Abstract
Minimal residual disease (MRD) quantification is an important predictor of outcome after treatment for acute lymphoblastic leukemia (ALL). Bone marrow ALL burden ≥ 10−4 after induction predicts subsequent relapse. Likewise, MRD ≥ 10−4 in bone marrow prior to the initiation of conditioning for allogeneic hematopoietic cell transplantation (allo-HCT) predicts transplant failure. Current methods for MRD quantification in ALL are not sufficiently sensitive for use with peripheral blood specimens and have not been broadly implemented in the management of adults with ALL. Consensus primed immunoglobulin (Ig) and T-cell receptor (TCR) amplification and high-throughput sequencing (HTS) permits use of a standardized algorithm for all patients and can detect leukemia at 10−6 or lower. We applied the Sequenta LymphoSIGHT™ HTS platform to quantification of MRD in 237 samples from 29 adult B-ALL patients before and after allo-HCT. Using primers for the IGH-VDJ, IGH-DJ, IGK, TCRB, TCRD, and TCRG loci, MRD could be quantified in 93% of patients. Leukemia-associated clonotypes at these loci were identified in 52%, 28%, 10%, 35%, 28%, and 41% of patients, respectively. MRD ≥ 10−4 before HCT conditioning predicted post-HCT relapse (HR 7.7, 95% CI 2.0–30, p=0.003). In post-HCT blood samples, MRD ≥ 10−6 had 100% positive predictive value for relapse with median lead-time of 89 days (HR 14; 95% CI 4.7–44, p<0.0001). The use of HTS-based MRD quantification in adults with ALL offers a standardized approach with sufficient sensitivity to quantify leukemia MRD in peripheral blood. Use of this approach may identify a window for clinical intervention prior to overt relapse.
INTRODUCTION
Tremendous progress has been made in the management of acute lymphoblastic leukemia (ALL) in children, in part, through the wide use of minimal residual disease (MRD) monitoring in bone marrow aspirates to guide therapeutic intensification before and after allogeneic hematopoietic cell transplantation (allo-HCT).1–7 Nonetheless, regional differences in standardization and the high costs of MRD testing have limited its use in the management of adult ALL. Similar to the significance of MRD positivity after induction therapy for pediatric ALL, MRD evaluation in adults with ALL has been shown to be useful for predicting clinical outcomes.8, 9 A broadly applicable MRD quantification method that addresses the limitations of currently available MRD technologies has the potential to significantly improve the management of ALL in adults.
At present, two prevailing technologies are available for quantification of MRD in ALL: real-time quantitative polymerase chain reaction (RQ-PCR) and multi-parametric flow cytometry (MPFC). MRD quantification in bone marrow specimens from patients with ALL using immunoglobulin (Ig) and T-cell receptor (TCR) RQ-PCR with allele-specific primers and amplification probes has achieved a high degree of standardization in Europe via the EuroMRD consortium.10 Unfortunately, this methodology has not become a standard of practice in the United States and elsewhere due to the significant expense and expertise required to develop such patient-specific genetic assays. Remission bone marrow specimens may alternatively be assessed by MPFC for aberrant blast immunophenotypes using standardized antibody panels;11 however, this method has decreased sensitivity in comparison with molecular disease quantification, requires assessment of fresh tissue for best results, and may be subject to inter-laboratory variability due to differing population gating strategies during flow cytometric analyses.
Although PCR-based and flow-based methods both have merits, molecular quantification of clonal Ig/TCR gene rearrangements in leukemic blasts has been repeatedly demonstrated to provide the most sensitive and specific MRD quantification with a detection limit of roughly 10−5 (i.e., one leukemic cell in 100,000 leukocytes). Post-therapy MRD burden ≥ 10−4 in BM aspirates, using either RQ-PCR or MPFC, has been demonstrated to be a more powerful prognostic marker for subsequent relapse than those typically used, including age, WBC count at diagnosis, and cytogenetic alterations.12, 13
To date, the potential advantages of higher sensitivity MRD quantification have remained somewhat theoretical. Some studies have shown, however, that patients who are MRD positive by a PCR-based method, but MRD negative by MPFC, are at increased risk for relapse compared with patients MRD negative with both techniques.14–16 This suggests higher sensitivity may indeed be clinically useful. Additionally, a largely unscrutinized potential benefit of higher sensitivity is the possibility of meaningful detection of MRD in peripheral blood (PB) instead of bone marrow (BM).17 In the present study, we applied a next-generation sequencing (NGS) based MRD assay, termed the LymphoSIGHT™ platform,18 which has a quantitative range to 10−5 and may have sensitivity to below 10−6 with adequately cellular specimens, to quantify ALL MRD in bone marrow and peripheral blood samples prior to and following allo-HCT.
Another challenge in ALL MRD quantification addressed by the HTS method we studied is the need for identifying clonal rearrangements in multiple Ig/TCR genes in ALL. Most B- and T-cell ALL patients exhibit clonal rearrangements of one or more immunoglobulin (heavy chain, IGH; kappa or lambda light chain, IGK/IGL) or T-cell receptor (beta, TCRB; delta, TCRD; gamma, TCRG) genes. Such gene rearrangements represent a genetic “barcode” within every lymphocyte that permits quantification of specific clonal populations and this phenomenon is the principle upon which RQ-PCR methods for MRD quantification are based. Fifty to 75 percent of B-ALL patients have a disease-associated IGH-VDJ clonotype detectable.19–21 Roughly 10–30% of B-ALL isolates lacking a stable IGH-VDJ rearrangement have a stable and unique IGH-DJ partial rearrangement detectable.20, 22, 23 Other immunoreceptor loci may also undergo rearrangement in precursor B and T lymphoblasts. TCRB rearrangements have been detected in up to 35% of B-ALLs and more than 75% of T-ALLs,24, 25 while the TCRD and TCRG loci are clonally rearranged in a significant minority of B-ALL isolates.20
In this study, we applied the LymphoSIGHT MRD quantification method to assess clonality at the IGH locus (complete IGH-VDJ rearrangements, and partial IGH-DJ rearrangements), the IGK locus, and the TCRB, TCRD, and TCRG loci, in diagnostic specimens from adult patients with B-ALL. The leukemia-associated clonotypes were then quantified in cryopreserved pre- and post-transplant peripheral blood and bone marrow samples and correlations between disease burden and clinical outcomes were evaluated.
