Summary
Cation/proton antiporter 1 (CPA1) genes encode cellular Na+/H+ exchanger proteins, which act to adjust ionic balance. Overexpression of CPA1s can improve plant performance under salt stress. However, the diversified roles of the CPA1 family and the various parameters used in evaluating transgenic plants over‐expressing CPA1s make it challenging to assess the complex functions of CPA1s and their physiological mechanisms in salt tolerance. Using meta‐analysis, we determined how overexpression of CPA1s has influenced several plant characteristics involved in response and resilience to NaCl stress. We also evaluated experimental variables that favour or reduce CPA1 effects in transgenic plants. Viewed across studies, overexpression of CPA1s has increased the magnitude of 10 of the 19 plant characteristics examined, by 25% or more. Among the ten moderating variables, several had substantial impacts on the extent of CPA1 influence: type of culture media, donor and recipient type and genus, and gene family. Genes from monocotyledonous plants stimulated root K+, root K+/Na+, total chlorophyll, total dry weight and root length much more than genes from dicotyledonous species. Genes transformed to or from Arabidopsis have led to smaller CPA1‐induced increases in plant characteristics than genes transferred to or from other genera. Heterogeneous expression of CPA1s led to greater increases in leaf chlorophyll and root length than homologous expression. These findings should help guide future investigations into the function of CPA1s in plant salt tolerance and the use of genetic engineering for breeding of resistance.
Keywords: antiporter, CPA1 gene family, meta‐analysis, overexpression, salt tolerance, transgenic
Introduction
Soil salinization is one of the chief abiotic stresses that constrain agriculture worldwide (Cramer et al., 2011), and at least 50% of global agricultural lands are at risk of salinization (Wang et al., 2003). Salinity impacts plants by causing osmotic stress, imbalance of ions, ion toxicity and excessive reactive oxygen species (Ruíz‐Lozano et al., 2012). Understanding the molecular mechanisms of salt stress responses and the functions of genes that regulate responses to salinity will help in designing strategies for development of salt‐resistant crop plants.
The most extensively studied gene category in relation to NaCl stress physiology is the family of cation/proton antiporter 1 (CPA1, Brett et al., 2005). The CPA1 gene family belongs to the CPA superfamily and includes the NHX type and Nhap type (Apse et al., 1999; Rodríguez et al., 2009). CPA1 genes encode Na+/H+ exchanger proteins, which modulate ion balance in plant cells (Feki et al., 2014; Fukuda et al., 2004). Many CPA1 genes have been isolated from various plants and several have been overexpressed in model and in crop plants, frequently leading to increased salinity tolerance (51 papers listed in Appendix S1).
Plants minimize the harmful ionic effects of Na+ stress by exclusion of Na+ from leaf tissues and by compartmentalization of Na+, mainly in vacuoles (Blumwald, 2000; Chanroj et al., 2012). CPA1 genes can alter ionic adjustment and other regulatory processes of transgenic plants under salt stress (Chinnusamy et al., 2005; Jiang et al., 2010). However, transgenic plants have varied markedly in degree of salt tolerance and physiological change. It has been difficult to elucidate the exact mechanisms involved in CPA1 transformation as overexpression of CPA1s affects many plant characteristics during normal growth as well as under salinity stress.
Phenotypes of overexpressed plants have not been assessed over consistent experimental conditions. Concentrations of NaCl and exposure time have varied greatly (Appendix S1). Gene donors and recipients have been both monocotyledonous and dicotyledonous, representing both herbaceous and woody lifestyles. Most genes used in transformation have been AtNHX7, AtNHX1 or their orthologs in other species, but the identity of the gene within the multigenic CPA1 family is another variable that has differed among studies.
Meta‐analysis refers to the statistical synthesis of results from multiple studies (Borenstein et al., 2009). The analysis allows us to characterize the consistency of a treatment effect and to estimate its magnitude much more precisely than from single studies alone. It also enables us to quantify the influence of various experimental factors on the treatment effect, factors that have been investigated across many studies and whose nuances go far beyond the capability of any one experiment. We performed a meta‐analysis on overexpression of CPA1 genes to summarize their effects on plant responses to NaCl stress. Additionally, we examined several experimental factors (often termed moderator variables, or simply, moderators) that may have influenced the extent of the CPA1 effect on plant response. We sought to answer the following questions: (i) What is the overall impact of CPA1 transformation across studies on plant response to NaCl stress? (ii) Has the CPA1 influence been more pronounced in NaCl‐stressed plants than in unstressed plants? (iii) How have particular experimental variables affected the size of CPA1 influence? The analysis points to several potential research areas which should help scholars better understand the roles of the CPA1 gene family in enhancing salt tolerance.
Results
Overall summary effects
Summary effect sizes represent the average ratio of transformed (TC) and nontransformed (NC) plants over all studies. Gene donor plants were represented by 30 species in 24 genera across the 284 studies (Appendix S1). The most highly studied CPA1 genes were from Arabidopsis (109 studies). Among gene donor plant types, the most information was available for dicotyledonous plants (228 studies). Eighteen species of recipient plants were studied. The data set included eight CPA1 gene family members with NHX1 most examined (181 studies), followed by SOS1 (52 studies). Single gene transformation was examined in 253 studies; the other 31 studies combined two or three genes, where one gene belonged to the CPA1 family.
Natural logs (ln R) of summary effect sizes are depicted in the plots (Figures 1, 2, 3, 4, 5, 6, 7, 8 and S1–S3). As the ratio of transformed/nontransformed treatment means (termed ‘response ratio’), the summary effect size reflects the relative magnitude of the treatment effect and its confidence intervals (CIs) indicate its precision. ‘Forest plot’ is the term used to refer to the graphical depiction of these unit‐less ratios and their CIs. When displayed as logs, positive values of the summary effect indicate promotive treatment effects and negative values inhibitive treatment effects. Raw percentage changes induced by CPA1 transformation are listed in Table 1.
Figure 1.
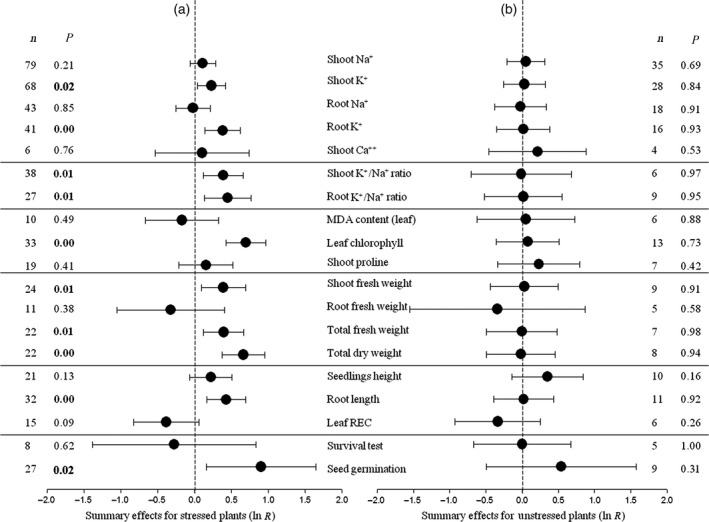
Weighted summary effect sizes (natural log of the transformed/control plant response ratios, ln R). Ratios are unit‐less. Horizontal bars associated with summary effects (closed circles) are 95% confidence intervals (CIs). Summary effects were analysed in plants exposed to NaCl stress (a) and unstressed plants (b). n is the number of studies contributing to each summary effect. P ≤ 0.05 indicates that the summary effect was significantly different than zero (same for Figures 2, 3, 4, 5, 6, 7, 8).
Figure 2.
