Abstract
Glutathione reductase (EC 1.6.4.2; CAS registry number 9001-48-3) and trypanothione reductase (CAS registry number 102210-35-5), which are related flavoprotein disulfide oxidoreductases, have marked specificities for glutathione and trypanothione, respectively. A combination of primary sequence alignments and molecular modeling, together with the high-resolution crystal structure of human glutathione reductase, identified certain residues as potentially being responsible for substrate discrimination. Site-directed mutagenesis of Escherichia coli glutathione reductase was used to test these predictions. The mutation of Asn-21 to Arg demonstrated that this single change was insufficient to generate the greater discrimination against trypanothione shown by human glutathione reductase compared with the E. coli enzyme. However, the mutation of Ala-18, Asn-21, and Arg-22 to the amino acid residues (Glu, Trp, and Asn, respectively) in corresponding positions in Trypanosoma congolense trypanothione reductase confirmed that this region of polypeptide chain is intimately involved in substrate recognition. It led to a mutant form of E. coli glutathione reductase that possessed essentially no activity with glutathione but that was able to catalyze trypanothione reduction with a kcat/Km value that was 10% of that measured for natural trypanothione reductases. These results should be of considerable importance in the design of trypanocidal drugs targeted at the differences between glutathione and trypanothione metabolism in trypanosomatids and their hosts.
Full text
PDF
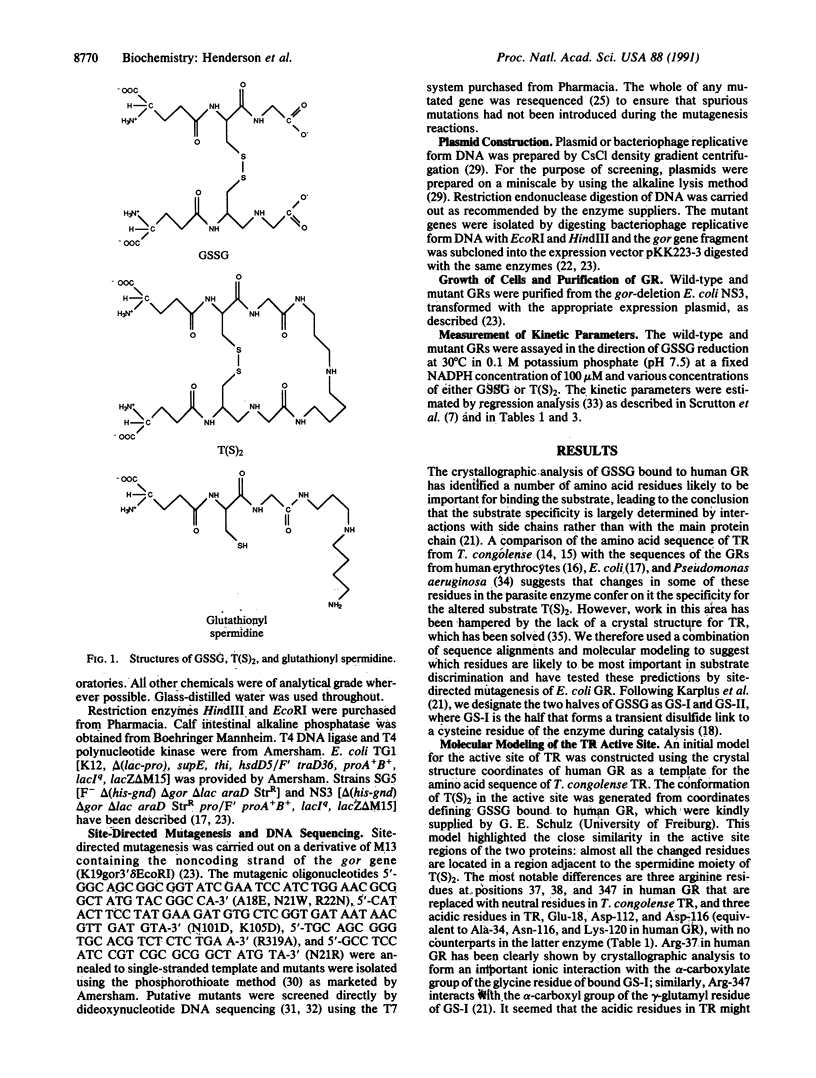



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berry A., Scrutton N. S., Perham R. N. Switching kinetic mechanism and putative proton donor by directed mutagenesis of glutathione reductase. Biochemistry. 1989 Feb 7;28(3):1264–1269. doi: 10.1021/bi00429a047. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone R., Silen J. L., Agard D. A. Structural plasticity broadens the specificity of an engineered protease. Nature. 1989 May 18;339(6221):191–195. doi: 10.1038/339191a0. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Atkinson T., Holbrook J. J. From analysis to synthesis: new ligand binding sites on the lactate dehydrogenase framework. Part II. Trends Biochem Sci. 1989 Apr;14(4):145–148. doi: 10.1016/0968-0004(89)90147-3. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Deonarain M. P., Berry A., Scrutton N. S., Perham R. N. Alternative proton donors/acceptors in the catalytic mechanism of the glutathione reductase of Escherichia coli: the role of histidine-439 and tyrosine-99. Biochemistry. 1989 Dec 12;28(25):9602–9607. doi: 10.1021/bi00451a008. [DOI] [PubMed] [Google Scholar]
- Ermler U., Schulz G. E. The three-dimensional structure of glutathione reductase from Escherichia coli at 3.0 A resolution. Proteins. 1991;9(3):174–179. doi: 10.1002/prot.340090303. [DOI] [PubMed] [Google Scholar]
