Abstract
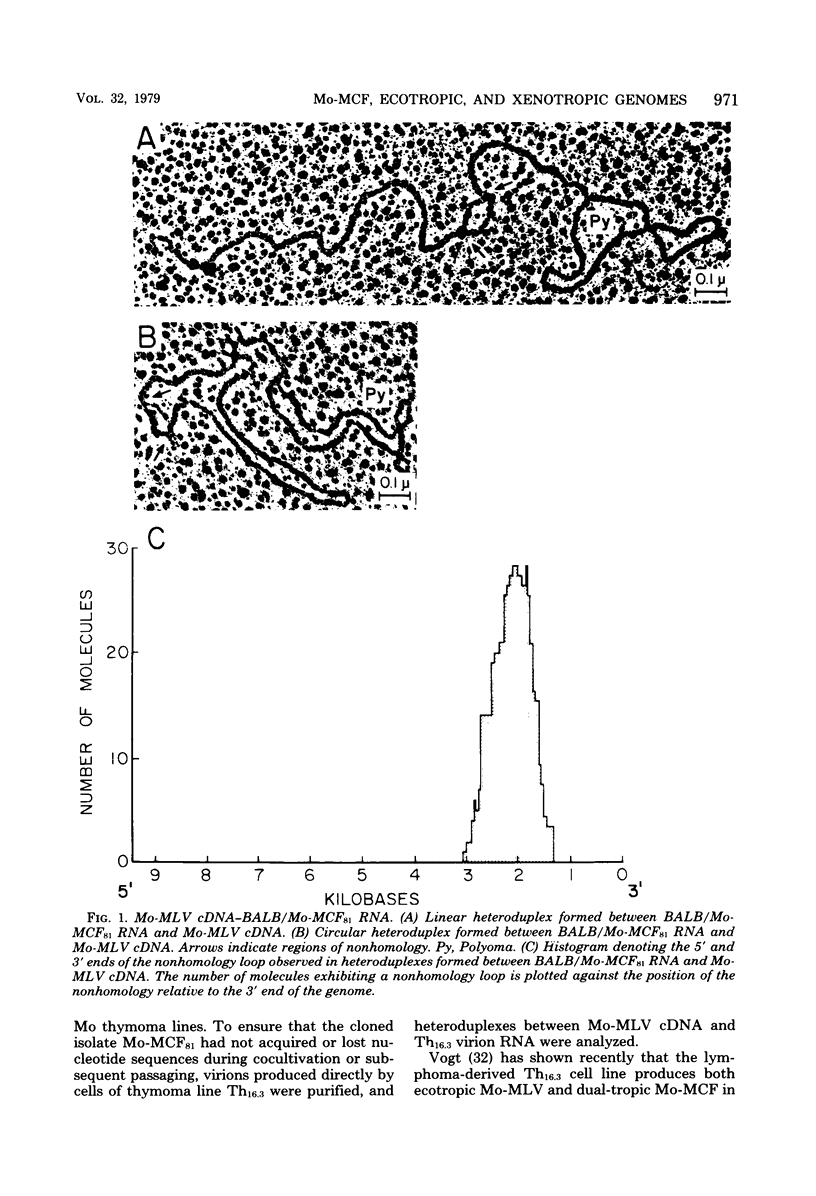
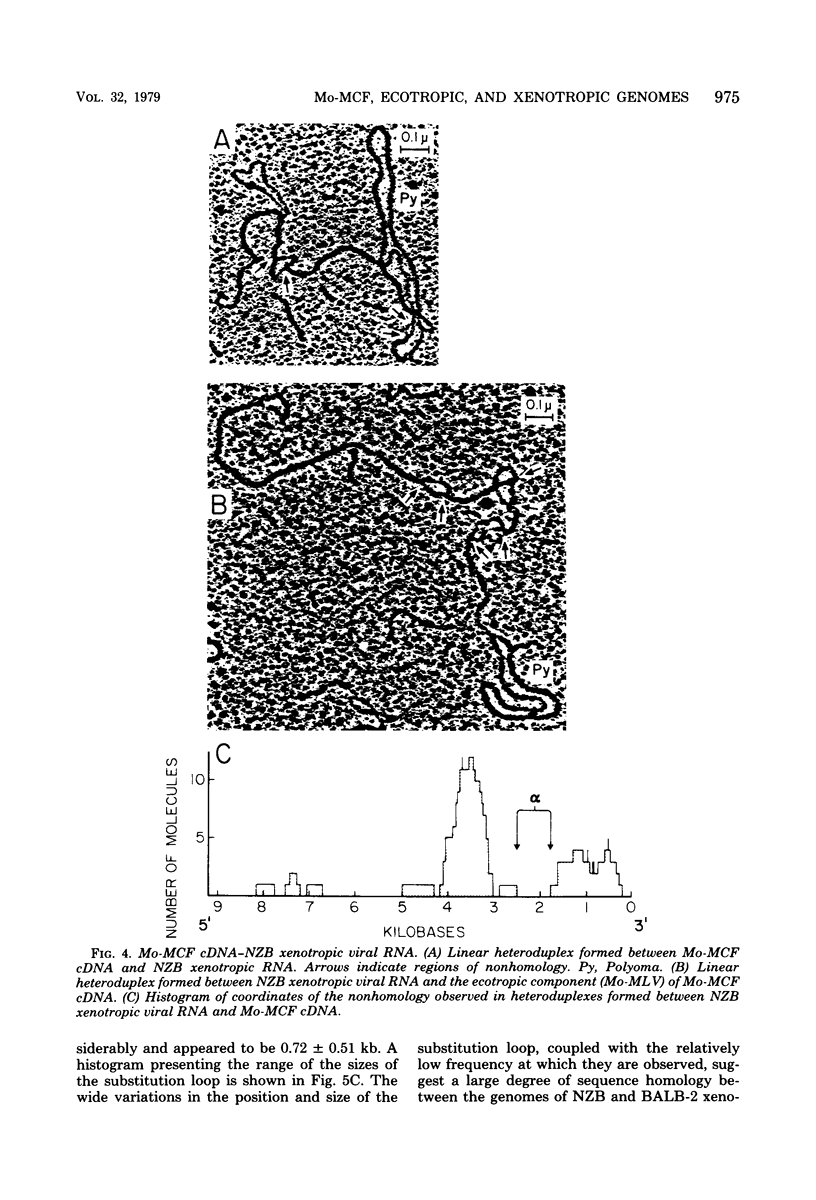
The genome of a recombinant murine leukemia virus capable of inducing focal areas of morphological alteration in mink lung fibroblasts was studied by heteroduplex analysis. The dual-tropic recombinant virus was isolated from a thymoma cell line (Th16.3) and is referred to as BALB/Moloney mink cell focus-inducing virus (BALB/Mo-MCF virus). The nucleic acid sequences of RNA from virions obtained from either a thymoma cell line (Th16.3) or a clonal isolate (BALB/Mo-MCF81) were compared with the genomes of ecotropic and xenotropic viruses. The following inferences were drawn (i) A single nonhomologous region (substitution loop alpha) of about 0.7 kilobase was observed in a heteroduplex formed between Moloney murine leukemia virus complementary DNA (cDNA) and BALB/MoMCF81 RNA. This nonhomology region was mapped between 1.71 and 2.40 kilobases from the 3' end of the genome. (ii) The predominant class of heteroduplexes formed between virion RNA obtained from the thymoma cell line (Th16.3) and Moloney murine leukemia virus cDNA showed a substitution loop similar to that observed with the RNA obtained from a cloned isolate, BALB/Mo-MCF81. However, there were other molecules with additional regions of nonhomology. (iii) Heteroduplexes formed between NZB xenotropic RNA and ecotropic Moloney murine leukemia virus cDNA exhibited four major nonhomology regions extending 0.75 to 1.46, 2.0 to 2.8, 3.6 to 4.3, and 7.4 to 7.9 kilobases from the 3' end of the genome. (iv) The MCF-specific substitution loop alpha (1.71 to 2.40 kilobases) appeared as a duplex region when NZB xenotropic RNA was hybridized to cDNA transcripts synthesized by virions obtained from thymoma cell line Th16.3. The position of the other substitution loops observed in a heteroduplex formed between NZB xenotropic RNA and Moloney murine leukemia virus cDNA was not affected. (v) Heteroduplexes formed between xenotropic BALB virus 2 cDNA and NZB xenotropic RNA demonstrated a large degree of nucleic acid sequence homology. Of the 29 heteroduplexes examined, 24 appeared to be homoduplexes, and in the remaining 5 heteroduplexes only one region of nonhomology located between 3.2 and 3.8 kilobases from the 3' end of the genome could be identified. Hybridization of BALB virus 2 xenotropic RNA to NZB xenotropic cDNA followed by digestion with single-strand-specific nuclease S1 showed an 80% sequence homology.