PATIENTS AND METHODS
Study subjects
Forty-two allografted ALL patients were eligible for this retrospective study based on availability of diagnostic samples believed to contain leukemic cells and minimum of 3 years post-HCT follow-up. Cryopreserved RNA isolates from diagnostic specimens in 13 patients were found to have unsuitable quality for comprehensive clone identification and no DNA or tissue samples were available, thus 29 patients with genomic DNA samples taken at diagnosis or relapse were studied for immunoreceptor clonality and pre- and post-HCT molecular MRD. Patients provided informed consent to tissue sample archival and this study was approved by the Stanford University School of Medicine Institutional Review Board.
Conditioning Regimens and GVHD Prophylaxis
Patients underwent allogeneic hematopoietic transplantation at Stanford University between May 2004 to March 2010. Pre-transplant conditioning regimens were myeloablative preparations with 1200 – 1320cGy total body irradiation (22 patients; 11 with cyclophosphamide, 8 with etoposide, and 3 with both) or chemotherapy (2 patients), or non-myeloablative preparation with total lymphoid irradiation and antithymocyte globulin (5 patients). Grafts were comprised of bone marrow (5 patients) or G-CSF mobilized peripheral blood apheresis products (24 patients), and originated from matched sibling donors (17 patients), matched unrelated donors (10 patients), or haploidentical donors (2 patients). All patients undergoing myeloablative preparation received standard immune prophylaxis with methotrexate and a calcineurin inhibitor. Recipients of non-myeloablative conditioning received prophylactic therapy with mycophenolate mofetil and a calcineurin inhibitor.
Tissue Sampling and MRD Quantification
Peripheral blood mononuclear cells (PBMC) from 10mL of whole blood or 5mL bone marrow mononuclear cell (BMMC) samples from marrow aspirates were collected and cryopreserved in liquid nitrogen vapor at diagnosis, prospectively planned post-transplant time points, and at relapse. Post-HCT samples were retrieved from storage, thawed, washed, and genomic DNA was isolated from cells using a Qiagen DNeasy kit according to manufacturer’s instructions.
Genomic DNA samples were then processed as described elsewhere.18 Briefly, using Sequenta’s LymphoSIGHT platform, we amplified and sequenced rearranged immunoreceptor loci from genomic DNA samples using V and J segment consensus primers for each gene (IGH, IGK, TCRB, TCRD, and TCRG) and, in some cases, D segment primers for incomplete IGH-DJ rearrangements.
Sequences were analyzed using standardized algorithms for clonotype determination. Leukemia-specific clonotypes were identified for each patient based on their high prevalence in a blood or marrow sample at a time of high disease burden. MRD levels were then determined in serial samples of PBMC or BMMC and quantified using spiked-in reference sequences in the reaction mixture to permit control for over-sequencing, thus enabling reporting of absolute clonotype frequency.18
Definition of Outcomes
We evaluated time to events defined as the interval between graft infusion (day 0) and MRD positivity or clinical relapse as determined by standard diagnostic criteria. Disease-free survival was defined as the interval between graft infusion and clinical relapse, the latter being defined by the recurrence of excess blasts exhibiting an immunophenotype congruent with the original diagnosis. Overall survival was defined as the interval between graft infusion and death from any cause.
Statistical Analyses
Disease-free survival and OS were estimated by the Kaplan-Meier method. GraphPad Prism (GraphPad Software, La Jolla, CA) was used to generate Kaplan-Meier curves and other figures after primary analysis in R or Spotfire (Tibco, Somerville, MA).
RESULTS
Patient and Sample Characteristics
Forty-two ALL patients who underwent allo-HCT were considered for this retrospective study. The LymphoSIGHT MRD quantification technique is dependent upon availability of a diagnostic specimen with adequate disease burden for identification of the Ig/TCR clonotypes specific to the leukemia cell population. For 13 patients, only RNA samples isolated from trizol lysates obtained at diagnosis were available. These samples were unfortunately degraded to an unacceptable quality and were not suitable for this study. We thus limited this survey to 29 patients with B-ALL based on availability of genomic DNA from a diagnostic sample (PB or BM) with adequate disease burden for identification of the leukemia-associated immunoreceptor clonotype(s) to be quantified in pre- and post-HCT samples (Table 1). Eleven patients (38%) had a t(9;22) translocation consistent with Philadelphia chromosome positive disease. Twelve patients (41%) were in first complete remission (CR), 9 (31%) in second CR, 2 (7%) in third CR, and 6 (21%) had relapsed/refractory disease at the time of conditioning. Twenty-four patients (83%) underwent myeloablative preparation, while 5 (17%) received non-myeloablative therapy. In total, 237 samples (176 PB and 61 BM) were analyzed by the LymphoSIGHT platform, including 29 leukemia-bearing samples and 208 pre- and post-HCT samples for MRD quantification.
Table 1. Patient features.