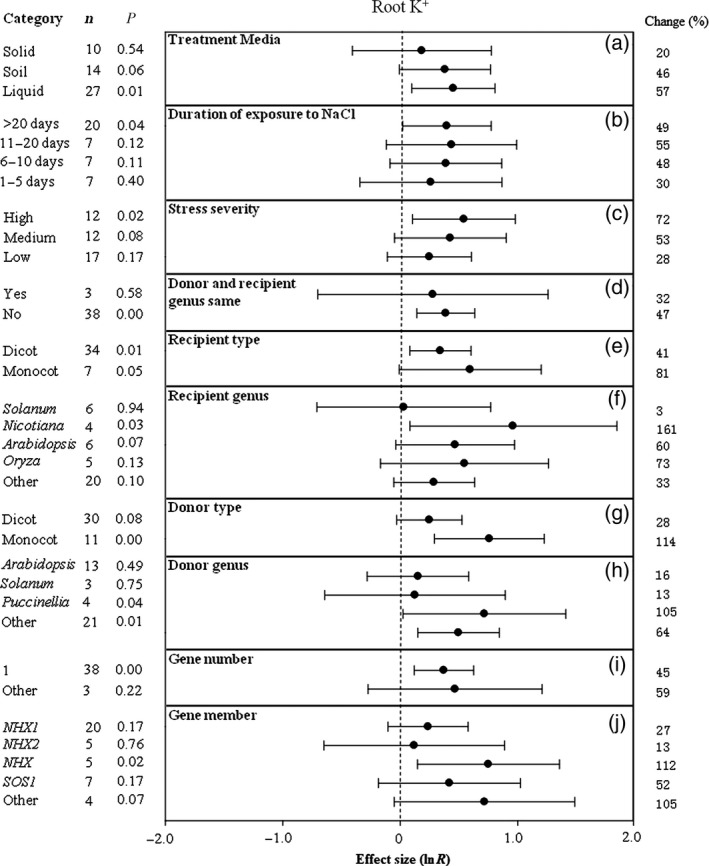
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on concentration of potassium ions in roots (root K+). Summary effects were analysed in plants exposed to NaCl stress, with the impacts of ten moderator variables on the magnitude of the treatment effect portrayed (a–j). Category lists levels (categories or subgroups) of each moderator. Change (to the right of plots) refers to raw percentage increase in root K+ induced by overexpression of CPA1.
Figure 3.
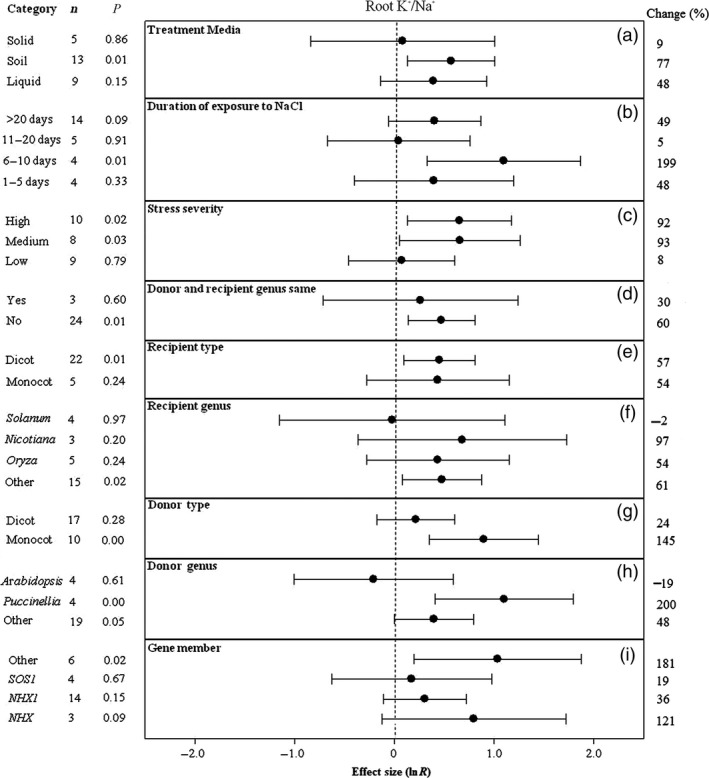
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on the ratio of K+/Na+ concentrations in roots of plants exposed to NaCl. The impacts of nine moderator variables on the magnitude of the treatment effect are portrayed (a–i; gene number not analysed due to insufficient studies). Category lists levels of each moderator. Change refers to raw percentage increase in root K+/Na+ induced by overexpression of CPA1.
Figure 4.
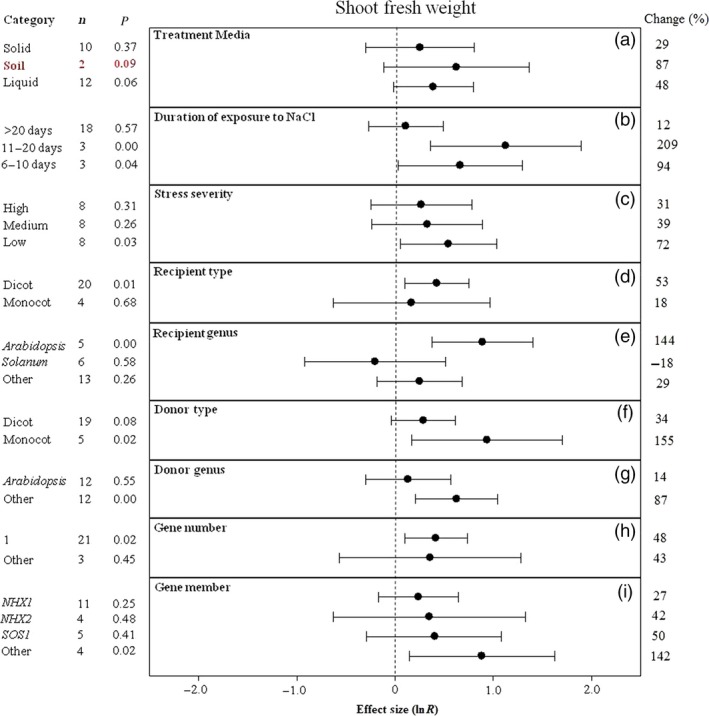
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot fresh weight in plants exposed to NaCl. The impacts of nine moderator variables on the magnitude of the treatment effect are portrayed (a–i; ‘same donor and recipient genus’ not analysed due to insufficient studies). Category lists levels of each moderator. Change refers to raw percentage increase in shoot fresh weight induced by overexpression of CPA1.
Figure 5.
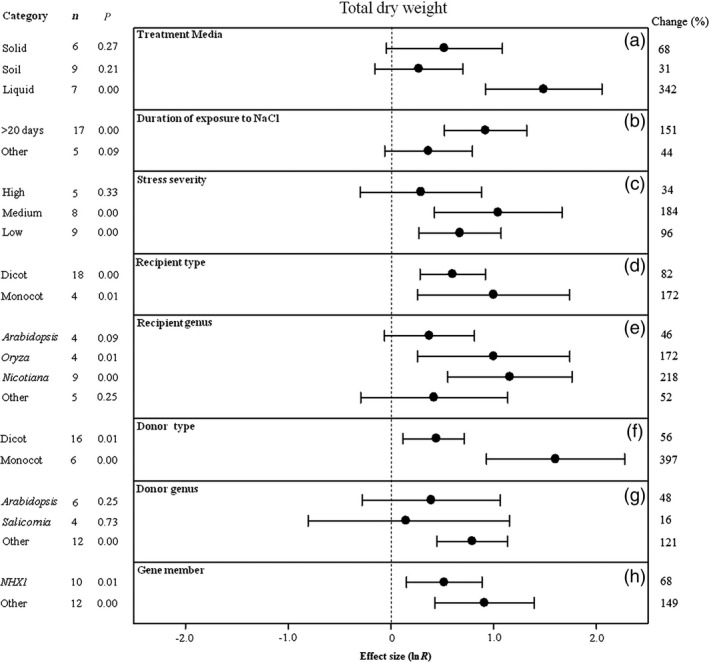
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on total dry weight in plants exposed to NaCl. The impacts of eight moderator variables on the magnitude of the treatment effect are portrayed (a–h; ‘same donor and recipient genus’ and gene number not analysed due to insufficient studies). Category lists levels of each moderator. Change refers to raw percentage increase in total fresh weight induced by overexpression of CPA1.
Figure 6.
