Abstract
Helicobacter pylori remains one of the most common bacterial infections worldwide. Clarithromycin resistance is the most important cause of H. pylori eradication failures. Effective antibiotic therapies in H. pylori infection must be rapidly adapted to local resistance patterns. We investigated the prevalence of clarithromycin resistance due to mutations in positions 2142 and 2143 of 23SrRNA gene of H. pylori by fluorescence in situ hybridisation (FISH), and compared with culture and antimicrobial susceptibility testing in 234 adult patients with dyspepsia who were enrolled. Antrum and corpus biopsy specimens were obtained for rapid urease test, histopathology and culture. Epsilometer test was used to assess clarithromycin susceptibility. H. pylori presence and clarithromycin susceptibility were determined by FISH in paraffin‐embedded biopsy specimens. We found that 164 (70.1%) patients were positive for H. pylori based on clinical criteria, 114 (69.5% CI 62.5–76.6%) were culture positive, and 137 (83.5% CI 77.8–89.2%) were FISH positive. Thus the sensitivity of FISH was significantly superior to that of culture. However specificity was not significantly different (91.4 versus 100.0%, respectively). The resistance rate to clarithromycin for both antrum and corpus was detected in H. pylori‐positive patients; 20.2% by FISH and 28.0% by E‐test.The concordance between E‐test and FISH was only 89.5% due to the presence of point mutations different from A2143G, A2142G or A2142C. We conclude that FISH is significantly more sensitive than culture and the E‐test for the detection of H. pylori and for rapid determinination of claritromycin susceptibility. The superior hybridisation efficiency of FISH is becoming an emerging molecular tool as a reliable, rapid and sensitive method for the detection and visualisation of H. pylori, especially when the management of H. pylori eradication therapy is necessary. This is particularly important for the treatment of patients with H. pylori eradication failure.
Keywords: clarithromycin resistance, FISH, H. pylori detection
Introduction
Helicobacter pylori is the aetiological agent of gastritis, gastric and duodenal ulcers, gastric adenocarcinoma and mucosa‐associated lymphoid tissue (MALT) lymphoma 1, 2, 3, 4, 5, 6, 7, 8, and its eradication depends on the choice of antibiotics to which the organism is susceptible 1, 4, 9. Disease outcome in H. pylori is associated with several virulence factors 10 including CagA, which is present in 60% of the strains 11. Triple therapy includes a proton pump inhibitor (PPI) in combination with two antibiotics: amoxicillin, clarithromycin or metranidazole 8, 12, 13, 14, 15, 16, 17, 18, 19. Some authors recommend the use of metranidazole instead of clarithromycin in regions where the resistance to this antibiotic exceeds 15–20% 13, 16, 19. The recent Maastricht IV Consensus reports and other studies recommended more than seven days of triple therapy for eradication of H. pylori. Treatment of H. pylori is important since gastric cancer risk decreases significantly in patients without pre‐malignant lesions who receive treatment 1, and also has a low relapse rate in patients with duodenal ulcer 20. Because H. pylori eradication failure is mainly associated with clarithromycin resistance, it is important to know the prevalence of resistance to this antibiotic in the different regions of the world 7, 21, 22. The prevalence of clarithromycin resistant H. pylori is as high as 10% in France and Belgium, 27% in Italy 23 and 24.2% in Turkey 24.
Clarithromycin binds to the 50S ribosomal subunit in the 23S rRNA and inhibits protein synthesis 4, 18, 23, 25, 26, 27, 28. Resistance to clarithromycin is associated with three main point mutations at positions A to G at 2142, 2143 and A to C at 2142 of the 23S rRNA gene 4, 6, 17, 23, 25, 29, 30, 31, 32, 33, 34. All antibiotic resistance mechanisms in H. pylori seem to be chromosomally mediated 27. Novel technologies that include in situ hybridisation for clarithromycin resistance on gastric biopsies are excellent options if culture is not possible 35, 36, 37.