| n (%) | ||
|---|---|---|
| Total patients | 29 | |
| Recipient gender, M/F | 16/13 | |
| Age, median (range) | 34 (16 – 67) | |
| Subtype | B-ALL | 29 (100) |
| Risk feature | Ph+ | 11 (38) |
| Status at HCT | CR1 | 12 (41) |
| CR2 | 9 (31) | |
| CR3 | 2 (6.9) | |
| Relapse/Refractory | 6 (21) | |
| Regimen | Ablative | 24 (83) |
| Non-myeloablative | 5 (17) | |
| Graft | PBSC | 24 (83) |
| BM | 5 (17) | |
| Donor | Matched sibling | 17 (59) |
| Matched unrelated | 10 (34) | |
| Haploidentical | 2 (6) | |
| Status at last f/u | Alive | 7 (24) |
| Dead | 22 (76) | |
| DFS, days, median (range) | 252 (74 – 3110) | |
| OS, days, median (range) | 572 (85 – 3110) |
BM=bone marrow; CR=complete remission; DFS=disease-free survival; PBSC=peripheral blood stem cells; OS=overall survival
Ig and TCR Locus Rearrangements in ALL
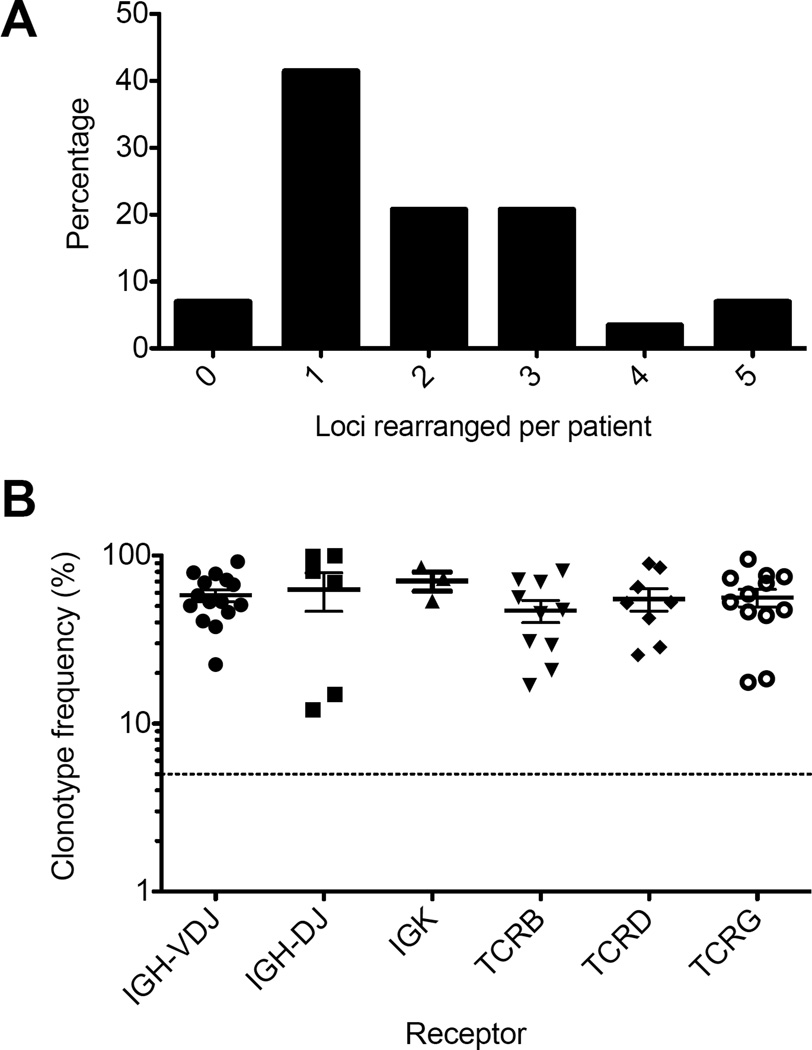
A clonal IGH sequence with a complete VDJ rearrangement was identified in 15/29 (52%) patients, whereas partial IGH-DJ rearrangements were observed in 8 (28%) patients. Other immunoreceptor loci exhibiting clonality included IGK in 3 (10%), TCRB in 10 (35%), TCRD in 8 (28%), and TCRG in 12 (41%) (Table 2). Amongst patients who did not have a detectable IGH clonotype, 12/14 (86%) had clonal sequences at one or more other loci, including partial IGH-DJ rearrangements (5/14; 38%), TCRB (4; 31%), TCRD (2; 15%), or TCRG (5; 38%). Leukemia-associated IGK clonotypes were identified only in leukemia isolates also possessing an IGH clonotype. In total, 27 of 29 (93%) patients had one or more clonal Ig/TCR sequences suitable for MRD quantification using LymphoSIGHT. Thirty-eight percent had just one rearranged locus suitable for MRD quantification, whereas 21% had two, 21% had three, 3.4% had four, and 6.9% had five (Figure 1A).
Table 2. Clonotype identification in diagnostic samples.
In this 29 patient cohort, 15 (52%) had identifiable IGH-VDJ clonotypes. 12/14 (86%) who did not have an IGH-VDJ clonotype were found to have a clonotype at one or more other loci as indicated. Coincident rearrangements are shows in columns.
| IGH-VDJ | IGH-DJ | IGK | TCRB | TCRD | TCRG | |
|---|---|---|---|---|---|---|
| IGH-VDJ | 15 | 3 | 3 | 6 | 7 | 7 |
| IGH-DJ | 3 | 8 | 1 | 0 | 2 | 2 |
| IGK | 3 | 1 | 3 | 3 | 3 | 2 |
| TCRB | 6 | 0 | 3 | 10 | 6 | 5 |
| TCRD | 7 | 2 | 3 | 6 | 8 | 4 |
| TCRG | 7 | 2 | 2 | 5 | 4 | 12 |
Figure 1. Frequency of disease clonotype in diagnostic samples.
At least one immunoreceptor clonotype suitable for use in MRD quantification was identified in 93% of patients with ALL. The number of leukemia-associated immunoreceptor locus rearrangements per patient are shown (A). For each of the immunoreceptors tested, the frequency of dominant clonotypes in diagnostic specimens is shown (B). Clonotype frequency greater than 5% (dotted line) was required to identify disease-associated immunoreceptor rearrangements.
When clonal rearrangements of specific loci were identified in diagnostic specimens, they comprised 58+/−18% of the IGH-VDJ repertoire, 63+/−40% of the IGH-DJ repertoire, 71+/−16% of the IGK repertoire, 47+/−22% of the TCRB repertoire, 55+/−24% of the TCRD repertoire, and 56+/−23% of the TCRG repertoire, all of which were well above the 5% frequency cut-off required for identification of leukemia-associated clonotypes (Figure 1B).