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Stephenson J. R. Independent segregation of loci for activation of biologically distinguishable RNA C-type viruses in mouse cells. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2055–2058. doi: 10.1073/pnas.70.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976 Apr;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Benveniste R. E., Lieber M. M., Todaro G. J. A distinct class of inducible murine type-C viruses that replicates in the rabbit SIRC cell line. Proc Natl Acad Sci U S A. 1974 Mar;71(3):602–606. doi: 10.1073/pnas.71.3.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Callahan R., Lieber M. M., Todaro G. J. Nucleic acid homology of murine xenotropic type C viruses. J Virol. 1975 Jun;15(6):1378–1384. doi: 10.1128/jvi.15.6.1378-1384.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y. H., Verma I. M., Shih T. Y., Scolnick E. M., Davidson N. Heteroduplex analysis of the sequence relations between the RNAs of mink cell focus-inducing and murine leukemia viruses. J Virol. 1978 Oct;28(1):352–360. doi: 10.1128/jvi.28.1.352-360.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue D. J., Rothenberg E., Hopkins N., Baltimore D., Sharp P. A. Heteroduplex analysis of the nonhomology region between Moloney MuLV and the dual host range derivative HIX virus. Cell. 1978 Aug;14(4):959–970. doi: 10.1016/0092-8674(78)90350-1. [DOI] [PubMed] [Google Scholar]
- Elder J. H., Gautsch J. W., Jensen F. C., Lerner R. A., Hartley J. W., Rowe W. P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. H., Jensen F. C., Bryant M. L., Lerner R. A. Polymorphism of the major envelope glycoprotein (gp70) of murine C-type viruses: virion associated and differentiation antigens encoded by a multi-gene family. Nature. 1977 May 5;267(5606):23–28. doi: 10.1038/267023a0. [DOI] [PubMed] [Google Scholar]