Traditional culture methodology is expensive and rarely available, therefore, antibiotic susceptibility testing is not performed routinely. Agar dilution, broth dilution, disc diffusion test and Epsilometer test (E‐test) are phenotypic methods used for assessment of clarithromycin susceptibility, but there is a need to obtain fast and more sensitive results using molecular tests rather than phenotypic methods 35, 36. Fluorescence in situ hybridisation (FISH) is a molecular technique that combines the detection of H. pylori and the determination of clarithromycin susceptibility, and correlates well with the results obtained by traditional culture methodology and clarithromycin susceptibility assay by E‐test as recommended in the Maastricht IV Consensus Report 2, 4, 19, 38.
FISH allows the morphology of whole bacteria to be seen 5. FISH can be performed on formalin‐fixed paraffin embedded tissue, on frozen antrum and corpus gastric biopsies, or isolated H. pylori colonies using fluorescence‐labeled oligonucleotide probes which hybridise to specific rRNA sequences 2, 5, 6, 8, 15, 22, 39. A major limitation is that the molecular basis for clarithromycin resistance may differ by country so the system needs to be individualised and occasionally checked against culture 36. However, more than 90% of the clarithromycin resistant H. pylori isolates have been associated with the three common point mutations mentioned above and which are included in FISH 40. In this study, we evaluated the efficacy of FISH for the detection of H. pylori, and for the determination of clarithromycin resistance due to mutations in the 2142 and 2143 positions of 23S rRNA gene. The results were compared with traditional culture and antimicrobial susceptibility testing results.
Methods
Patients
Two hundred and thirty four patients with dyspepsia (65 male, 169 female; mean age 43.8 ± 14.0 years, age range 17–83) were admitted to the Outpatient Gastroenterology Clinic and Endoscopy Unit at Dokuz Eylül University Hospital and referred to upper endoscopy between April 2006 and February 2011. The patients included in the study were treatment naïve before endoscopy was performed. Patients were excluded if they had received antibiotics in the previous month, had received proton pump inhibitors (PPIs), or had prior gastric surgery. Patients were also excluded if they were pregnant or had gastrointestinal malignancy, alcohol abuse, drug addiction, or chronic use of corticosteroids or nonsteroidal anti‐inflammatory drugs. All patients provided written informed consent to participate in this study and demographic information was obtained.
Endoscopy and gastric biopsy sampling
Three antrum and corpus biopsies were taken from each patient: one set of antrum and corpus biopsies was used for rapid urease test (RUT); a second set was fixed and transported in 10% formalin solution for histopathological examination. Finally the last set of biopsies was immediately transported to the Medical Microbiology Laboratory and processed for culture.
Rapid urease test
One biopsy from the antrum and one biopsy from the corpus were used for RUT. Antrum and corpus biopsy specimens were placed separately in tubes containing urea solution, and then two drops of 1% (vol/vol) phenol red solution were added. If the indicator solution changed from yellow to pink, it was considered a positive result. If the indicator did not change, it was considered a negative result. The results of this house‐made RUT were recorded in less than 24 h.
Histopathological examination of biopsy specimens
Paraffin‐embedded gastric biopsy specimens were routinely processed. Haematoxylin and eosin, Alcian blue and Giemsa stains were used for grading bacterial density and gastritis activity according to the updated Sydney System 41. A four‐point grade scale of none (grade 0), mild (grade 1), moderate (grade 2) and severe (grade 3) was used to score the presence of chronic superficial gastritis, active gastritis, gastric atrophy, intestinal metaplasia (IM) and Helicobacter‐like organisms (HLO). Histopathology evaluation for all patients was performed by a single pathologist (SS) for consistency and she was unaware of the results of culture and FISH.
H. pylori status criteria used
We used the test‐and‐treat criteria, which dictate that any positive test for H. pylori should be interpreted as patient positive for H. pylori infection 19.
H. pylori status was defined as positive when one of two diagnostic tests – RUT and/or histopathology – were positive. A patient was classified as being H. pylori negative when histopathological examination and RUT were both negative.
