using zinc‐finger nucleases. Nature, 459, 437–441. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Zhang, X. , Wu, C. , He, Y. , Ma, Y. , Hou, H. , Guo, X. et al (2016) Engineering herbicide‐resistant rice plants through CRISPR/Cas9‐mediated homologous recombination of acetolactate synthase. Mol. Plant, 9, 628–631. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene editing and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Yu, C. , Zuo, T. , Zhang, J. , Weber, D.F. and Peterson, T. (2015) Alternative transposition generates new chimeric genes and segmental duplications at the maize p1 locus. Genetics, 201, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Chopra, S. and Peterson, T. (2000) A segmental gene duplication generated differentially expressed myb‐homologous genes in maize. Plant Cell, 12, 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Lu, H.H. , Chung, W.Y. , Yang, J. and Li, W.H. (2005) Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 22, 135–141. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. , et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Liu, B. , Weeks, D.P. , Spalding, M.H. and Yang, B. (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Peng, Z. , Long, J. , Sosso, D. , Liu, B. , Eom, J.‐S. , Huang, S. et al (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Maize ARGOS8 gene expression. The transcript abundance of ARGOS8 in various tissues of maize inbred PH184C was measured by RNA sequencing. Samples were taken from the plants at the developmental stage of V10, VT/R1 and R4. TPTM, transcript per ten million.
Figure S2 Comparison of ARGOS8 gene expression among 419 maize inbred lines. The ARGOS8 transcript levels in leaves of 3‐week‐old seedlings were measured by RNA sequencing. TPTM, transcript per ten million.
Figure S3 Single guide RNA genes used in editing the genomic sequence of ARGOS8. (a) Scheme illustrating the structure of a single guide RNA (sgRNA) gene. (b) DNA sequences of the transcription region in the sgRNA genes CRISPR RNA1 (CR1), CR2 and CR3. U6 PRO, maize U6 polymerase III promoter; crRNA, CRISPR RNA (in red font) functioning as a guide; tracrRNA, transactivating CRISPR RNA, the sgRNA scaffold.
Figure S4 DNA sequences of the junction PCR products amplified from ARGOS8 variants.
Table S1 Frequency of the GOS2 promoter insertion with one target site and the promoter swap with two target sites in T0 maize plants.
Table S2 Primers and probes used for generation and characterization of ARGOS8 genome editing variants.
Table S3 Plant height (PLTHT), ear height (EARHT) and grain moisture (MST) of ARGOS8 genome‐edited variants and wild type under flowering stress, grain‐filling stress and optimal (well‐watered) conditions.


