Abstract
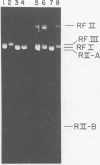
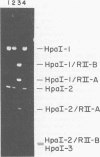
A mutant (designated mec−) has been isolated from Escherichia coli C which has lost DNA-cytosine methylase activity and the ability to protect phage λ against in vivo restriction by the RII endonuclease. This situation is analogous to that observed with an E. coli K-12 mec− mutant; thus, the E. coli C methylase appears to have overlapping sequence specificity with the K-12 and RII enzymes; (the latter methylases have been shown previously to recognize the same sequence). Covalently closed, supertwisted double-standed DNA (RFI) was isolated from C mec+ and C mec− cells infected with bacteriophage φX174. φX· mec− RFI is sensitive to in vitro cleavage by R·EcoRII and is cut twice to produce two fragments of almost equal size. In contrast, φX·mec+ RFI is relatively resistant to in vitro cleavage by R·EcoRII. R·BstI, which cleaves mec+/RII sites independent of the presence or absence of 5-methylcytosine, cleaves both forms of the RFI and produces two fragments similar in size to those observed with R· EcoRII. These results demonstrate that φX·mec+ RFI is methylated in vivo by the host mec+ enzyme and that this methylation protects the DNA against cleavage by R·EcoRII. This is consistent with the known location of two mec+/ RII sequences (viz., [Formula: see text]) on the φX174 map. Mature singlestranded virion DNA was isolated from φX174 propagated in C mec+ or C mec− in the presence of l-[methyl-3H]methionine. Paper chromatographic analyses of acid hydrolysates revealed that φX·mec+ DNA had a 10-fold-higher ratio of [3H]5-methylcytosine to [3H]cytosine compared to φX·mec−. Since φX·mec+ contains, on the average, approximately 1 5-methylcytosine residue per viral DNA, we conclude that methylation of φX174 is mediated by the host mec+ enzyme only. These results are not consistent with the conclusions of previous reports that φX174 methylation is mediated by a phage-induced enzyme and that methylation is essential for normal phage development.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARBER W., MORSE M. L. HOST SPECIFICITY OF DNA PRODUCED BY ESCHERICHIA COLI. VI. EFFECTS ON BACTERIAL CONJUGATION. Genetics. 1965 Jan;51:137–148. doi: 10.1093/genetics/51.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché J. P., Dubert J. M. DNA methylases of Escherichia coli K12. Evidence for changes in their state of association following purification. Eur J Biochem. 1972 May;27(1):53–59. doi: 10.1111/j.1432-1033.1972.tb01810.x. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Fujimoto D., Srinivasan P. R., Borek E. On the nature of the deoxyribonucleic acid methylases. Biological evidence for the multiple nature of the enzymes. Biochemistry. 1965 Dec;4(12):2849–2855. doi: 10.1021/bi00888a041. [DOI] [PubMed] [Google Scholar]
- GOLD M., HURWITZ J. THE ENZYMATIC METHYLATION OF RIBONUCLEIC ACID AND DEOXYRIBONUCLEIC ACID. V. PURIFICATION AND PROPERTIES OF THE DEOXYRIBONUCLEIC ACID-METHYLATING ACTIVITY OF ESCHERICHIA COLI. J Biol Chem. 1964 Nov;239:3858–3865. [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methylation of T-even bacteriophages and of their nonglucosylated mutants: its role in P1-directed restriction. Virology. 1970 Oct;42(2):359–367. doi: 10.1016/0042-6822(70)90279-5. [DOI] [PubMed] [Google Scholar]
- Hattman S., Gold E., Plotnik A. Methylation of cytosine residues in DNA controlled by a drug resistance factor (host-induced modification-R factors-N 6 -methyladenine-5-methylcytosine). Proc Natl Acad Sci U S A. 1972 Jan;69(1):187–190. doi: 10.1073/pnas.69.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Partial purification of the Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase: in vitro methylation completely protects bacteriophage lambda deoxyribonucleic acid against cleavage by R-EcoRII. J Bacteriol. 1977 Mar;129(3):1330–1334. doi: 10.1128/jb.129.3.1330-1334.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in bacteriophage lambda deoxyribonucleic acid. J Virol. 1972 Sep;10(3):356–361. doi: 10.1128/jvi.10.3.356-361.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in single-stranded DNA bacteriophages M13 and fd. J Mol Biol. 1973 Mar 15;74(4):749–752. doi: 10.1016/0022-2836(73)90064-8. [DOI] [PubMed] [Google Scholar]
- Hattman S., Schlagman S., Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973 Sep;115(3):1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. G. The sensitivity of bacteriophage lambda DNA to restriction endonuclease RII. J Mol Biol. 1975 Nov 5;98(3):645–647. doi: 10.1016/s0022-2836(75)80093-3. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. Location of the 5-methylcytosine group on the bacteriophage phi X174 genome. J Virol. 1974 Oct;14(4):872–877. doi: 10.1128/jvi.14.4.872-877.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattman S. Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host- and R-factor-controlled enzymes. J Bacteriol. 1975 Aug;123(2):768–770. doi: 10.1128/jb.123.2.768-770.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. DNA methylase induced by bacteriophage phiX174. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3773–3775. doi: 10.1073/pnas.70.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Goren D., Friedman J. Studies on the biological role of DNA methylation: inhibition of methylation and maturation of the bacteriophage phichi174 by nicotinamide. Nucleic Acids Res. 1975 Oct;2(10):1967–1974. doi: 10.1093/nar/2.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Sedat J. W., Sinsheimer R. L. In vivo methylation of replicating bacteriophage phi chi174 DNA. J Mol Biol. 1973 Aug 15;78(3):417–425. doi: 10.1016/0022-2836(73)90465-8. [DOI] [PubMed] [Google Scholar]
- Razin A., Sedat J. W., Sinsheimer R. L. Structure of the DNA of bacteriophage phiX174. VII. Methylation. J Mol Biol. 1970 Oct 28;53(2):251–259. doi: 10.1016/0022-2836(70)90298-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S., Hattman S. Mutants of the N-3 R-factor conditionally defective in hspII modification and deoxyribonucleic acid-cytosine methylase activity. J Bacteriol. 1974 Oct;120(1):234–239. doi: 10.1128/jb.120.1.234-239.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Endonuclease R-EcoRII restriction of bacteriophage f1 DNA in vitro: ordering of genes V and VII, location of an RNA promotor for gene VIII. J Virol. 1975 Sep;16(3):674–684. doi: 10.1128/jvi.16.3.674-684.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takano T., Arai T., Nishida H., Sato S. Episome-mediated Transfer of Drug Resistance in Enterobacteriaceae X. Restriction and Modification of Phages by fi R Factors. J Bacteriol. 1966 Aug;92(2):477–486. doi: 10.1128/jb.92.2.477-486.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]