H. pylori culture
Antrum and corpus biopsies were cultured either onto (1) Tripticase soy agar containing 5% (vol/vol) sheep blood (TSA) (Beckton Dickinson, [BD] Sparks MD, USA), and Skirrow Medium (BD) containing antibiotics (Trimethoprim, Vancomycin, and Polymixin B) supplemented with 5% (vol/vol) horse blood (BD), or (2) Columbia Blood Agar (Oxoid, Basingstoke, Hampshire, England) containing 7% (vol/vol) defibrinated horse blood (Oxoid) and H. pylori selective supplement (DENT) (Oxoid). Plates were incubated at 37 °C in an anaerobic jar containing GasPak Campy Container System (BD) for 3–7 days under microaerophilic conditions. In the case of no growth at the seventh day, all cultures were held for up to 14 days which is required for initial isolation. After incubation, colony morphology, microscopic examination for motility, and Gram staining for morphology were documented and urease, catalase, and oxidase tests were performed to identify H. pylori. H. pylori strains were stored at −80 °C in brain heart infusion (BHI) broth (Oxoid) containing 20% (vol/vol) glycerol.
H. pylori NCTC 11637 reference strain was used in this study as a positive control for culture and antimicrobial susceptibility testing in the E‐test.
Antimicrobial susceptibility testing
Gradient diffusion test (E‐test) (AB Biodisk, Solna, Sweden) was used to assess clarithromycin susceptibility in isolated H. pylori strains. The bacterial inoculum was prepared in Brucella Broth (BBL) from subcultures grown on Columbia Blood Agar (Oxoid) containing 7% (vol/vol) defibrinated horse blood (Oxoid) and H. pylori selective supplement (DENT) (Oxoid). The Mueller‐Hinton agar (Oxoid) supplemented with 5% (vol/vol) defibrinated sheep blood (Oxoid) was used to inoculate 100 µl of H.pylori culture suspension of a McFarland standard 3 (∼109 CFU/ml) turbidity in Brucella broth (BBL). Plates with clarithromycin E‐test strips were incubated at 37 °C in a jar including the GasPak Campy Container System (BD) under microaerophilic conditions for 72 h. The minimum inhibitory concentration (MIC) was considered the lowest concentration of the drug that inhibited visible growth of H. pylori. Isolates were defined as clarithromycin resistant when the MIC was ≥1 µg/ml, susceptible when MIC was <0.5 µg/ml, and intermediate when MIC was 0.5–1 µg/ml according to Clinical Laboratory Standard Institutes (CLSI) recommendation 42. However, these breakpoints were defined only for the agar dilution method and they have not yet been established for the E‐test. A plate without antibiotic strip was used to confirm the purity of the culture and the lack of contamination.
FISH
Paraffin‐embedded antrum and corpus biopsy sections were examined by FISH (BactFISH H. pylori Combi‐Kit IZINTA Trading Co. Ltd., Budapest, Hungary) as previously described 22. This kit detects several point mutations of the 23S rRNA gene of H. pylori at positions A2143G, A2144G, A2143C. In brief, formalin‐fixed, paraffin‐embedded biopsy sections were deparaffinised 6, 15, 17, 22, 39. After air‐drying, 40 μl of DNA hybridisation solution containing FISH probes 5′ labeled with FITC or Cy3 was added at 46 °C for 90 min for hybridisation. The hybridised slide was rinsed with wash buffer to prevent non‐specific hybridisation and washed for 15 min at 48 °C in a high‐density polyethylene coplin staining jar with plastic screw cap (SIGMA Co. Saint Louis MO, USA). Air‐dried slides were counterstained with 10 µl of 4′, 6′‐diamidino‐2‐phenylindole (DAPI/Antifade‐MC QBIOgene) to visualise DNA for 5 min at room temperature 7, 22. Probe specificity was confirmed using control slides as previously reported 22. H. pylori was visualised with a Nikon Eclipse E600 epifluorescence microscope (Nikon Corp., Tokyo, Japan) that included five different filter sets for DAPI, FITC, rhodamine, dual band (FITC/Rhodamine), triple band (DAPI/FITC/Rhodamine) automatised inter‐changeable filters. Individual single‐colour images were captured through a high‐sensitivity monochrome charge‐coupled device (CCD) camera (Figure 1). Quantification of H. pylori was performed using a semiquantitative grading system (1+ to 3+). Digitally captured individual image photographs were overlaid and processed with MacProbe imaging software (PSI Scientific Systems, USA).
Figure 1.