Blood Versus Bone Marrow MRD Quantification
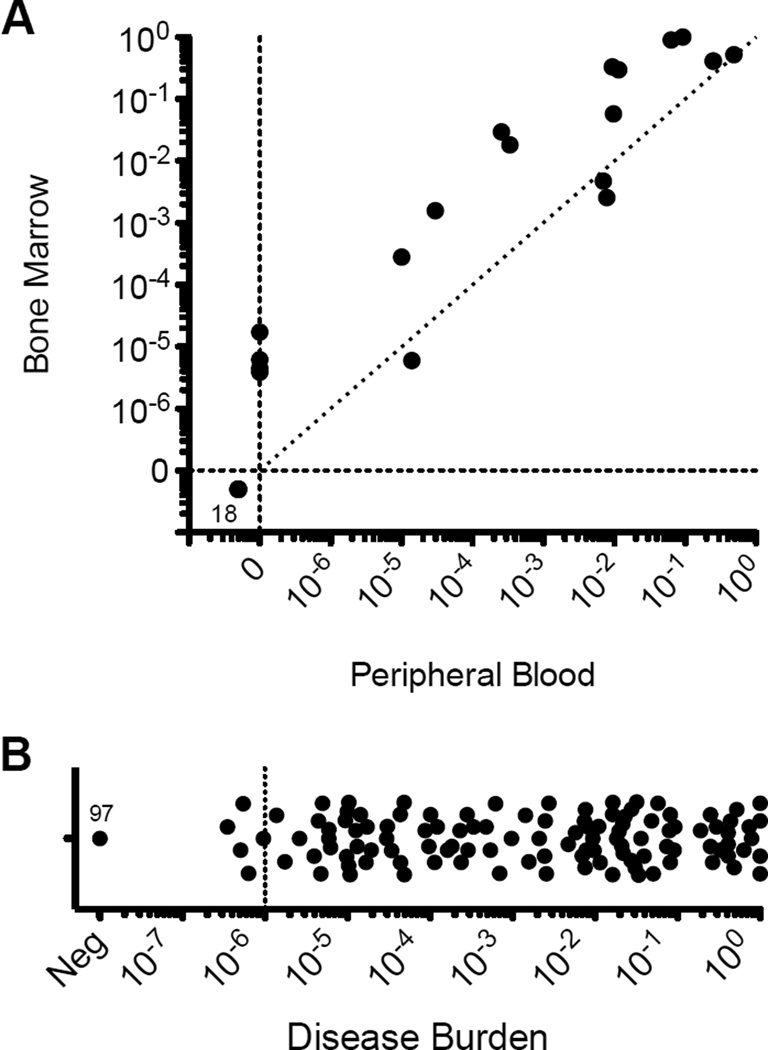
Amongst 37 contemporaneous PB and BM samples evaluated by the LymphoSIGHT platform, 18 (49%) were concordantly MRD negative. Four (11%) were MRD positive in BM but negative in PB, and the median disease burden in these samples was 5.5×10−6 (range 4.5×10−6 – 1.7×10−5). Twelve (32%) exhibited higher MRD in BM (0.18; range 2.8×10−3 – 1.0) versus PB (9.6×10−3; range 1.0×10−5 – 0.49) and 3 (10%) were equivalent or slightly lower in BM (2.6×10−3; range 6×10−6 – 4.7×10−3) than PB (7.0×10−3; range 1.4×10−5 – 7.8×10−3), representing a median 8.4-fold higher (range 0.3 – 115-fold) disease burden in BM (Figure 2A). Additional PB samples with low disease burden (10−6 range) were present in this study, but did not have concurrent marrow samples for this comparison. Amongst the 134 PB specimens without a contemporaneous BM specimen, disease burden was quantifiable in 63 with a disease burden ranging 2.8×10−7 to 1.0 (Figure 2B).
Figure 2. Comparison of molecular MRD sensitivity in bone marrow aspirates and peripheral blood.
In 37 paired bone marrow (BM) aspirate and peripheral blood (PB) samples, MRD quantification was concordantly negative (0) in 18 (49%). MRD quantification was discordantly positive in BM while negative in PB in 4 (11%), and the median disease burden in these BM samples was 5.5×10−6 (range 4.5×10−6 – 1.7×10−5). When MRD was detected in both sample sources, disease burden was higher (median 8.4-fold) in BM than PB (A). MRD quantification in all samples in this study is demonstrated (B).
Prognostic Value of Pre- and Post-HCT MRD Quantification in Blood and Bone Marrow
The leukemia specific clonotypes identified in diagnostic materials were used to screen the Ig/TCR repertoires of pre- and post-HCT PBMC samples for MRD. When IGH-VDJ was clonal, this immunoreceptor was used for MRD quantification, and other Ig/TCR loci, even if found to be rearranged on initial screening, were not queried in remission samples. In patients not possessing a leukemia-specific IGH-VDJ clonotype, MRD samples were screened for the clonotype exhibiting the highest result from the diagnostic sample.
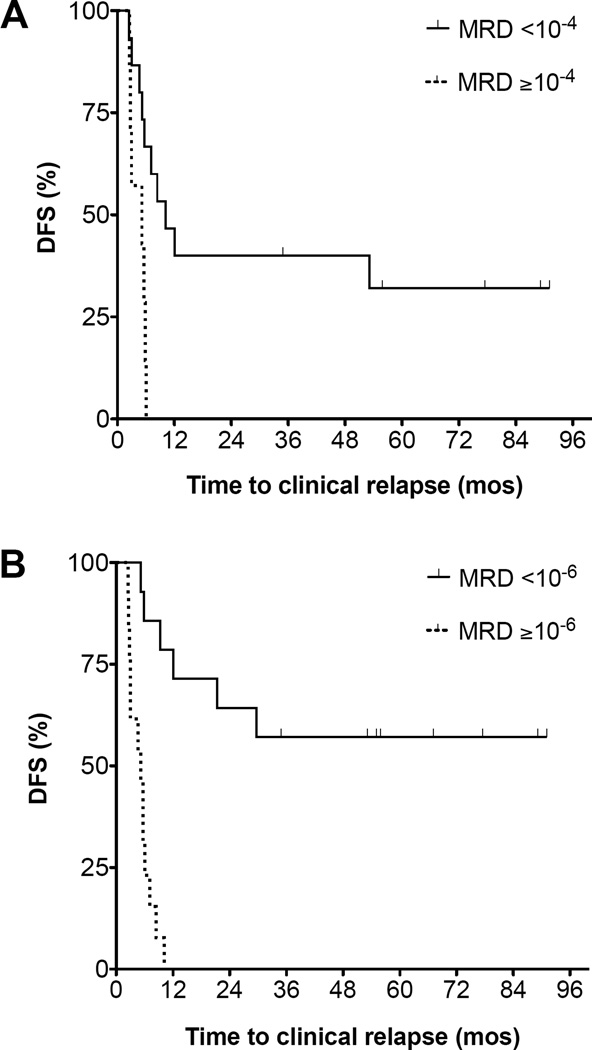
Of the 27 patients with leukemia clonotypes to quantify for MRD assessment, 22 had a blood sample available within 30 days prior to initiation of transplant conditioning. Nine patients (41%) had no MRD detected at the 10−6 detection threshold of the LymphoSIGHT assay. Two patients (9.1%) had disease burden in the 10−6 range, 4 patients (18.2%) had disease burden in the 10−5 range, and MRD was ≥ 10−4 in 7 patients (32%). Disease burden ≥ 10−4 within 30 days prior to HCT was significantly associated with post-HCT relapse (HR 7.7; 95% CI 2.0 – 30; p=0.003) (Figure 3A). The presence of MRD <10−4 was not a significant predictor of post-HCT relapse (p=0.24); however, the number of samples with MRD between 10−4 and 10−6 was limited in this data set.