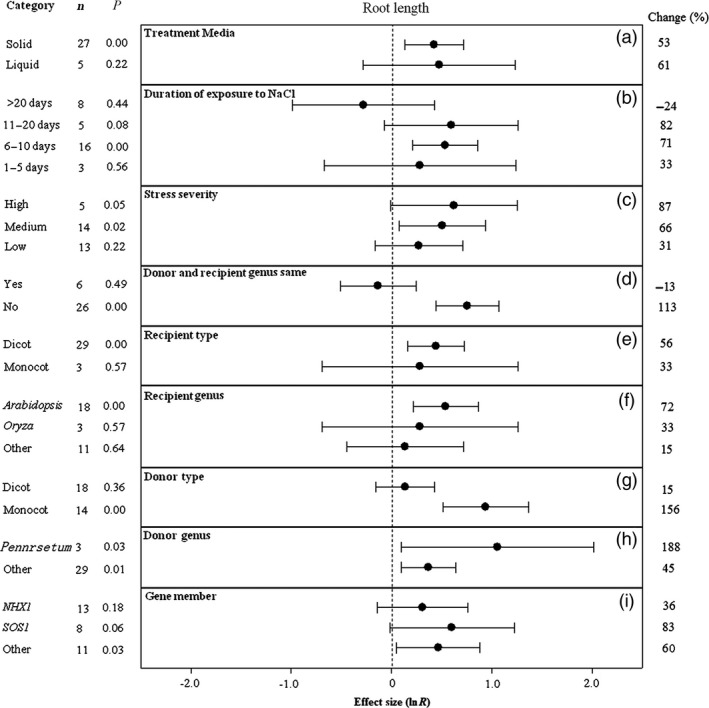
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on root length in plants exposed to NaCl. The impacts of nine moderator variables on the magnitude of the treatment effect are portrayed (a–i; gene number not analysed due to insufficient studies). Category lists levels of each moderator. Change refers to raw percentage increase in root length induced by overexpression of CPA1.
Figure 7.
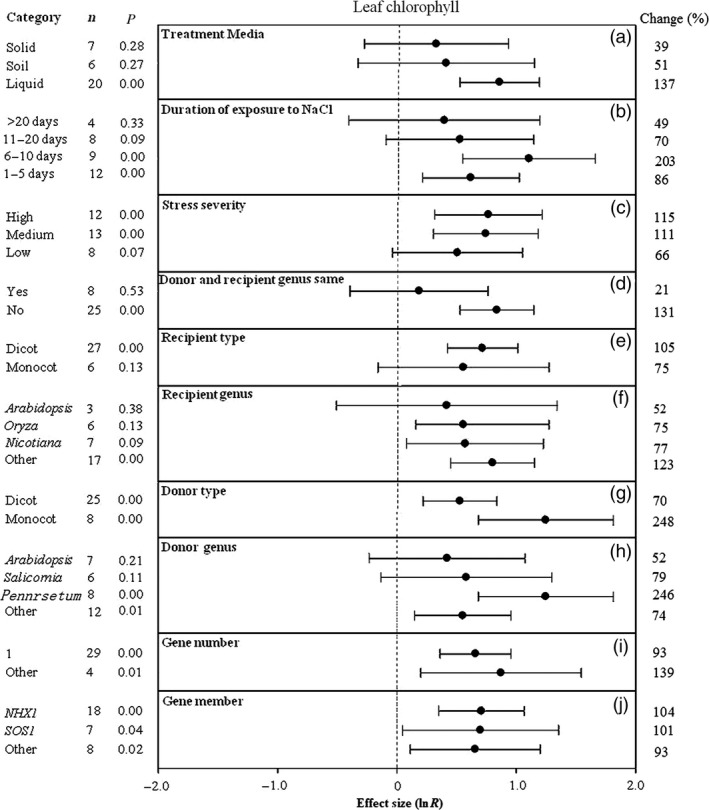
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on leaf chlorophyll content in plants exposed to NaCl. The impacts of ten moderator variables on the magnitude of the treatment effect are portrayed (a–j). Category lists levels of each moderator. Change refers to raw percentage increase in leaf chlorophyll induced by overexpression of CPA1.
Figure 8.
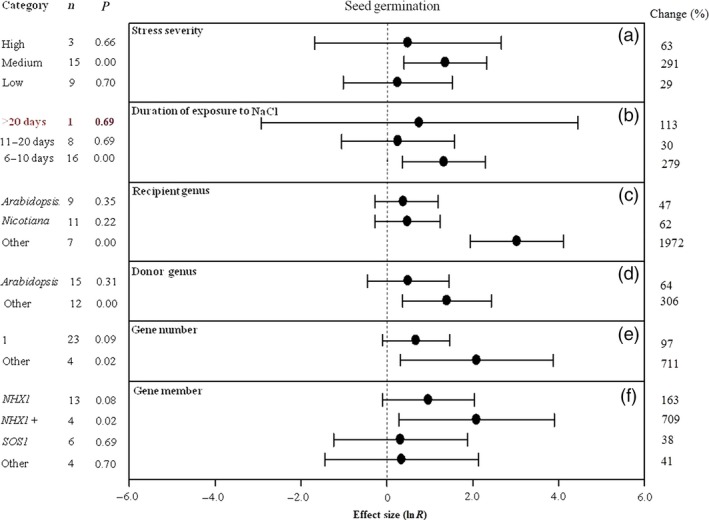
Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on seed germination in plants exposed to NaCl. The impacts of six moderator variables on the magnitude of the treatment effect are portrayed (a–f; some moderators not analysed due to insufficient studies). Category lists levels of each moderator. Change refers to raw percentage increase in seed germination induced by overexpression of CPA1.
Table 1.
Heterogeneity statistics for the 19 summary effect sizes under NaCl stress: Qt, total observed variation among studies; P hetero, probability that Qt was due entirely to sampling error and not to real variation among studies; changes to summary effects caused by foreign CPA1 genes, transformed from ln R to raw percentages. Analysis was conducted on log‐transformed values (ln R) from each study, and raw values were provided below as percentage changes
| Summary effect size | Qt | P hetero | TC‐induced change (%) |
|---|---|---|---|
| Shoot Na+ | 34.169 | 0.0 | 12 |
| Shoot K+ | 18.685 | 0.0 | 26 |
| Root Na+ | 16.556 | 0.0 | −2 |
| Root K+ | 11.686 | 0.0 | 46 |
| Shoot Ca2+ | 0.546 | 0.0 | 11 |
| Shoot K+/Na+ ratio | 7.937 | 0.0 | 47 |
| Root K+/Na+ ratio | 13.300 | 0.0 | 57 |
| MDA (leaf) | 1.985 | 0.0 | −16 |
| Leaf chlorophyll | 15.559 | 0.0 | 100 |
| Shoot proline | 8.720 | 0.0 | 17 |
| Shoot fresh weight | 10.573 | 0.0 | 48 |
| Root fresh weight | 7.917 | 0.0 | −28 |
| Total fresh weight | 11.256 | 0.0 | 48 |
| Total dry weight | 18.627 | 0.0 | 94 |
| Seedlings height | 3.720 | 0.0 | 25 |
| Root length | 30.680 | 0.0 | 53 |
| Leaf REC | 1.934 | 0.0 | −32 |
| Survival test | 6.833 | 0.0 | −24 |
| Seed germination | 16.585 | 0.0 | 146 |
For TC‐induced changes (changes induced by CPA1 transformation), bold text signifies change was significant (P ≤ 0.05), with positive values indicating TC‐induced promotion and negative values TC‐induced inhibition. The magnitude and precision of each effect size listed below is illustrated in Figure 1.
In meta‐analysis, greater emphasis should be placed on the magnitude and precision of the summary effects than on tests of their statistical significance and P values (Cooper, 2009; Cumming, 2012). Scientific significance of the size of the treatment is of greater importance than its statistical significance, owing often to lack of adequate statistical power. However, many meta‐analysts in plant biology and ecology rely on P values in interpreting their analyses. We have endeavoured to combine these perspectives in summarizing our findings.
CPA1 transformation had statistically significant impacts for 10 of the 19 plant characters examined under salt stress (Figure 1a: P ≤ 0.05, CIs did not cross‐zero), increasing values by 25% or more (Table 1). Seed germination has been most affected by transformation, about 2.5x higher in TC than in NC plants. Chlorophyll content and total dry weight were doubled by transformation. Root K+/Na+ ratio and root length were also markedly increased in TC plants, by over 50%. Transformation had negligible impact on root Na+ concentration and leaf MDA content. Root fresh weight, plant survival and leaf REC were each substantively reduced in TC plants relative to their NC counterparts, by 24%–31%; small numbers of studies led to relatively large CIs and statistical insignificance for these summary effects. CPA1 transformation has had no statistically significant impacts (P ≥ 0.05) on these plant characteristics in the absence of NaCl (Figure 1b), likely due to imprecision (low numbers of studies) in some instances (seed germination).