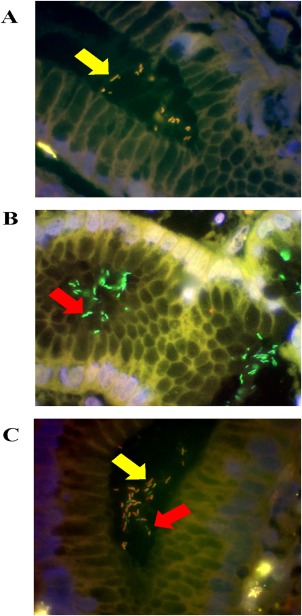
Detection of H. pylori and determination of clarithromycin susceptibility in gastric biopsies from three different patients by FISH. Probes were visualised using a triple filter. Green fluorescence indicates clarithromycin sensitive H. pylori (B and C); yellow fluorescence indicates clarithromycin resistant H. pylori (A and C). The DAPI counterstain produces blue fluorescence (A, B and C). Mixed infection is present within the same biopsy specimen in panel C. Arrows indicate the presence of H. pylori infecting the gastric mucosa (Magnification, X100).
Extraction of H. pylori genomic DNA
Bacterial genomic DNA was obtained from isolated H. pylori strains. DNA was extracted using the Wizard Genomic DNA isolation kit (Promega Madison, WI, USA), or the High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany). All DNA products were measured using a Nanodrop 1000 Spectrophotometer (Thermo Scientific Pittsburgh, PA, USA).
Determination of H. pylori and cagA positivity by PCR
H. pylori strains were determined for the presence of the ureA gene by HPU PCR with the primers HPU‐1 (5′‐GCCAATGGTAAATTAGTT‐3′) and HPU‐2 (5′‐CTCCTTAATTGTTTTTAC‐3′) 33. The presence of the cagA gene was determined by CagA PCR with the primers cagA2530S (5′‐GTTAARAATRGTGTRAAYGG‐3′) and cagA3000AS (5′‐TTTAGCTTCTGATACCGC‐3′) 43. PCR conditions have been previously reported 43, 44.
Statistical analysis
The McNemar's χ2 test was used. The sensitivity, specificity, positive predictive values (PPV) and negative predictive values (NPV) were calculated (SPSS version 15.0). The results were considered to be significant at a p‐value of <0.05. Fisher exact test and Kappa confidence were also used.
Ethics
The research protocol for this study was approved by the Institutional Review Board, and the Ethical Committee, of the Dokuz Eylül University, Faculty of Medicine (13.07.2006/166).
Results
One hundred sixty four of 234 patients (70.1%) were diagnosed as H. pylori positive by histopathology and/or RUT methods. The descriptive demographic information of H. pylori positive and H. pylori negative patients is shown in Table 1. Amongst 164 H.pylori positive patients, 93.9% were positive by both histology and RUT (Table 2). By culture, 114 (69.5%) patients were positive. The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for culture were 69.5, 100.0, 100.0, and 58.3%, respectively (κ = 0.577). We compared the two different culture methods for H. pylori. For the first part of the study 64 patients or 128 biopsies of antrum and corpus were processed in NYU School of Medicine and the last 340 gastric biopsies of 170 patients were processed at Izmir, Dokuz Eylül University Faculty of Medicine. We found no significant differences in the recovery of H. pylori by location (56.3% in New York versus 53.5% in Izmir).
Table 1.
Demographic characteristics of 234 dyspeptic Turkish patients according to H. pylori status
| H. pylori infection (n = 234) | ||
|---|---|---|
| Negative | Positive | |
| Number | 70 | 164 |
| Mean age | 43.1 + 15.6 | 44.0 + 13.3 |
| Median age | 43.5 | 46 |
| Age range | 20–83 | 17–75 |
| F:M ratio | 2.89 | 2.49 |
Table 2.
Biopsy‐based results of the 164 patients identified as H. pylori positive
| Histology | RUT | Number (%) | FISH (%) | Culture (%) |
|---|---|---|---|---|
| Negative | Positive | 4 (2.4) | 2 (50.0) | 4 (100.0) |
| Positive | Negative | 7 (4.3) | 6 (85.7) | 6 (85.7) |
| Positive | Positive | 153 (93.3) | 129 (84.3) | 104 (68.0) |
RUT = rapid urease test; FISH = fluorescence in situ hybridisation.