- Fairlamb A. H., Blackburn P., Ulrich P., Chait B. T., Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985 Mar 22;227(4693):1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- Fairlamb A. H., Henderson G. B., Cerami A. Trypanothione is the primary target for arsenical drugs against African trypanosomes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf L., Craik C. S., Patthy A., Roczniak S., Fletterick R. J., Rutter W. J. Selective alteration of substrate specificity by replacement of aspartic acid-189 with lysine in the binding pocket of trypsin. Biochemistry. 1987 May 5;26(9):2616–2623. doi: 10.1021/bi00383a031. [DOI] [PubMed] [Google Scholar]
- Henderson G. B., Fairlamb A. H., Ulrich P., Cerami A. Substrate specificity of the flavoprotein trypanothione disulfide reductase from Crithidia fasciculata. Biochemistry. 1987 Jun 2;26(11):3023–3027. doi: 10.1021/bi00385a011. [DOI] [PubMed] [Google Scholar]
- Henderson G. B., Ulrich P., Fairlamb A. H., Rosenberg I., Pereira M., Sela M., Cerami A. "Subversive" substrates for the enzyme trypanothione disulfide reductase: alternative approach to chemotherapy of Chagas disease. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5374–5378. doi: 10.1073/pnas.85.15.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Jockers-Scherübl M. C., Schirmer R. H., Krauth-Siegel R. L. Trypanothione reductase from Trypanosoma cruzi. Catalytic properties of the enzyme and inhibition studies with trypanocidal compounds. Eur J Biochem. 1989 Mar 15;180(2):267–272. doi: 10.1111/j.1432-1033.1989.tb14643.x. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Pai E. F., Schulz G. E. A crystallographic study of the glutathione binding site of glutathione reductase at 0.3-nm resolution. Eur J Biochem. 1989 Jan 2;178(3):693–703. doi: 10.1111/j.1432-1033.1989.tb14500.x. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Kim J., Ruzicka F., Frey P. A. Remodeling hexose-1-phosphate uridylyltransferase: mechanism-inspired mutation into a new enzyme, UDP-hexose synthase. Biochemistry. 1990 Nov 27;29(47):10590–10593. doi: 10.1021/bi00499a003. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Blatterspiel R., Saleh M., Schiltz E., Schirmer R. H., Untucht-Grau R. Glutathione reductase from human erythrocytes. The sequences of the NADPH domain and of the interface domain. Eur J Biochem. 1982 Jan;121(2):259–267. doi: 10.1111/j.1432-1033.1982.tb05780.x. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Enders B., Henderson G. B., Fairlamb A. H., Schirmer R. H. Trypanothione reductase from Trypanosoma cruzi. Purification and characterization of the crystalline enzyme. Eur J Biochem. 1987 Apr 1;164(1):123–128. doi: 10.1111/j.1432-1033.1987.tb11002.x. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Kong X. P., Krishna T. S., Sweet R. M., Murgolo N. J., Field H., Cerami A., Henderson G. B. X-ray structure of trypanothione reductase from Crithidia fasciculata at 2.4-A resolution. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8764–8768. doi: 10.1073/pnas.88.19.8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg R. L., Negishi M. Alteration of mouse cytochrome P450coh substrate specificity by mutation of a single amino-acid residue. Nature. 1989 Jun 22;339(6226):632–634. doi: 10.1038/339632a0. [DOI] [PubMed] [Google Scholar]