Figure 3. Disease-free survival impaired by pre- or post-transplant MRD.
ALL disease burden ≥10−4 during the 30 days prior to transplant strongly predicts likelihood of DFS after transplant (HR 7.7, 95% CI 2.0–30, p=0.003) (A). MRD positivity (≥10−6) at any time through day +100 post-transplant predicts subsequent relapse (HR 14; 95% CI 4.7 – 44, p<0.0001) (B).
Samples collected before day 100 post-HCT in 24 patients were evaluated for MRD. Twelve patients (50%) achieved or maintained molecular remission (<10−6) within the first 90 days following HCT and 12 (50%) did not. In the MRD negative group, 6/12 patients (50%) ultimately relapsed with a median time to clinical progression of 320 days post-HCT (range 77–889), whereas 12/12 patients (100%) in the MRD positive group (3 with ≥ 10−4 MRD, 5 between 10−4 to 10−5, and 4 between 10−5 to 10−6) relapsed with a median time to clinical progression of 162 days post-HCT (range 77–304) (HR 14; 95% CI 4.7 – 44; p<0.0001) (Figure 3B).
Time Interval From Molecular Failure to Clinical Events
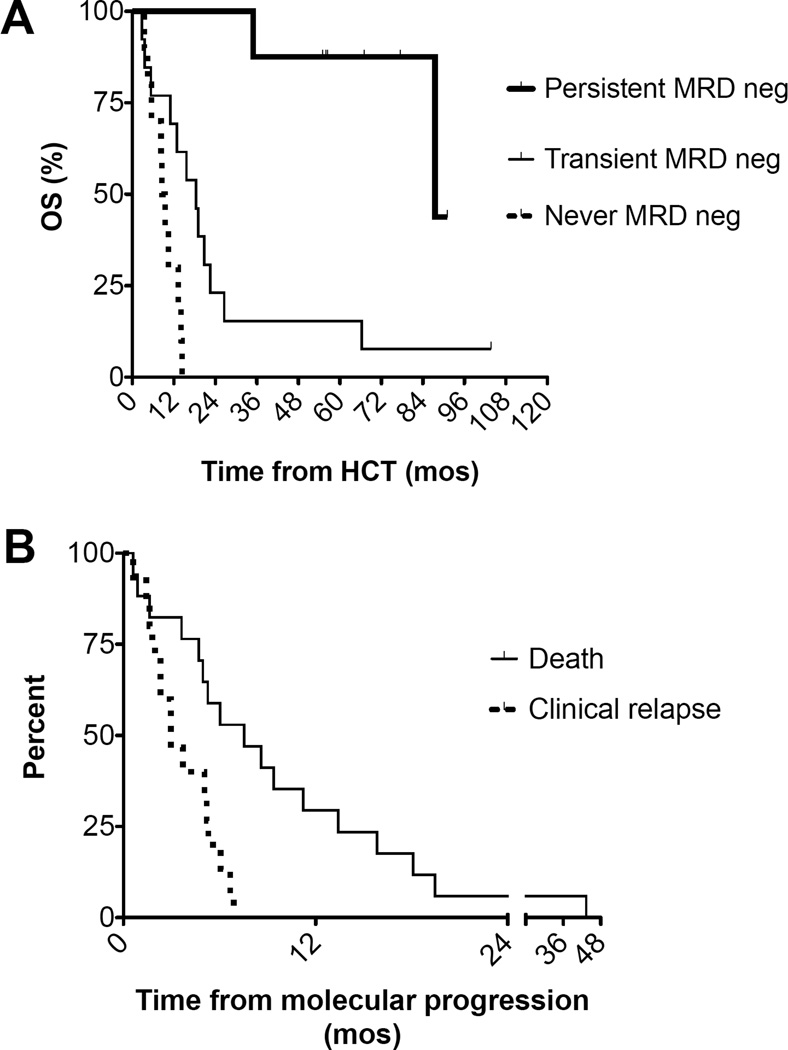
Of the 8 patients who maintained MRD negativity (<10−6) in PB following HCT, 6 (75%) remain alive at a median 1853 days (range 1651–2732 days), and 2 (25%) died in remission from complications of chronic graft-versus-host disease (at 1047 and 2675 days post-HCT). One patient was MRD negative at 34 days post-HCT, and did not have subsequent samples stored prior to relapsing 176 days post-HCT. All 17 patients with MRD ≥ 10−6 detected at any time following HCT relapsed and all died (median survival 359 days; range 85–1991 days) (Figure 4A). The lead time between molecular disease detection by Ig/TCR-HTS and clinical relapse was a median 89 days (range 0–207 days) (Figure 4B).
Figure 4. Overall survival and time to event analysis.
Patients with persistent MRD negativity experienced significantly higher overall survival after hematopoietic cell transplantation (HCT) than patients with transient MRD negativity and failure to achieve MRD negativity (p<0.0001) (A). The Kaplan-Meier estimate of time from molecular progression to clinical relapse and death for patients who relapsed is shown (B). The median time from molecular progression to relapse was 89 days.
DISCUSSION
Minimal residual disease quantification has been successfully incorporated into the majority of pediatric treatment regimens through concerted actions of cooperative study groups. Remarkably, the routine assessment of MRD in adult patients with ALL, who generally have worse prognoses than their pediatric counterparts, remains uncommon. There are likely many factors contributing to this current state of the art, not least of which is the distribution of adult ALL patient care throughout a large number of institutions with very heterogeneous diagnostic capabilities. For many patients, routine MPFC assessment of MRD is not possible; likewise, the development of patient-specific RQ-PCR assays is not feasible. As a consequence of the disparate infrastructure capabilities at centers where adult ALL patients receive treatment, the availability of an assay that does not require patient-specific reagents, yet can be performed on archived or fresh mononuclear cell isolates, offers the potential for innovation in adult ALL management.