Of 164 H. pylori positive patients, 137 (83.5%) were H. pylori positive by FISH method. The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of FISH were 83.5, 91.4, 95.8 and 70.3%, respectively (κ = 0.690). Despite the high specificity and positive predictive value of culture, the FISH method was able to detect significantly more H. pylori positive subjects than culture (83.5 versus 69.5%, p =0.004). The sensitivity of FISH and the patchy distribution of H. pylori infection allowed the identification of two positive FISH subjects who were negative by traditional histology (Table 2).
Susceptibility testing was only possible in 114 (69.5%) patients with positive cultures. In contrast, we evaluated 137 (83.5%) patients by FISH due to the higher sensitivity of this method. We assessed clarithromycin susceptibility by E‐test in the 114 culture positive patients, and found 32 patients (28.0%) carrying H. pylori strains resistant to clarithromycin. On the other hand, when we assessed H. pylori clarithromycin susceptibility by FISH in the same patients, resistance was demonstrated in 23 (20.2%) of those patients. The difference in patients with resistant strains between the two methods was not significant. The concordance between E‐test and FISH was 89.5% (Table 3).
Table 3.
Comparison of E‐test and FISH results for both antrum and corpus biopsy specimens in 114 culture positive patients
| Results obtainedby FISH | Number of strains (%) | Clarithromycin by E‐test | |
|---|---|---|---|
| Resistant (%) | Sensitive (%) | ||
| Positive for clarithromycin resistance | 23 (20.2) | 19 (82.6) | 4 (17.4) |
| Positive for clarithromycin sensitive | 87 (76.3) | 12 (13.8) | 75 (86.2) |
| Negative for H. pylori | 4 (3.5) | 1 (25.0) | 3 (75.0) |
FISH = fluorescence in situ hybridisation.
Next, we compared the prevalence of clarithromycin resistant strains by specific location in the stomach. H. pylori isolates were recovered from 102 antrum biopsies (89.5%) and from 96 corpus biopsies (84.2%) in the 114 culture positive patients. Amongst antrum positive H. pylori isolates, we identified 26 (25.5%) clarithromycin‐resistant strains by E‐test but only 15 (14.7%) by FISH. This result suggests that FISH has lower sensitivity for the detection of clarithromycin resistant H. pylori strains in antrum biopsies although the difference was not significant (p‐value = 0.06). Amongst the corpus biopsy results, we identified 20 (20.8%) clarithromycin‐resistant H. pylori strains by E‐test and 15 (15.6%) by FISH. The differences observed between the methods in the corpus were also not significant (Table 4). An interesting observation was that in two patients with FISH positive results but negative culture results, we observed clarithromycin‐resistant coccoid forms. However, similar coccoid forms were observed in four patients positive for both FISH and culture that were either sensitive or resistant to clarithromycin. Therefore, bacterial morphology may not explain why some culture negative patients had positive FISH results. H. pylori strains were confirmed by PCR of the ureA gene with the primers HPU‐1 and HPU‐2. Confirmed H. pylori strains were assessed for the presence of the cagA gene. Amongst 114 H.pylori‐isolated strains, 53 (46.5%) were cagA positive. CagA status was compared with clarithromycin susceptibility in antrum and corpus biopsy sections. There were no differences between cagA positivity and clarithromycin susceptibility in antrum and corpus biopsy sections (Fisher exact test, p = NS)
Table 4.
The comparison of E‐test and FISH results for specific biopsy specimens in H. pylori positive patients
| Results by FISH | Antrum biopsy (n = 102) | Corpus biopsy (n = 96) | ||||
|---|---|---|---|---|---|---|
| Number of strains (%) | Clarithromycin by E‐test | Number of strains (%) | Clarithromycin by E‐test | |||
| Resistant (%) | Sensitive (%) | Resistant (%) | Sensitive (%) | |||
| Positive for clarithromycin resistance | 15 (14.7) | 13 (86.7) | 2 (13.3) | 15 (15.6) | 12 (80.0) | 3 (20.0) |
| Positive for clarithromycin sensitive | 83 (81.4) | 11 (13.3) | 72 (86.7) | 73 (76.1) | 7 (9.6) | 66 (90.4) |
| Negative for H. pylori | 4 (3.9) | 2 (50.0) | 2 (50.0) | 8 (8.3) | 1 (12.5) | 7 (87.5) |
FISH = fluorescence in situ hybridisation.