Moderator variables
Heterogeneity refers to true differences among studies in treatment outcomes (as opposed to variation due to sampling error within studies). Moderator analysis is performed to understand heterogeneity: to determine which experimental variables (moderating variables, or ‘moderators’) have had particularly large impacts on the treatment outcomes and which have had trivial or no effects. We conducted moderator analysis of the ten summary effects that were significantly affected by CPA1 transformation (Table 2, Appendix S1). Figures 2, 3, 4, 5, 6, 7, 8 depict the influence of the moderator levels (categories) on those summary effects that have shown the most sensitivity to the moderators. None of the summary effects exhibited statistically significant heterogeneity (P hetero > 0.10, Table 1); a P hetero value for the Q‐test of <0.1 has been used to indicate significant heterogeneity in the summary effect sizes (Iacovelli et al., 2014). This led to values of zero in I 2 for each summary effect. I 2 is a statistic that indicates the percentage of variation due to real differences in outcomes among studies. Here, too, we note the cautions of meta analyst (Borenstein et al., 2009), who point out that while a significant heterogeneity P value or positive I 2 provides evidence that subgroups differed among trials (that there were real differences among studies), the converse does not hold. A P value above 0.1 does not provide evidence that moderator level studies are consistent among studies. Again, lack of significance may frequently be due to low statistical power. Even substantial real dispersion of true effects might yield P hetero > 0.1 with a small number of studies or large within‐study variance, often the case in plant biology meta‐analyses.
Table 2.
Heterogeneity P values (P hetero): probability that moderators (experimental variables) accounted for real differences in treatment effects among studies
| Moderator | Effect size | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Shoot K+ | Root K+/Na+ | Root K+ | Shoot K+/Na+ | Leaf chlorophyll | Shoot fresh weight | Total fresh weight | Total dry weight | Root length | Seed germination | |
| Treatment media | 0.59 | 0.62 | 0.75 | 0.61 | 0.24 | 0.73 | 0.25 | <0.01 | 0.90 | – |
| Duration of exposure to NaCl | 0.71 | 0.26 | 0.98 | 0.83 | 0.38 | 0.04 | 0.39 | 0.06 | 0.21 | 0.61 |
| Stress severity | 0.37 | 0.23 | 0.58 | 0.83 | 0.74 | 0.72 | 0.27 | 0.23 | 0.61 | 0.36 |
| Recipient type | 0.78 | 0.97 | 0.45 | 0.37 | 0.69 | 0.56 | – | 0.33 | 0.76 | – |
| Recipient genus | 0.85 | 0.83 | 0.54 | 0.64 | 0.81 | 0.04 | 0.30 | 0.14 | 0.47 | <0.001 |
| Donor type | 0.20 | 0.05 | 0.07 | 0.10 | 0.03 | 0.13 | – | <0.01 | <0.01 | – |
| Donor genus | 0.76 | 0.05 | 0.42 | 0.15 | 0.18 | 0.18 | 0.26 | 0.35 | 0.17 | 0.21 |
| Donor and recipient genus same | 0.88 | 0.69 | 0.83 | 0.78 | 0.05 | – | – | – | <0.001 | – |
| Gene number | 0.91 | – | 0.81 | – | 0.57 | 0.95 | – | – | – | 0.16 |
| Gene member | 0.50 | 0.34 | 0.51 | 0.79 | 0.99 | 0.52 | 0.99 | 0.21 | 0.74 | – |
P hetero ≤ 0.1 signifies that observed variation is not due solely to sampling error (expected variation). Dashed entries indicate that insufficient data were available to perform an analysis. Each moderator was examined in relation to the ten summary effect sizes (ratios of treatment/control plants) significantly impacted by CPA1 overexpression. Appendix S1 provides associated Qm values (between‐study variation); n (sample size); df (degrees of freedom, levels within a moderator).
Cations
The influence of several moderators on the extent to which CPA1 transformation affected root K+ in salt‐stressed plants is illustrated in Figure 2. Greater root K+ has been observed when experiments used liquid media than when plants were grown on soil or solid media (Figure 2a). It has generally taken a few days for root K+ to increase and then plateau after exposure to NaCl stress; the average TC‐induced increase over 1–5 days is 30% with the increase levelling off at 50–55% (Figure 2b). Greater stress severity has been associated with higher TC‐induced root K+, with low, moderate and severe stress giving average increases of 28%, 53% and 72%, respectively (Figure 2c). Monocotyledonous plants have been much more sensitive to the process than dicots; root K+ was twice as high in recipient monocot than dicot plants and 4x as high when gene donors were monocots than when they were dicots (Figure 2e, g). Taxonomy has also mattered; recipient and donor genera have had marked effects on the outcome of transformation. Root K+ has shown the greatest response in transformed Nicotiana, and response has been largest when genes came from Puccinellia (Figure 2f, h). NHX has had several‐fold greater impact than NHX1 or NHX2 and about twice the impact of SOS1 (Figure 2j). Study numbers are low for most of these comparisons and so summary effects are imprecise (large CIs) and in most cases not statistically significant.
Unlike root K+, K+/Na+ in roots of transformed plants has been higher when experiments were conducted in soil than on solid or liquid laboratory media (Figure 3a). Also unlike root K+, root K+/Na+ has peaked dramatically in TC plants after 6–10 days of stress then subsided (Figure 3b). The impact of CPA1 transformation on root K+/Na+ has been over 10× greater at medium and high stress than at low stress (Figure 3c). When gene donors and recipients have been of different genera, the impact of transformation has been about twice as large as when donor and recipient were of the same genus (Figure 3d). Transformed monocot and dicot recipient plants have had similar root K+/Na+, but this cation ratio has been increased substantially when donors were monocots (Figure 3e, g). As for root K+, Nicotiana and Puccinella have also been the most responsive genera to transformation for root K+/Na+ increases (Figure 2f, h) and NXH the most influential CPA1 gene member (Figure 3i).
Size characteristics
The effect of transformation on increasing shoot fresh weight and total dry weight of salt‐stressed plants has not been consistent across treatment media, with shoot fresh weight higher in soils (only two studies) and total dry weight considerably higher in liquid media (Figures 4a, 5a). Shoot fresh weight has been at least twice as large at moderate stress durations (11–20 days) than at shorter or longer durations (Figure 4b). Averaged across studies, both shoot dry weight and total dry weight have been almost 3× larger at low stress severity than at the most severe stress, with variable response at medium severity (Figures 4c, 5c). Monocot recipients exhibited lower shoot fresh weight but higher total dry weight with CPA1 overexpression (Figures 4d, 5d). Biomass of salt‐stressed plants has been much more affected by transformation when donors were monocots (Figures 4f, 5f). The TC‐induced growth increases differed among recipient and donor genera but inconsistently (Figures 4e, g, 5e, g). Transformation effects on biomass have been mostly similar among the NHX and SOS1 genes (Figures 4i, 5h). Across moderators, study numbers were small for these biomass effect sizes, resulting in large confidence intervals and low precision. There was much overlap among levels of most moderators; more studies are needed to resolve moderator influences.
Media type has not affected the magnitude of the TC‐induced increase in root length (Figure 6a). Intermediate exposure times have caused transformation to increase root length much more than the shortest or longest durations (Figure 6b). Also as for most effect sizes portrayed in earlier figures, greater stress severity has increased the TC‐induced impact, with the increases associated with high severity averaging about 3x greater than those for low severity (Figure 6c). CPA1 transformation has more than doubled root length when gene donor and recipient plants subjected to NaCl stress have been of different genera (Figure 6d). When donors and recipients were of the same genus, transformation has had a slightly negative effect. Recipient dicots and monocots have not differed in TC‐induced impact on root length (Figure 6e), while monocots have clearly had much more impact than dicots as gene donors (Figure 6g). Root length of recipient Arabidopsis appears to have been somewhat more responsive than for other genera (Figure 6f). Pennisetum appears to be a promising CPA1 donor genus, imparting a dramatic TC‐induced 188% increase in root length to recipient plants (Figure 6h). Among CPA1 gene family members, SOS1 has tended to have the most influence on root length of salt‐stressed plants (Figure 6i).