- Murgolo N. J., Cerami A., Henderson G. B. Biomedical science and the third world. Under the volcano. Trypanothione reductase. Ann N Y Acad Sci. 1989;569:193–200. doi: 10.1111/j.1749-6632.1989.tb27369.x. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Karplus P. A., Schulz G. E. Crystallographic analysis of the binding of NADPH, NADPH fragments, and NADPH analogues to glutathione reductase. Biochemistry. 1988 Jun 14;27(12):4465–4474. doi: 10.1021/bi00412a038. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Schulz G. E. The catalytic mechanism of glutathione reductase as derived from x-ray diffraction analyses of reaction intermediates. J Biol Chem. 1983 Feb 10;258(3):1752–1757. [PubMed] [Google Scholar]
- Perry A. C., Ni Bhriain N., Brown N. L., Rouch D. A. Molecular characterization of the gor gene encoding glutathione reductase from Pseudomonas aeruginosa: determinants of substrate specificity among pyridine nucleotide-disulphide oxidoreductases. Mol Microbiol. 1991 Jan;5(1):163–171. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Engineering of an intersubunit disulphide bridge in glutathione reductase from Escherichia coli. FEBS Lett. 1988 Dec 5;241(1-2):46–50. doi: 10.1016/0014-5793(88)81028-7. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Purification and characterization of glutathione reductase encoded by a cloned and over-expressed gene in Escherichia coli. Biochem J. 1987 Aug 1;245(3):875–880. doi: 10.1042/bj2450875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Shames S. L., Fairlamb A. H., Cerami A., Walsh C. T. Purification and characterization of trypanothione reductase from Crithidia fasciculata, a newly discovered member of the family of disulfide-containing flavoprotein reductases. Biochemistry. 1986 Jun 17;25(12):3519–3526. doi: 10.1021/bi00360a007. [DOI] [PubMed] [Google Scholar]
- Shames S. L., Kimmel B. E., Peoples O. P., Agabian N., Walsh C. T. Trypanothione reductase of Trypanosoma congolense: gene isolation, primary sequence determination, and comparison to glutathione reductase. Biochemistry. 1988 Jul 12;27(14):5014–5019. doi: 10.1021/bi00414a010. [DOI] [PubMed] [Google Scholar]
- Sullivan F. X., Shames S. L., Walsh C. T. Expression of Trypanosoma congolense trypanothione reductase in Escherichia coli: overproduction, purification, and characterization. Biochemistry. 1989 Jun 13;28(12):4986–4992. doi: 10.1021/bi00438a013. [DOI] [PubMed] [Google Scholar]
- Sullivan F. X., Sobolov S. B., Bradley M., Walsh C. T. Mutational analysis of parasite trypanothione reductase: acquisition of glutathione reductase activity in a triple mutant. Biochemistry. 1991 Mar 19;30(11):2761–2767. doi: 10.1021/bi00225a004. [DOI] [PubMed] [Google Scholar]
- Tabor H., Tabor C. W. Isolation, characterization, and turnover of glutathionylspermidine from Escherichia coli. J Biol Chem. 1975 Apr 10;250(7):2648–2654. [PubMed] [Google Scholar]
- Taylor J. W., Schmidt W., Cosstick R., Okruszek A., Eckstein F. The use of phosphorothioate-modified DNA in restriction enzyme reactions to prepare nicked DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8749–8764. doi: 10.1093/nar/13.24.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington D. J., Rosemeyer M. A. Glutathione reductase from human erythrocytes. Catalytic properties and aggregation. Eur J Biochem. 1976 Aug 1;67(1):231–238. doi: 10.1111/j.1432-1033.1976.tb10654.x. [DOI] [PubMed] [Google Scholar]