The current RQ-PCR method for MRD quantification requires identification of clonal rearrangements via heteroduplex gel electrophoresis, traditional Sanger sequencing of each diagnostic clone, and a laborious process of RQ-PCR primer/probe development and validation for each patient-specific assay. When multiple clonotypes at multiple Ig/TCR genes are present at diagnosis, as is frequently the case in ALL, developing RQ-PCR assays for each clonotype may be unfeasible. As an alternative to this resource-intensive approach, we have developed methods using multiplexed PCR and massively parallel HTS to identify leukemia-marker clones and quantify them in post-treatment clinical specimens. Using this approach, clonal immunoreceptor gene rearrangements may be quantified in a mixture of polyclonal B or T cells, enabling highly sensitive MRD quantification. We previously demonstrated methods for quantifying MRD in CLL using IGH-VDJ clonotypes amplified by consensus-primer multiplex PCR followed by IGH high-throughput sequencing (IGH-HTS) and bioinformatic quantification.26–28
The LymphoSIGHT platform, a robust method for absolute MRD quantification using HTS, has been developed and provides highly sensitive and specific MRD quantification in lymphoid malignancies.18, 27 The LymphoSIGHT platform has also demonstrated adequate sensitivity for detection of NHL-associated clonotypes in cell-free DNA circulating in the peripheral blood.29 The LymphoSIGHT method uses multiplexed PCR and thus has the benefit of being applicable to all patients without the reagent customization required for allele-specific RQ-PCR techniques. Furthermore, due to advantages of scalability and automation of large dataset analysis, the detection threshold of this method is 10−6 with adequately cellular specimens, which cannot easily be achieved using RQ-PCR or flow cytometry. This additional degree of sensitivity permits the assessment of PB for MRD, even in diseases where malignant cells concentrate in the BM.
Whereas most CLL isolates have a stable IGH clonotype exhibiting limited clonal evolution,26, 30 the IGH locus is not clonally rearranged in up to half of patients with ALL. The IGH locus may also be prone to clonal evolution in ALL lymphoblasts by VH segment replacement, meaning the locus may evolve as the malignancy progresses or relapses following therapy.31–33 This phenomenon cannot easily be accounted for by standard RQ-PCR techniques, since subclones may not be captured by the heteroduplex gel electrophoresis method, and any RQ-PCR assay will only detect a single clonotype. Using massively parallel IGH-HTS, it is possible to identify and quantify clonotypes that are related by D-N-J sequence identity, but differ in VH segment sequences.33
From a methodological perspective, ALL presents challenges to the universal application of Ig/TCR-HTS that are important to resolve. For one, several studies have reported 25–50% of B-ALL patients do not have a disease-associated IGH-VDJ clonotype.19–21 In addition, as discussed above, clonal switch at the IGH locus may limit its usefulness as a post-therapy disease marker,23, 31 although IGH oligoclonality appears to be less common in adult patients than in pediatric patients.34 Fortunately, other genetic targets for clonotyping ALL exist which may overcome the limitations imposed by lack of stable IGH clonotypes, including partial IGH-DJ rearrangements, which likely capture an additional 10–30% of patients,20, 22, 23 or rearrangements of the IGL or IGK loci which may be detectable in up to 50% of patients in some series.19 Additionally, TCRB, TCRD or TCRG loci are rearranged in a significant percentage of both B-cell (30–50%) and T-cell (up to 75%) ALL isolates.20, 24, 25 Performing consensus PCR at all these loci is feasible, but the routine development of RQ-PCR assays given the multitude of possible targets is not scalable to widespread use.
In the work presented here, we have demonstrated application of the LymphoSIGHT platform to ALL MRD quantification before and after allo-HCT. The advantages of the LymphoSIGHT method are manifold, including: 1) wide access to MRD quantification via a central reference lab (or labs, depending on assay volume within a region) with standardized processes for sample handling and clone identification, 2) the use of multiplexed PCR to capture relevant immunoreceptor clonotypes associated with the leukemic clone to permit the complexity of leukemic oligoclonality to be quantified, and 3) the high degree of sensitivity that can be achieved, provided a suitable sample with adequate leukocyte genome input.
Using the LymphoSIGHT platform for clonotype identification, we identified Ig/TCR rearrangements suitable for MRD quantification in 93% of patients in this series. Of the 27 patients with leukemic isolates exhibiting molecular clonality, over 50% had more than one clonal locus suitable for MRD quantification, which correlates well with other studies using standard techniques for clonality identification.13 Although LymphoSIGHT can be used to address oligoclonality and clonal evolution, we elected not to address clonal evolution during the treatment courses of patients in this cohort since for some cases the only sample available for identification of leukemia-associated clonotypes was a relapse sample. Comparison of Ig/TCR-HTS-quantified leukemia burdens in PB and BM revealed the latter to have a median 8.4-fold higher disease burden. This finding suggests that the deep sensitivity of the LymphoSIGHT method may be useful for quantifying MRD in PB, even when BM disease burden is in the 10−4 range. This will require additional validation in a larger cohort of patients, however, and we note that it is sometimes not possible to achieve 10−6 sensitivity when fewer than one million recoverable cell genomes are present in cytopenic blood samples at early post-transplant time points in some patients.
Pre-HCT MRD quantification using Ig/TCR-HTS analysis of PB or BM samples is predictive of post-HCT relapse when disease burden before conditioning is ≥ 10−4. Post-HCT MRD ≥ 10−6 is highly associated with relapse and poor survival. In this study, only 5 patients were treated with non-myeloablative (NMA) conditioning, so the vast majority received myeloablative conditioning, a setting in which detectable residual disease burden, even at the 10−6 level, within the first 3 months post-HCT appears to be associated with relapse and extremely limited salvageability. Although additional study in the non-myeloablative setting will be required since the kinetics of disease clearance may differ in comparison with patients undergoing myeloablative conditioning, we note that none of the NMA conditioned patients in this series who were MRD positive during the first 100 days post-HCT subsequently became MRD negative.