Other characteristics
The outcomes of transformation on leaf chlorophyll content have been most evident in association with liquid media, with the TC‐induced impact averaging 39%, 51% and 137% for solid, soil and liquid media, respectively (Figure 7a). Again, the most dramatic impact has occurred at intermediate durations. Transformed plants exposed to NaCl stress for 6–10 days showed threefold (200%) increases in leaf chlorophyll, as opposed to more modest increases at longer time periods and at 1–5 days (Figure 7b). TC‐induced stimulation of leaf chlorophyll at the lowest stress severity was about half that at the two higher levels of severity (Figure 7c). As in root length and other effect sizes, transformation has had much greater effect on leaf chlorophyll when donor and recipient genera have differed relative to experiments in which donor and recipient were of the same genus (Figure 7d). Dicot and monocot recipient plants have not differed greatly in their TC‐induced chlorophyll response (Figure 7e), but monocots have tended to be much more influential as donors (Figure 7g). Again, Arabidopsis has tended to be among the least responsive species to CPA1 transformation (Figure 7f, h). Multiple gene numbers have tended to impart more salt tolerance than single gene transformation (Figure 7i). NHX1, SOS1 and other CPA1 genes have affected leaf chlorophyll similarly, doubling it relative to controls (Figure 7j).
Seed germination data have been reported less frequently than other effect sizes, allowing less moderator analysis as well as resulting in large CIs. NaCl stress of medium severity has resulted in much greater TC‐induced impact on seed germination than high and low severity (Figure 8a). Again, a much greater impact of CPA1 transformation has occurred at the moderate stress exposure (6–10 days) compared to longer duration, 11–20 days and >20 days (Figure 8b). Donor and recipient genera other than Arabidopsis have been much more affected by the CPA1 genes (Figure 8c, d); other recipient genera combined showed a phenomenal TC‐induced increase in seed germination of 21‐fold (1972%). Transfer of multiple genes has resulted in about 8× (711%) greater impact than single gene transfers (97%) (Figure 8e), with the NHX1 gene again having substantially greater effect than SOS1 and other genes (Figure 8f).
Publication bias
Publication bias is the term applied to research in the published literature that is systematically unrepresentative of all completed studies (Rothstein et al., 2005). A concern about bias stems from the possibility that significant treatment differences may be more likely to be published than nonsignificant findings. Several statistical methods developed to test for potential bias involve exploring the relationship between study effect size and precision. The idea is that studies with smaller sample sizes (and hence higher variance) will tend to have larger effect sizes than larger studies with greater precision; greater treatment differences are required for statistical significance as sample size decreases.
We found little evidence of publication bias in the parameters commonly used to test for it, for any of the summary effects. Funnel plots indicated that studies displayed the expected dispersion around the overall summary effect. Within the Begg and Mazumbar rank correlation test, all summary effects except shoot Ca2+ had absolute Kendall tau values below 0.3, indicating little concern for bias (no tendency for effect sizes to increase as study size decreases). The Duvall and Tweedie trim and fill analysis indicated no concern that publication bias has resulted in an inflated summary effect. The purpose of the fail‐safe calculation is to estimate whether publication biases (if they exist) can be safely ignored (Rosenberg, 2005). In all cases, the Orwin's failsafe N was much higher than the classic Rosenthal threshold number of 5k + 10. In other words, a very large (and very unlikely) number of missing studies would be needed to reduce the treatment effect to <4%, for any of the summary effects. The Egger's P values did suggest the possibility of bias for shoot Na+, shoot Ca2+, shoot fresh weight and total fresh weight. This one test should be viewed within the context of the several other measures that do not indicate a concern for bias. Additional details and tests are provided in Appendix S2.
Discussion
The involvement of NHX1, SOS1 and other CPA1 genes in plant response to NaCl stress has been well documented (An et al., 2007; Ma et al., 2015). Sodium toxicity disrupts ion homoeostasis and consequently cellular metabolism (Ahmad and Maathuis, 2014). Many cytosolic enzymes that are activated by K+ are inhibited by Na+ (Flowers et al., 1977), and high cytosolic K+/Na+ ratio helps sustain normal metabolism. Na+/H+ antiporters move sodium ions across membranes by exchanging H+ for Na+ (Adem et al., 2015). Genes in the CPA1 family encode Na+/H+ exchangers that act in compartmentalizing Na+ and thereby reducing its accumulation in the cytosol (Apse et al., 1999; Serrano and Rodriguez‐Navarro, 2001). Maintaining high K+ is important in sodium tolerance, and NHX genes have been found to assist K+ homoeostasis (Barragán et al., 2012). CPA1 gene members also act in control of long‐distance transport of Na+ (Shi et al., 2002), by retrieving sodium from root xylem and sequestering it in root vacuoles (Martínez‐Alcántara et al., 2015) and potentially by exporting it back to the soil solution (Nie et al., 2015). Favourable ion regulation protects cells in several ways, for example by enhancing proline accumulation, preserving membrane integrity and increasing the scavenging of reactive oxygen species (Fan et al., 2015). The value of the meta‐analysis, beyond the confirming the overall promotive influence of ectopic CPA1 overexpression on salt tolerance, gives the average magnitude of the influence across studies and enables us to see which plant attributes have been most influenced by overexpression. Perhaps more importantly, the analysis gives perspective on which experimental circumstances and plant taxa have been associated with the greatest impacts of overexpression and which with minimal impact.
The positive TC‐induced effects on phenology have occurred at all durations but have been greatest for several plant characteristics when measured with exposures of intermediate duration. The CPA1 impact is usually substantial in the first days of exposure, becomes highest at 6–20 days then subsides, often becoming negligible after about three weeks. This suggests that overexpression of CPA1 genes may provide only moderate salt tolerance because the amount of Na+ that can be moved into vacuoles has limits; ultimately, compartmentation is swamped. When Na+ accumulation reaches the limit of vacuolar capacity, likely the case under severe salinity or long‐term exposure to moderate salinity, the impact of CPA1 overexpression subsides. This may explain why no highly salt‐resistant crop varieties have been developed by overexpression of CPA1 genes. Still, there are many instances in which modest boosts in resilience may prove very worthwhile. For example, achieving maximum protection at 2–3 weeks may have particular advantage for transplants, which are especially vulnerable during the first weeks after transplantation to the field or into containers.
Adem et al. (2015) suggested that several genes need to be pyramided for antiporter transformation to achieve a meaningful effect. The meta‐analytic perspective across the literature provides some support for this idea, in that the TC‐induced improvement was largest for some plant characteristics when more than one gene was transferred to recipient plants relative to transformation with a single gene. Low study numbers for these comparisons resulted in imprecise summaries (large CIs), but three of the four characteristics examined root K+, leaf chlorophyll and seed germination clearly displayed this trend. The functions of CPA1 genes for plant salt tolerance can be classified into two main categories based on the locations of proteins and the different pathways in the molecular evolution of stress tolerance (Bassil and Blumwald, 2014; Pires et al., 2013). Therefore, it seems reasonable that transferring two types of genes into one recipient plant may potentially have an additive effect in reducing Na+ toxicity. This should be a fruitful area for additional research.
Why has CPA1 transformation failed to improve stress tolerance in some studies? Adem et al. (2015) pointed to several possibilities for why the phenology or behaviour of transformed plants may not differ from controls: low activity of vacuolar enzymes resulting in an insufficient proton gradient; inability of transgenic plants to prevent a passive leak of sodium from vacuoles back into cytosol; insufficient ATP to support proton pumping; and improper folding or incorrect targeting of antiporter proteins. It is possible that one or more of these processes have been associated with those moderator subgroups that have shown relatively low TC‐induced impact. This is another avenue that warrants future research.
Other promising investigation involves plant taxa. The identity of the donor plant has influenced the efficacy of CPA1 transformation on each effect size. The Na+/H+ antiporter gene in A. thaliana was the first NHX homolog to be cloned (Gaxiola et al., 1999), and much subsequent overexpression work has used Arabidopsis as a model plant, providing pioneering breakthroughs (Khan et al., 2015). Yet the meta‐analysis shows that Arabidopsis is among the least effective donor genera and only slightly more efficacious when it has been the transgenic recipient. The inconstancy of impact of Arabidopsis in CPA1 overexpression studies raises the question of whether it is the best model species for evaluating CPA1 gene function in other species. The reliability of endogenous versus ectopic overexpression also warrants more investigation.