- Faller D. V., Rommelaere J., Hopkins N. Large T1 oligonucleotides of Moloney leukemia virus missing in an env gene recombinant, HIX, are present on an intracellular 21S Moloney viral RNA species. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2964–2968. doi: 10.1073/pnas.75.6.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Baltimore D. RNA metabolism of murine leukemia virus: detection of virus-specific RNA sequences in infected and uninfected cells and identification of virus-specific messenger RNA. J Mol Biol. 1973 Oct 15;80(1):93–117. doi: 10.1016/0022-2836(73)90235-0. [DOI] [PubMed] [Google Scholar]
- Fan H., Paskind M. Measurement of the sequence complexity of cloned Moloney murine leukemia virus 60 to 70S RNA: evidence for a haploid genome. J Virol. 1974 Sep;14(3):421–429. doi: 10.1128/jvi.14.3.421-429.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Verma I. M. Size analysis and relationship of murine leukemia virus-specific mRNA's: evidence for transposition of sequences during synthesis and processing of subgenomic mRNA. J Virol. 1978 May;26(2):468–478. doi: 10.1128/jvi.26.2.468-478.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger P. J., Ihle J. N., de Noronha F., Bolognesi D. P. Oncogenic and immunogenic potential of cloned HIX virus in mice and cats. Med Microbiol Immunol. 1977;164(1-3):119–129. doi: 10.1007/BF02121308. [DOI] [PubMed] [Google Scholar]
- Fischinger P. J., Nomura S., Bolognesi D. P. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Rowe W. P. Naturally occurring murine leukemia viruses in wild mice: characterization of a new "amphotropic" class. J Virol. 1976 Jul;19(1):19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S., Stephenson J. R., Aaronson S. A. Radiommunoassays for the 70,000-molecular-weight glycoproteins of endogenous mouse type C viruses: viral antigen expression in normal mouse tissues and sera. J Virol. 1976 Jun;18(3):933–941. doi: 10.1128/jvi.18.3.933-941.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Davidson N. A heteroduplex study of the sequence relationships between the RNAs of M-MSV and M-MLV. Cell. 1977 Mar;10(3):469–477. doi: 10.1016/0092-8674(77)90034-4. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration of moloney leukemia virus: effect of homozygosity at the m-mulV locus. Cell. 1977 Nov;12(3):691–696. doi: 10.1016/0092-8674(77)90269-0. [DOI] [PubMed] [Google Scholar]