Discussion
The diagnosis of H. pylori is based on non‐invasive and invasive tests. However, a major limitation of invasive tests is that H. pylori infection has a patchy distribution, and the likelihood of recovering or visualising the bacteria increases with each additional biopsy specimen studied from the same patient 35. When gastric biopsies are evaluated by a specialised pathologist, the sensitivity and specificity of histopathology for the diagnosis of H. pylori can be very high 36. For other tests such as RUT, its sensitivity depends on bacterial load. It has been published that at least 105 bacteria are necessary for a positive RUT. Furthermore, there is a possibility that other urease positive gastric Helicobacter and non Helicobacter species may be responsible for the positive urease test, but their prevalence is <1% in gastric biopsies 45, 46. The success of H. pylori recovery depends on several factors that affect the sensitivity of H. pylori isolation, including slow in vitro growth of the bacteria, the sensitivity to PPIs and other medications that affect bacteria load 35. All these factors may explain the small number of specific laboratories that are capable of isolating the organism from clinical gastric biopsies 36. In the success of H. pylori eradication, we need to obtain correct information on the susceptibility of strains to antimicrobials and this represents a big challenge for clinical microbiologists and gastroenterologists.
In many cases it is imperative to culture H. pylori and determine its antimicrobial susceptibility to guarantee the success of the therapy.
In this study, we compared FISH and traditional culture methods and, according to our results, FISH was a more sensitive method than culture for the detection of H. pylori (83.5 versus 69.5%, respectively, p = 0.004). Furthermore, the specificity of FISH was 91.4%. Our specificity result is comparable with the Samarbaf‐Zadeh et al study in which the specificity of FISH for the detection of H. pylori was 100% 5, and with other reports on specificity of FISH for the detection of H. pylori that were >92 and 97%, respectively 12, 15.
We found 32 patients with negative cultures in whom H. pylori was detected by FISH. Failure to culture H. pylori may be due to contamination with other bacteria or the patchy distribution of H. pylori infection in the stomach 2. Negative culture due to low bacterial load, contamination, coccoid forms or transport delay is also possible. This is an important problem in the clinical setting 7. The stability of H.pylori in biopsy material during transport is a limiting factor for culture that does not affect FISH results 17, and it is possible to detect H. pylori by FISH in culture negative patients. For this reason, we believe that FISH is better than culture methods for the management of treatment therapy. The fact that we found four culture positive but FISH negative patients was an intriguing finding that may be explained by the patchy distribution of H. pylori infection or a low level of colonisation in the biopsy used for FISH. Rüssmann et al reported that FISH may fail to detect H. pylori in biopsies with fewer than 10 colony forming units (CFU) (4.0 ±1.4 CFU) whilst the site‐specific culture was positive 17.
The results of clarithromycin susceptibility by FISH were compared with the results of clarithromycin susceptibility by E‐test (Table 3). We detected four strains as clarithromycin resistant with FISH but clarithromycin sensitive by E‐test. Discrepant results between FISH and E‐test can be explained by the fact that the E‐test, and other traditional methods, base their results on a single isolated colony whilst FISH tests multiple bacterial strains by in situ hybridisation, making it a more nuanced, accurate methodology. We also observed 12 other discrepant results, in which H. pylori strains were clarithromycin resistant by E‐test but clarithromycin‐susceptible by FISH. One explanation of these results is that the FISH method used in this study only detected clarithromycin resistance with point mutations in the 2143 and 2142 positions of 23S rRNA. Therefore, it is possible that the 12 strains resistant to clarithromycin by E‐test contain point mutations different from the mutations in the 2142 and 2143 positions. When we sequenced the 23S rRNA region between 2060 and 2690 bp in the discrepant strains, our results showed that two strains had the same A2143G or A2142G point mutations and should have been identified. For three other strains, we identified point mutations at C2131T, C2310A and C2428T, but none of those point mutations has been implicated in clarithromycin resistance. However, since we only sequenced a partial region of the 23S rRNA gene, our results do not exclude the possibility of point mutations in the rest of the 23S rRNA that was not sequenced. Based on this observation, we need to emphasis to clinicians the limitations of FISH because not all the clarithromycin resistance can be explained by the three point mutations that we investigated here.