The association of Arabidopsis with lower effectiveness of CPA1 transformation may at least partially relate to plant type. Arabidopsis, a dicotyledonous plant, is the most frequently studied donor genus. For each of the six plant characteristics for which plant types were compared, transformation had a markedly greater effect when the donor plant was monocotyledonous than when it was dicotyledonous. That this was not the case for recipient plants is likely fortunate for agriculture. Both dicot and monocot recipient plants have benefited by transformation, maintaining an open field for improvement of salt tolerance in both crop types.
The meta‐analysis indicates that the transgenic effect on K+ has generally been more pronounced than on Na+; CPA1 overexpression has increased shoot and root K+ concentrations while maintaining shoot and root Na+ concentrations. The TC‐induced increase in shoot and root K+/Na+ ratio, a well‐known finding (Garciadeblás et al., 2007; Qi and Spalding, 2004), has been statistically significant and dramatic when viewed across the literature. Calcium has been less studied. SOS1 is the most crucial gene in the SOS pathway associated with plant salt‐induced responses, and Ca2+ has been identified as the key signal in the pathway (Ditta, 2013). However, less than ten studies on SOS1‐transformed plants included measurements of Ca2+ concentration in plant tissues. Previous physiological studies have shown that the SOS1 gene is related to Na+ exclusion, especially in the root tissue (Bassil and Blumwald, 2014; Bassil et al., 2012; Jannesar et al., 2014; ), but Ca2+ concentration in the root has rarely been measured in these CPA1‐overexpressed plants. The CPA1 gene family belongs to the CPA super‐family (Brett et al., 2005; Mäser et al., 2001); therefore, overexpression of these genes should potentially influence many ions in addition to Na+ and K+, such as Ca2+ and Mg2+ (Kurusu et al., 2013; Pandey et al., 2013). Only two of 51 studies of root behaviour (Bao et al., 2014; Yadav et al., 2012) provided root Ca2+ measurements (which did not meet our effect size inclusion criterion of a minimum of ten studies). Mg2+, another bivalent ion, plays an important role in chlorophyll function (Walker and Weinstein, 1994). The meta‐analysis confirmed that CPA1 transformation has led to dramatically higher chlorophyll concentration when plants were exposed to salinity. Few studies have reported Mg2+ concentrations, and it is unknown whether the impact on chlorophyll relates to maintained Mg2+ concentration or to other factors. The measurement of bivalent ion changes in future studies can clarify the contribution of these ions to salt tolerance in relation to overexpression of CPA1 genes.
Among transgenic strategies examined thus far, Na+/H+ antiporters offer the best mechanism for ion homoeostasis in plants subjected to NaCl stress (Khan et al., 2015; Rozema and Schat, 2013). The weighted average view across the literature confirms that using CPA1 overexpression to fortify crop plants against stress is a promising biotechnology, especially as its impact has tended to increase as salt stress severity increases. The analysis points to several areas that warrant further inquiry, in addition to those noted above. Seed germination may represent an especially profitable area for investigation, as it has been the attribute most affected by overexpression, not only in salt‐stressed plants but in unstressed plants. CPA1 overexpression has had its greatest impact during intermediate durations, roughly coinciding with germination times. Across the 27 studies which measured seed germination, all of the genes were isolated from dicotyledonous plants, and most of the recipient plants were also dicotyledonous. Given that monocots have tended to be more responsive to CPA1 overexpression than dicots, it will be beneficial to learn whether germination under salt stress is impacted even more in monocot seeds. Salt can cause several kinds of physiological stress, and quite a few parameters have been evaluated in traditional physiological studies, for example antioxidant enzymes, MDA, REC, and proline and other compatible solutes (Song et al., 2014; Wang et al., 2011). However, many of these parameters have not been examined in CPA1 overexpression studies, so it is unclear whether overexpression of a CPA1 gene would affect these parameters; this may also be an area of promising research. Another important future research area is cropping situations. As Khan et al. (2015) noted, more field studies are needed to determine the extent to which laboratory findings will translate to increased yield potential in actual cropping situations.
Materials and methods
Data collection
Data were located through a systematic search of 12 electronic databases using Endnote and ISI Web of Science (Thompson Reuters; additional search details provided in Appendix S1). The search was conducted on 3 October 2014 for articles dated through September 2014 using the search terms (‘CPA1 gene family’ or ‘NHX gene*’ or ‘SOS1 gene*’) AND (‘overexpress*’). The search returned 612 unique articles, all from refereed journals. Upon examination, 573 of the articles were excluded because they did not meet our inclusion criteria: data were unrelated to plants (81); CPA1 data were not reported (329); CPA1 genes were not overexpressed (99); they were review articles (60); either treatment or control mean was not reported (1); same data had been reported in another article (1); or the plant parameters in which we were interested were not reported (1). Thirty‐nine articles from the Web of Science search met our screening criteria. We located an additional 12 articles by examining article reference lists. In all, 51 articles were included in the meta‐analysis, written in English and Chinese and spanning 15 years (citations provided in Appendix S1). Treatment and control means with sample sizes were obtained for each study. Where sample size was not reported, we defined it conservatively as n = 1 if no mean statistics were provided (one study) and n = 2 if mean statistics were reported (12 studies). We used the final time point for studies that included data for multiple time points. Data from graphs were extracted using GetData Graph Digitizer (http://getdata-graph-digitizer.com).
Multiple treatments from one article were treated as independent studies and represented individual units in the meta‐analysis. For example, An et al.(2007) examined the behaviour of transformed and nontransformed plants subjected to four different NaCl concentrations, which resulted in four studies. Wang et al. (2007) included five different concentration NaCl (0, 150, 175, 200 and 225 mm), which resulted in five studies. Although including more than one study from an article has the disadvantage of potentially increasing the statistical dependence among studies, which for the purposes of meta‐analysis are assumed to be independent (Gurevitch and Hedges, 1999), the larger number of studies maximizes the analysis’ statistical power (Lajeunesse and Forbes, 2003). This approach has been used commonly in plant biology meta‐analyses (Klümper and Qaim, 2014; Wujeska et al., 2013), especially when studies address the moderators being evaluated (Lehmann and Rillig, 2015).
Effect sizes and moderators
We analysed several response variables commonly investigated in plant NaCl stress research (Figure 1). Each was incorporated into the analysis as the ratio of transgenic to nontransgenic (control) plant means. This response ratio is referred to as the effect size, and its natural logarithm, ln R, is used in the meta‐analysis (Borenstein et al., 2009):
where Y TC and Y NC are the means of plants that were transformed (TC) and nontransformed (NC) with a CPA1 gene (including transformed with an empty vector), respectively. Response ratios are commonly used in meta‐analyses of plant behaviours (Schaeffer et al., 2013; Worchel et al., 2013), as they provide a standardized, unit‐less expression of treatment‐induced change. The log transformation properly balances positive and negative treatment effects across response ratios (maintains symmetry in the analysis, Borenstein et al., 2009). Ln R values above 0 indicate a TC‐induced increase in the parameter, values below 0 indicate a TC‐induced decrease in the parameter, and a value of 0 signifies a lack of effect of CPA1 overexpression. An overall, summary effect size was computed for each of the 19 plant characteristics as the weighted mean of effect sizes from the primary studies. Chlorophyll concentration refers to leaves except for three studies in which shoot chlorophyll was reported. K+/Na+ ratios were computed when papers reported respective treatment means for K+ and Na+ concentration and did not directly provide the K+/Na+ ratio. Root and shoot data were analysed for those plant characteristics for which these were commonly reported for both organs. Generally, only leaf/shoot data were reported for relative electric conductivity (REC) and concentrations of malondialdehyde, proline and Ca2+.
In addition to effect size data, we collected information from each study on ten experimental variables or ‘moderators’ that may modify response to salinity stress. Moderators were of three varieties: (i) experimental conditions: treatment medium, stress severity and duration; (ii) experimental materials: type and genus of gene donor and recipient, whether donor and recipient were the same genus; and (iii) character of foreign genes: identity of the CPA1 genes, number of genes transferred. Moderators were examined to determine whether the effect of CPA1 overexpression has been more pronounced in some experimental contexts than in others. Each moderator contained at least two levels (categories), and a level of a moderator was included in the analysis if data were available for it from at least three studies from more than one article. Categorical levels not meeting these criteria were grouped into a level analysed as ‘other’ and included in the analyses if ‘other’ contained at least three studies from more than one article.