We found that quantification of MRD in PB gives a lead time to clinical relapse of roughly 3 months, which is similar to the findings of Uzunel and colleagues using RQ-PCR-based MRD quantification in BM aspirates after allo-HCT.35 Thus, the higher sensitivity of this HTS approach for MRD quantification is a clinically useful assay characteristic that enables assessment of PB samples on a more frequent schedule than would likely be feasible for most patients if only BM is assessed. Increasing the frequency of molecular disease measurement will likely improve the diagnostic lead-time prior to clinical relapse and could provide an opportunity to apply additional therapeutic maneuvers while disease burden is low. Topp and colleagues recently demonstrated, for example, the utility of treating ALL patients with MRD >10−4 after induction chemotherapy with an immune targeting therapy, blinatumomab, which directs T cell eradication of CD19+ B cells, including ALL lymphoblasts.36, 37
Our study confirms that in the post-allogeneic-HCT setting, ALL MRD positivity is associated with extremely high risk of relapse and uniformly poor outcomes. Bone marrow assessment is not always feasible, and frequent BM aspiration is generally undesirable for most patients, so quantifying MRD in PB using highly sensitive Ig/TCR-HTS may yield significantly improved feasibility for post-HCT MRD monitoring plans. The dismal outcome of patients with ALL who relapse after allo-HCT demands further clinical study of both post-transplant MRD monitoring and novel methods for treating ALL relapse in evolution.
Acknowledgments
This work was supported, in part, by American Society of Hematology Research Training Award (to ACL), National Cancer Institute P01 CA049605, Leukemia and Lymphoma Translational Research Award (R618-09) (to DBM), and Stanford University Cancer Institute-supported grant 1P030 CA124435-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
ACL performed laboratory and informatics work for the study and wrote the paper. DBM was the principal investigator and wrote the paper. MF performed informatics work for the study and wrote the paper. NV, VC, KK, IB, JZ, MM, MK, BZ, AW, and JL performed laboratory or informatics work for the study. MF, VC, KK, JZ, MM, and MK are employees and holders of equity in Sequenta, Inc.
Disclosures
Other authors report no potential conflicts of interest.
REFERENCES
- 1.Bruggemann M, Gokbuget N, Kneba M. Acute lymphoblastic leukemia: monitoring minimal residual disease as a therapeutic principle. Semin. Oncol. 2012;39(1):47–57. doi: 10.1053/j.seminoncol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Bunin N, Johnston DA, Roberts WM, Ouspenskaia MV, Papusha VZ, Brandt MA, et al. Residual leukaemia after bone marrow transplant in children with acute lymphoblastic leukaemia after first haematological relapse or with poor initial presenting features. Br. J. Haematol. 2003;120(4):711–715. doi: 10.1046/j.1365-2141.2003.04135.x. [DOI] [PubMed] [Google Scholar]
- 3.Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115(16):3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 4.Bader P, Hancock J, Kreyenberg H, Goulden NJ, Niethammer D, Oakhill A, et al. Minimal residual disease (MRD) status prior to allogeneic stem cell transplantation is a powerful predictor for post-transplant outcome in children with ALL. Leukemia. 2002;16(9):1668–1672. doi: 10.1038/sj.leu.2402552. [DOI] [PubMed] [Google Scholar]
- 5.Knechtli CJ, Goulden NJ, Hancock JP, Grandage VL, Harris EL, Garland RJ, et al. Minimal residual disease status before allogeneic bone marrow transplantation is an important determinant of successful outcome for children and adolescents with acute lymphoblastic leukemia. Blood. 1998;92(11):4072–4079. [PubMed] [Google Scholar]
- 6.Knechtli CJ, Goulden NJ, Hancock JP, Harris EL, Garland RJ, Jones CG, et al. Minimal residual disease status as a predictor of relapse after allogeneic bone marrow transplantation for children with acute lymphoblastic leukaemia. Br. J. Haematol. 1998;102(3):860–871. doi: 10.1046/j.1365-2141.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 7.van Dongen JJ, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet. 1998;352(9142):1731–1738. doi: 10.1016/S0140-6736(98)04058-6. [DOI] [PubMed] [Google Scholar]
- 8.Bruggemann M, Raff T, Flohr T, Gokbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107(3):1116–1123. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 9.Gokbuget N, Kneba M, Raff T, Trautmann H, Bartram CR, Arnold R, et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood. 2012;120(9):1868–1876. doi: 10.1182/blood-2011-09-377713. [DOI] [PubMed] [Google Scholar]
- 10.EuroMRD. EuroMRD: A division of European Scientific Foundation for Laboratory HematoOncology (ESHLO) [Accessed November 13, 2013]; url: http://www.euromrd.org. [Google Scholar]
- 11.van Dongen JJ, Orfao A, EuroFlow C. EuroFlow: Resetting leukemia and lymphoma immunophenotyping. Basis for companion diagnostics and personalized medicine. Leukemia. 2012;26(9):1899–1907. doi: 10.1038/leu.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruggemann M, Raff T, Kneba M. Has MRD monitoring superseded other prognostic factors in adult ALL? Blood. 2012;120(23):4470–4481. doi: 10.1182/blood-2012-06-379040. [DOI] [PubMed] [Google Scholar]
- 13.Scrideli CA, Queiroz RG, Bernardes JE, Valera ET, Tone LG. PCR detection of clonal IgH and TCR gene rearrangements at the end of induction as a non-remission criterion in children with ALL: comparison with standard morphologic analysis and risk group classification. Med. Pediatr. Oncol. 2003;41(1):10–16. doi: 10.1002/mpo.10154. [DOI] [PubMed] [Google Scholar]
- 14.Ryan J, Quinn F, Meunier A, Boublikova L, Crampe M, Tewari P, et al. Minimal residual disease detection in childhood acute lymphoblastic leukaemia patients at multiple time-points reveals high levels of concordance between molecular and immunophenotypic approaches. Br. J. Haematol. 2009;144(1):107–115. doi: 10.1111/j.1365-2141.2008.07429.x. [DOI] [PubMed] [Google Scholar]