Furthermore, clarithromycin‐resistant strains without mutation in 23S rRNA may exist, therefore unknown genes outside 23S rRNA could be involved in clarithromycin resistance 47.
Results of clarithromycin susceptibility by biopsy location yielded 26 (25.5%) clarithromycin‐resistant H. pylori isolates by E‐test but only 15 (14.7%) by FISH in antrum biopsies. Amongst corpus biopsies, we found 20 (20.8%) clarithromycin‐resistant H. pylori strains by E‐test and 15 (15.6%) by FISH. Although the differences observed between the methods in the corpus were not significant, we found a significant difference in the sensitivity between E‐test and FISH in the antrum. Despite that, no topographic site differences in the stomach have been reported. Several authors have indicated that specific babAB genotype correlates with colonisation of the antrum 48, or specific chemotaxis to the antrum is controlled by some genes (TLpD) 49.
When antrum and corpus results were combined, we detected 10 (8.8%) clarithromycin‐susceptible and –resistant strains (mixed infections) by E‐test and 14 (12.3%) clarithromycin‐susceptible and ‐resistant strains (mixed infections) by FISH. Therefore, the presence of mixed infection in this study was estimated to be higher than 10% and this phenomenon may help to explain some failures in treatment.
We found that clarithromycin resistance was 28.0% by E‐test and 20.2% by FISH. We also found a good correlation for sensitive strains (89.5%) between both methods. Our results are not comparable with the results of Vega et al 7, who utilised frozen gastric biopsies to compare E‐test with FISH, and found that sensitivity and specificity of FISH was 90 and 100%; respectively. The authors found only two discrepant results between FISH and E‐test and not mixed infections. However, they did not perform FISH in culture negative patients 7. Rüssmann et al found no discrepancies between FISH, E‐test and disc diffusion 39. In our study, we observed discrepancies in four patients in whom we isolated clarithromycin‐susceptible strains by E‐test, but FISH detected the presence of clarithromycin‐susceptible and –resistant strains. Therefore, these discrepant results may be explained by the fact that mixed populations were present or sensitive strains were present at low concentrations and could not be detected by the E‐test method. In this study, if susceptible and resistant strains were present in the same patient, he or she was considered colonised with a resistant strain. Similar discrepant results were found by Cerqueira et using a novel method of peptide nucleic acid probes to detect clarithromycin resistant H. pylori strains when compared with E‐test 50.
In 137 H. pylori FISH positive patients, we found 111 (81.0%) patients infected with a clarithromycin‐susceptible strain, 11 (8.0%) patients infected with a clarithromycin‐resistant strain and 15 (11.0%) patients infected with both clarithromycin‐susceptible and –resistant strains, as determined by FISH. One possible explanation for the results in the last group is that there are two copies of the 23S rRNA gene in H. pylori. Both copies of the mutated 23S rRNA gene frequently exhibit the same mutation, but heterozygotes (one copy of the wild‐type and one copy of the mutated gene) have sometimes been observed 18, 23. In addition, Toracchio et al reported the colonisation by clarithromycin sensitive and resistant H. pylori strains in the same geographic area and in the same patients 28.
Trebesius et al found that FISH could be a powerful tool to successfully hybridise ‘culture resistant’ coccoid forms of H. pylori in the stomach 6. In our study, we found two patients with FISH positive but culture negative results showing clarithromycin‐resistant coccoid forms. However, we also found FISH and culture positive patients who showed clarithromycin‐sensitive coccoid forms. Thus, morphology (coccoid forms) does not explain FISH positive but culture negative results in those patients.
Finally, the prevalence of cagA‐positive strains was 46.5%. A previous report from Turkey showed a CagA prevalence of 82% 51, which is almost twice the prevalence found in this study. We do not have an explanation for this difference.
In conclusion, FISH appears to be a good technique that simultaneously determines H. pylori status and clarithromycin susceptibility with better sensitivity and specificity than traditional culture methods. FISH can help clinicians to manage the treatment of H. pylori infected patients. When clarithromycin resistant strains are detected by FISH, clinicians should be able to select the best treatment for H. pylori eradication. Because of the alarming increase in clarithromycin and other antibiotic resistance in H. pylori strains, antimicrobial susceptibility testing is imperative.