- Levy J. A. Xenotropic viruses: murine leukemia viruses associated with NIH Swiss, NZB, and other mouse strains. Science. 1973 Dec 14;182(4117):1151–1153. doi: 10.1126/science.182.4117.1151. [DOI] [PubMed] [Google Scholar]
- Rasheed S., Gardner M. B., Chan E. Amphotropic host range of naturally occuring wild mouse leukemia viruses. J Virol. 1976 Jul;19(1):13–18. doi: 10.1128/jvi.19.1.13-18.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J., Faller D. V., Hopkins N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc Natl Acad Sci U S A. 1978 Jan;75(1):495–499. doi: 10.1073/pnas.75.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T. Y., Weeks M. O., Troxler D. H., Coffin J. M., Scolnick E. M. Mapping host range-specific oligonucleotides within genomes of the ecotropic and mink cell focus-inducing strains of Moloney murine leukemia virus. J Virol. 1978 Apr;26(1):71–83. doi: 10.1128/jvi.26.1.71-83.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Troxler D. H., Scolnick E. M. Rapid leukemia induced by cloned friend strain of replicating murine type-C virus. Association with induction of xenotropic-related RNA sequences contained in spleen focus-forming virus. Virology. 1978 Mar;85(1):17–27. doi: 10.1016/0042-6822(78)90408-7. [DOI] [PubMed] [Google Scholar]
- Troxler D. H., Yuan E., Linemeyer D., Ruscetti S., Scolnick E. M. Helper-independent mink cell focus-inducing strains of Friend murine type-C virus: potential relationship to the origin of replication-defective spleen focus-forming virus. J Exp Med. 1978 Sep 1;148(3):639–653. doi: 10.1084/jem.148.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. Genome organization of RNA tumor viruses. I. In vitro synthesis of full-genome-length single-stranded and double-stranded viral DNA transcripts. J Virol. 1978 Jun;26(3):615–629. doi: 10.1128/jvi.26.3.615-629.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M. Properties of "mink cell focus-inducing" (MCF) virus isolated from spontaneous lymphoma lines of BALB/c mice carrying Moloney leukemia virus as an endogenous virus. Virology. 1979 Feb;93(1):226–236. doi: 10.1016/0042-6822(79)90290-3. [DOI] [PubMed] [Google Scholar]