- 15.Malec M, van der Velden VH, Bjorklund E, Wijkhuijs JM, Soderhall S, Mazur J, et al. Analysis of minimal residual disease in childhood acute lymphoblastic leukemia: comparison between RQ-PCR analysis of Ig/TcR gene rearrangements and multicolor flow cytometric immunophenotyping. Leukemia. 2004;18(10):1630–1636. doi: 10.1038/sj.leu.2403444. [DOI] [PubMed] [Google Scholar]
- 16.Neale GA, Coustan-Smith E, Stow P, Pan Q, Chen X, Pui CH, et al. Comparative analysis of flow cytometry and polymerase chain reaction for the detection of minimal residual disease in childhood acute lymphoblastic leukemia. Leukemia. 2004;18(5):934–938. doi: 10.1038/sj.leu.2403348. [DOI] [PubMed] [Google Scholar]
- 17.van der Velden VH, Jacobs DC, Wijkhuijs AJ, Comans-Bitter WM, Willemse MJ, Hahlen K, et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukemia (ALL), but not in precursor-B-ALL. Leukemia. 2002;16(8):1432–1436. doi: 10.1038/sj.leu.2402636. [DOI] [PubMed] [Google Scholar]
- 18.Faham M, Zheng J, Moorhead M, Carlton VE, Stow P, Coustan-Smith E, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120(26):5173–5180. doi: 10.1182/blood-2012-07-444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assumpcao JG, Ganazza MA, de Araujo M, Silva AS, Scrideli CA, Brandalise SR, et al. Detection of clonal immunoglobulin and T-cell receptor gene rearrangements in childhood acute lymphoblastic leukemia using a low-cost PCR strategy. Pediatr. Blood Cancer. 2010;55(7):1278–1286. doi: 10.1002/pbc.22709. [DOI] [PubMed] [Google Scholar]
- 20.Dawidowska M, Jolkowska J, Szczepanski T, Derwich K, Wachowiak J, Witt M. Implementation of the standard strategy for identification of Ig/TCR targets for minimal residual disease diagnostics in B-cell precursor ALL pediatric patients: Polish experience. Arch. Immunol. Ther. Exp. (Warsz.) 2008;56(6):409–418. doi: 10.1007/s00005-008-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toubai T, Tanaka J, Ota S, Fukuhara T, Hashino S, Kondo T, et al. Minimal residual disease (MRD) monitoring using rearrangement of T-cell receptor and immunoglobulin H gene in the treatment of adult acute lymphoblastic leukemia patients. Am. J. Hematol. 2005;80(3):181–187. doi: 10.1002/ajh.20461. [DOI] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Csinady E, van der Velden VH, Joas R, Fischer S, de Vries JF, Beverloo HB, et al. Chromosome 14 copy number-dependent IGH gene rearrangement patterns in high hyperdiploid childhood B-cell precursor ALL: implications for leukemia biology and minimal residual disease analysis. Leukemia. 2009;23(5):870–876. doi: 10.1038/leu.2008.390. [DOI] [PubMed] [Google Scholar]
- 24.van der Velden VH, Bruggemann M, Hoogeveen PG, de Bie M, Hart PG, Raff T, et al. TCRB gene rearrangements in childhood and adult precursor-B-ALL: frequency, applicability as MRD-PCR target, and stability between diagnosis and relapse. Leukemia. 2004;18(12):1971–1980. doi: 10.1038/sj.leu.2403505. [DOI] [PubMed] [Google Scholar]
- 25.Wu D, Sherwood A, Fromm JR, Winter SS, Dunsmore KP, Loh ML, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 2012;4(134):134ra63. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 26.Logan AC, Gao H, Wang C, Sahaf B, Jones CD, Marshall EL, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc. Natl. Acad. Sci. U. S. A. 2011;108(52):21194–21199. doi: 10.1073/pnas.1118357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logan AC, Zhang B, Narasimhan B, Carlton V, Zheng J, Moorhead M, et al. Minimal residual disease quantification using consensus primers and high-throughput IGH sequencing predicts post-transplant relapse in chronic lymphocytic leukemia. Leukemia. 2013;27(8):1659–1665. doi: 10.1038/leu.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armand P, Oki Y, Neuberg DS, Faham M, Cummings C, Klinger M, et al. Detection of circulating tumour DNA in patients with aggressive B-cell non-Hodgkin lymphoma. Br. J. Haematol. 2013;163(1):123–126. doi: 10.1111/bjh.12439. [DOI] [PubMed] [Google Scholar]
- 30.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl. Acad. Sci. U. S. A. 2008;105(35):13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imashuku S, Terui K, Matsuyama T, Asami K, Tsuchiya S, Ishii E, et al. Lack of clinical utility of minimal residual disease detection in allogeneic stem cell recipients with childhood acute lymphoblastic leukemia: multi-institutional collaborative study in Japan. Bone Marrow Transplant. 2003;31(12):1127–1135. doi: 10.1038/sj.bmt.1704067. [DOI] [PubMed] [Google Scholar]
- 32.Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 2009;206(8):1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gawad C, Pepin F, Carlton VE, Klinger M, Logan AC, Miklos DB, et al. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood. 2012;120(22):4407–4417. doi: 10.1182/blood-2012-05-429811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brisco MJ, Latham S, Sutton R, Hughes E, Wilczek V, van Zanten K, et al. Determining the repertoire of IGH gene rearrangements to develop molecular markers for minimal residual disease in B-lineage acute lymphoblastic leukemia. J Mol Diagn. 2009;11(3):194–200. doi: 10.2353/jmoldx.2009.080047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzunel M, Jaksch M, Mattsson J, Ringden O. Minimal residual disease detection after allogeneic stem cell transplantation is correlated to relapse in patients with acute lymphoblastic leukaemia. Br. J. Haematol. 2003;122(5):788–794. doi: 10.1046/j.1365-2141.2003.04495.x. [DOI] [PubMed] [Google Scholar]
- 36.Topp MS, Gokbuget N, Zugmaier G, Degenhard E, Goebeler ME, Klinger M, et al. Long-term follow-up of hematologic relapse-free survival in a phase 2 study of blinatumomab in patients with MRD in B-lineage ALL. Blood. 2012;120(26):5185–5187. doi: 10.1182/blood-2012-07-441030. [DOI] [PubMed] [Google Scholar]
- 37.Klinger M, Brandl C, Zugmaier G, Hijazi Y, Bargou RC, Topp MS, et al. Immunopharmacologic response of patients with B-lineage acute lymphoblastic leukemia to continuous infusion of T cell-engaging CD19/CD3-bispecific BiTE antibody blinatumomab. Blood. 2012;119(26):6226–6233. doi: 10.1182/blood-2012-01-400515. [DOI] [PubMed] [Google Scholar]