When an experiment included three or more levels of a NaCl stress treatment, we scored severity of the stress as ‘low’, ‘medium’ and ‘high’, in an effort to evaluate whether the summary effects were impacted differently by different degrees of stress. For example, if an experiment employed four levels of NaCl stress, 100, 200, 300 and 400 mm NaCl, we defined the 100 mm treatment as low, 400 mm treatment as high and 200 and 300 mm treatments as medium.
Meta‐analysis
Meta‐analyses were performed using Comprehensive Meta‐Analysis (CMA) software (Version 3.0, Biostat, Englewood, NJ; 2014). Individual studies were weighted using nonparametric variance:
where V ln R is the variance of the natural log of the response ratio R and n TC and n NC are the sample sizes of the TC and NC treatments. Standard errors or standard deviations were not reported for most studies nor was sufficient information given in many instances to estimate these from ANOVA mean separation test values. It is not uncommon for measures of dispersion to have been omitted from publications in plant biology, which makes calculating weight based solely on sample size (nonparametric variance) a necessity. Treatment summary effects were considered significant if their 95% confidence intervals did overlap zero. Presence or absence of heterogeneity (real or true variation in effects) was assessed with the Q statistic (a measure of weighted squared deviations) and estimated using I 2 (a descriptive index that estimates the ratio of true heterogeneity to total variation across the observed effect sizes) (Borenstein et al., 2009). Potential publication bias was assessed statistically with Begg and Mazumbar rank (Kendall) correlation and Egger's linear regression (Begg and Mazumdar, 1994; Sterne and Egger, 2005). It was represented graphically with funnel plots of effect sizes versus their standard errors (Borenstein et al., 2009). A fail‐safe method was used to ask whether the entire summary effect may be attributed to bias. We employed the Orwin's fail‐safe N approach (Borenstein, 2005), considered an improvement on the original Rosenthal fail‐safe N method (Rosenthal, 1979). The Duval and Tweedie iterative trim and fill method was used to demonstrate how the summary effect size would shift if apparent bias were to be removed (Duval and Tweedie, 2000). Additional details about publication bias tests are provided in Appendix S1.
Supporting information
Figure S1 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+ concentration of plants exposed to NaCl.
Figure S2 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+/Na+ ratio of plants exposed to NaCl.
Figure S3 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+/Na+ ratio of plants exposed to NaCl.
Appendix S1 Details on the CPA1‐overexpression studies used in the meta‐analyses, including each of the moderators used for categorical analyses, the transgenic and untransformed control means, sample sizes (n) and the value of the natural log (ln) of the response ratio with corresponding non‐parametric variances.
Appendix S2 Analyses of publication bias.
Table S1 Measures used in characterizing publication bias for each effect size.
Acknowledgements
This project was supported by the Priority Academic Program Development of Modern Horticulture Science in Jiangsu Province, China, and the Tennessee Agricultural Experiment Station, USA.
References
- Adem, G.D. , Roy, S.J. , Plett, D.C. , Zhou, M. , Bowman, J.P. and Shabala, S. (2015) Expressing AtNHX1 in barley (Hordium vulgare L.) does not improve plant performance under saline conditions. Plant Growth Regul. 77, 289–297. [Google Scholar]
- Ahmad, I. and Maathuis, F.J.M. (2014) Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J. Plant Physiol. 171, 708–714. [DOI] [PubMed] [Google Scholar]
- An, R. , Chen, Q.J. , Chai, M.F. , Lu, P.L. , Su, Z. , Qin, Z.X. , Chen, J. et al (2007) AtNHX8, a member of the monovalent cation: proton antiporter‐1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J., 49, 718–728. [DOI] [PubMed] [Google Scholar]
- Apse, M.P. , Aharon, G.S. , Snedden, W.A. and Blumwald, E. (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis . Science, 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Bao, A.K. , Wang, Y.W. , Xi, J.J. , Liu, C. , Zhang, J.L. and Wang, S.M. (2014) Co‐expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1‐1 enhances salt and drought tolerance in transgenic lotus corniculatus by increasing cations accumulation. Funct. Plant Biol. 41, 203–214. [DOI] [PubMed] [Google Scholar]
- Barragán, V. , Leidi, E.O. , Andrés, Z. , Rubio, L. , De Luca, A. , Fernandez, J.A. , Cubero, B. et al (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis . Plant Cell, 24, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, E. and Blumwald, E. (2014) The ins and outs of intracellular ion homeostasis: NHX‐type cation/H+ transporters. Curr. Opin. Cell Biol. 22, 1–6. [DOI] [PubMed] [Google Scholar]
- Bassil, E. , Coku, A. and Blumwald, E. (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na/H antiporters in plant growth and development. J. Exp. Bot. 63, 5727–5740. [DOI] [PubMed] [Google Scholar]
- Begg, C.B. and Mazumdar, M. (1994) Operating characteristics of a bank correlation test for publication bias. Biometrics, 50, 1088–1101. [PubMed] [Google Scholar]
- Blumwald, E. (2000) Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434. [DOI] [PubMed] [Google Scholar]
- Borenstein, M. (2005) Software for publication bias In Publication Bias in Meta‐Analysis: Prevention, Assessment and Adjustments(Rothstein H.R., Sutton A.J. and Borenstein M., eds), pp. 193–220. England: Blackwell Science. [Google Scholar]
- Borenstein, M. , Hedges, L.V. , Higgins, J.P.T. and Rothstein, H.R. (2009) Introduction to Meta‐Analysis. Manhattan: John Wiley & Sons. [Google Scholar]
- Brett, C.L. , Donowitz, M. and Rao, R. (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239. [DOI] [PubMed] [Google Scholar]
- Chanroj, S. , Wang, G. , Venema, K. , Zhang, M.W. , Delwiche, C.F. and Sze, H. (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 3, 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy, V. , Jagendorf, A. and Zhu, J.K. (2005) Understanding and improving salt tolerance in plants. Crop Sci. 45, 437–448. [Google Scholar]
- Cooper, H.M. (2009) Research Synthesis and Meta‐Analysis: A Step‐by‐Step Approach. Los Angeles: Sage. [Google Scholar]
- Cramer, G.R. , Urano, K. , Delrot, S. , Pezzotti, M. and Shinozaki, K. (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming, C. (2012) Understanding the New Statistics: Effect Sizes, Confidence Intervals, and Meta‐Analysis. New York: Routledge Taylor & Francis Group. [Google Scholar]
- Ditta, A. (2013) Salt tolerance in cereals: molecular mechanisms and applications In Molecular Stress Physiology of Plants(Rout R.G. and Das B.A., eds), pp. 133–154. India: Springer. [Google Scholar]
- Duval, S. and Tweedie, R. (2000) A nonparametric “trim and fill” method of accounting for publication bias in meta‐analysis. J. Am. Stat. Assoc. 95, 89–98. [Google Scholar]
- Fan, W. , Deng, G. , Wang, H. , Zhang, H. and Zhang, P. (2015) Elevated compartmentalization of Na+ into vacuoles improves salt and cold stress tolerance in sweet potato (Ipomoea batatas). Physiol. Plant. 154, 560–571. [DOI] [PubMed] [Google Scholar]
- Feki, K. , Quintero, F.J. , Khoudi, H. , Leidi, E.O. , Masmoudi, K. , Pardo, J.M. and Brini, F. (2014) A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis . Plant Cell Rep. 33, 277–288. [DOI] [PubMed] [Google Scholar]
- Flowers, T.J. , Troke, P.F. and Yeo, A.R. (1977) Mechanism of salt tolerance in halophytes. Ann. Rev. Plant Physiol. Plant Mol. Biol. 28, 89–121. [Google Scholar]
- Fukuda, A. , Nakamura, A. , Tagiri, A. , Tanaka, H. , Miyao, A. , Hirochika, H. and Tanaka, Y. (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 45, 146–159. [DOI] [PubMed] [Google Scholar]
- Garciadeblás, B. , Haro, R. and Benito, B. (2007) Cloning of two SOS1 transporters from the seagrass Cymodocea nodosa. SOS1 transporters from Cymodocea and Arabidopsis mediate potassium uptake in bacteria. Plant Mol. Biol. 63, 479–490. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A. , Rao, R. , Sherman, A. , Grisafi, P. , Alper, S.L. and Fink, G.R. (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. 96, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitch, J. and Hedges, L.V. (1999) Statistical issues in ecological meta‐analyses. Ecology, 80, 1142–1149. [Google Scholar]
- Iacovelli, R. , Alesini, D. , Palazzo, A. , Trenta, P. , Santoni, M. , De Marchis, L. , Cascinu, S. et al (2014) Targeted therapies and complete responses in first line treatment of metastatic renal cell carcinoma. A meta‐analysis of published trials. Cancer Treat. Rev. 40, 271–275. [DOI] [PubMed] [Google Scholar]
- Jannesar, M. , Razavi, K. and Saboora, A. (2014) Effects of salinity on expression of the salt overly sensitive genes in Aeluropus lagopoides . Aust. J. Crop Sci. 8, 1–8. [Google Scholar]
- Jiang, X. , Leidi, E.O. and Pardo, J.M. (2010) How do vacuolar NHX exchangers function in plant salt tolerance?. Plant Signal. Behav. 5, 792–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.S. , Admad, D. and Khan, M.A. (2015) Trends in genetic engineering of plants with (Na+/H+) antiporters for salt stress tolerance. Biotechnol. Biotechnol. Equip. 29, 815–825. [Google Scholar]
- Klümper, W. and Qaim, M. (2014) A meta‐analysis of the impacts of genetically modified crops. PLoS One, 9, e111629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu, T. , Kuchitsu, K. , Nakano, M. , Nakayama, Y. and Iida, H. (2013) Plant mechanosensing and Ca2+ transport. Trends Plant Sci. 18, 227–233. [DOI] [PubMed] [Google Scholar]
- Lajeunesse, M.J. and Forbes, M.R. (2003) Variable reporting and quantitative reviews: a comparison of three meta‐analytical techniques. Ecol. Lett. 6, 448–454. [Google Scholar]
- Lehmann, A. and Rillig, M.C. (2015) Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops‐a meta‐analysis. Soil Biol. Biochem. 81, 147–158. [Google Scholar]
- Ma, Y.C. , Wang, J.Y. , Zhong, Y. , Geng, F. , Cramer, G.R. and Cheng, Z.M. (2015) Subfunctionalization of cation/proton antiporter 1 genes in grapevine in response to salt stress in different organs. Hortic. Res. 2, 15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Alcántara, B. , Martínez‐Cuenca, M.R. , Quiñones, A. , Iglesias, D.J. , Primo‐Millo, E. and Forner‐Giner, M.A. (2015) Comparative expression of candidate genes involved in sodium transport and compartmentation in citrus. Environ. Exp. Bot. 111, 52–62. [Google Scholar]
- Mäser, P. , Thomine, S. , Schroeder, J.I. , Ward, J.M. , Hirschi, K. , Sze, H. , Talke, I.N. et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis . Plant Physiol. 126, 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, W. , Xu, L. and Yu, B. (2015) A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1‐1 mutant. Protoplasma, 252, 127–134. [DOI] [PubMed] [Google Scholar]
- Pandey, N. , Ranjan, A. , Pant, P. , Tripathi, R.K. , Ateek, F. , Pandey, H.P. , Patre, U.V. et al (2013) CAMTA 1 regulates drought responses in Arabidopsis thaliana . BMC Genom. 14, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, I.S. , Negrao, S. , Pentony, M.M. , Abreu, I.A. , Oliveira, M.M. and Purugganan, M.D. (2013) Different evolutionary histories of two cation/proton exchanger gene families in plants. BMC Plant Biol. 13, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Z. and Spalding, E.P. (2004) Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+–H+ antiporter during salinity stress. Plant Physiol. 136, 2548–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, M.P. , Gálvez, F.J. , Huertas, R. , Aranda, M.N. , Baghour, M. , Cagnac, O. and Venema, K. (2009) Plant NHX cation/proton antiporters. Plant Signal. Behav. 4, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, M.S. (2005) The file‐drawer problem revisited: a general weighted method for calculating fail‐safe numbers in meta‐analysis. Evolution 59, 464–468. [PubMed] [Google Scholar]
- Rosenthal, R. (1979) The file drawer problem and tolerance for null results. Psychol. Bull. 86, 638–641. [Google Scholar]
- Rothstein, H.R. , Sutton, A.J. and Borenstein, M. (2005) Publication bias in meta‐analysis In Publication Bias in Meta‐Analysis: Prevention, Assessment and Adjustments(Rothstein H.R., Sutton A.J. and Borenstein M., eds), pp. 1–7. England: Blackwell Science. [Google Scholar]
- Rozema, J. and Schat, H. (2013) Salt tolerance of halophytes, research questions reviewed in the perspective of saline agriculture. Environ. Exp. Bot. 92, 83–95. [Google Scholar]
- Ruíz‐Lozano, J.M. , Porcel, R. , Azcón, C. and Aroca, R. (2012) Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J. Exp. Bot. 63, 695–709. [DOI] [PubMed] [Google Scholar]
- Schaeffer, R.N. , Manson, J.S. and Irwin, R.E. (2013) Effects of abiotic factors and species interactions on estimates of male plant function: a meta‐analysis. Ecol. Lett. 16, 399–408. [DOI] [PubMed] [Google Scholar]
- Serrano, R. and Rodriguez‐Navarro, A. (2001) Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13, 399–404. [DOI] [PubMed] [Google Scholar]
- Shi, H.Z. , Quintero, F.J. , Pardo, J.M. and Zhu, J.K. (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long‐distance Na+ transport in plants. Plant Cell, 14, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, A. , An, J. , Guan, Z. , Jiang, J. , Chen, F. , Lou, W. , Fang, W. et al (2014) The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 80, 114–120. [DOI] [PubMed] [Google Scholar]
- Sterne, J.A.C. and Egger, M. (2005) Regression methods to detect publication and other bias in meta‐analysis In Publication bias in Meta‐Analysis: Prevention, Assessment and Adjustments(Rothstein H.R., Sutton A.J. and Borenstein M., eds), pp. 99–110. England: Blackwell Science. [Google Scholar]
- Walker, C.J. and Weinstein, J.D. (1994) The magnesium‐insertion step of chlorophyll biosynthesis a 2‐stage reaction. Biochem. J. 299, 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.X. , Vinocur, B. and Altman, A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta, 218, 1–14. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Li, Y. , Zhang, Y. , Yang, C. , Zheng, N. and Xie, Q. (2007) Comparative expression analysis of three genes from the Arabidopsis vacuolar Na+/H+ antiporter (AtNHX) family in relation to abiotic stresses. Chin. Sci. Bull. 52, 1754–1763. [Google Scholar]
- Wang, X. , Yang, R. , Wang, B. , Liu, G. , Yang, C. and Cheng, Y. (2011) Functional characterization of a plasma membrane Na+/H+ antiporter from alkali grass (Puccinellia tenuiflora). Mol. Biol. Rep. 38, 4813–4822. [DOI] [PubMed] [Google Scholar]
- Worchel, E.R. , Giauque, H.E. and Kivlin, S.N. (2013) Fungal symbionts alter plant drought response. Microb. Ecol. 65, 671–678. [DOI] [PubMed] [Google Scholar]
- Wujeska, A. , Bossinger, G. and Tausz, M. (2013) Responses of foliar antioxidative and photoprotective defence systems of trees to drought: a meta‐analysis. Tree Physiol. 33, 1018–1029. [DOI] [PubMed] [Google Scholar]
- Yadav, N.S. , Rashmi, D. , Singh, D. , Agarwal, P.K. and Jha, B. (2012) A novel salt‐inducible gene SbSI‐1 from Salicornia brachiata confers salt and desiccation tolerance in E. coli . Mol. Biol. Rep. 39, 1943–1948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+ concentration of plants exposed to NaCl.
Figure S2 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+/Na+ ratio of plants exposed to NaCl.
Figure S3 Summary effects (as natural logs, ln R) and 95% confidence intervals (CIs) for the influence of CPA1 overexpression on shoot K+/Na+ ratio of plants exposed to NaCl.
Appendix S1 Details on the CPA1‐overexpression studies used in the meta‐analyses, including each of the moderators used for categorical analyses, the transgenic and untransformed control means, sample sizes (n) and the value of the natural log (ln) of the response ratio with corresponding non‐parametric variances.
Appendix S2 Analyses of publication bias.
Table S1 Measures used in characterizing publication bias for each effect size.