Author contributions
EDG, CG, SS, MS, ST, OA, IS and AZO participated actively in the acquisition of data, analysis and interpretation, participated also in revising the document and provided final approval of the version to be published. OY and GIPP participated in the conception and design of the study, drafting the article and provided final approval of the version to be published.
Acknowledgements
This research was supported by Dokuz Eylül University Research Foundation grant 2006.KB.SAG.042 and also supported in part by R01 GM63270 by the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. EDG was the recipient of a PhD fellowship from the Scientific and Technological Research Council of Turkey (TUBITAK). We thank Isabel Teitler and Kartik Raju for their revision of the English style of this manuscript.
The authors report no conflicts of interest. The authors are all responsible for the content and writing of the paper.
References
- 1. Ahmad N, Zakaria WR, Mohamed R. Analysis of antibiotic susceptibility patterns of Helicobacter pylori isolates from Malaysia. Helicobacter 2011; 16: 47–51. [DOI] [PubMed] [Google Scholar]
- 2. Feydt‐Schmidt A, Rüssmann H, Lehn N, et al Fluorescence in situ hybridization vs. epsilometer test for detection of clarithromycin‐susceptible and clarithromycin‐resistant Helicobacter pylori strains in gastric biopsies from children. Aliment Pharmacol Ther 2002; 16: 2073–2079. [DOI] [PubMed] [Google Scholar]
- 3. Kanada R, Uchida T, Tsukamoto Y, et al Genotyping of the cagA gene of Helicobacter pylori on immunohistochemistry with East Asian CagA‐specific antibody. Pathol Int 2008; 58: 218–225. [DOI] [PubMed] [Google Scholar]
- 4. Rimbara A, Fischbach LA, Graham DY. Optimal therapy for Helicobacter pylori infections. Nat Rev Gastroenterol Hepatol 2011; 8: 79–88. [DOI] [PubMed] [Google Scholar]
- 5. Samarbaf‐Zadeh AR, Tajbakhsh S, Moosavian SM, et al Application of fluorescent in situ hybridization (FISH) for the detection of Helicobacter pylori . Med Sci Monit 2006; 12: CR426–CR430. [PubMed] [Google Scholar]
- 6. Trebesius K, Panthel K, Strobel S, et al Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridization. Gut 2000; 46: 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vega AE, Alarcon T, Domingo D, et al Detection of clarithromycin‐resistant Helicobacter pylori in frozen gastric biopsies from pediatric patients by a commercially available fluorescent in situ hybridization. Diagn Microbiol Infect Dis 2007; 59: 421–423. [DOI] [PubMed] [Google Scholar]
- 8. Yılmaz Ö, Demiray E. Clinical role and importance of fluorescence in situ hybridization method in diagnosis of H. pylori infection and determination of clarithromycin resistance in H. pylori eradication therapy. World J Gastroenterol 2007; 13: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buta N, Tanih NF, Ndip RN. Increasing trend of metronidazole resistance in the treatment of Helicobacter pylori infection: A global challenge. Afr J Biotechnol 2010; 9: 1115–1121. [Google Scholar]
- 10. Yamaoka Y. Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis. World J Gastroenterol 2008; 14: 4265–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudi J, Kolb C, Maiwald M, et al Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol 1998; 36: 944–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jüttner S, Vieth M, Miehlke S, et al Reliable detection of macrolide‐resistant Helicobacter pylori via fluorescence in situ hybridization in formalin‐fixed tissue. Mod Pathol 2004; 17: 684–689. [DOI] [PubMed] [Google Scholar]
- 13. Malfertheiner P, Megraud F, O'Morain C, et al Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut 2007; 56: 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Megraud FH. pylori antibiotic resistance: prevalance, importance, and advances in testing. Gut 2004; 53: 1374–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris JM, Reasonover AL, Bruce MG, et al Evaluation of seaFAST, a rapid fluorescent in situ hybridization test, for detection of Helicobacter pylori and resistance to clarithromycin in paraffin‐embedded biopsy sections. J Clin Microbiol 2005; 43: 3494–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Connor A, Gisbert J, O'Morain C. Treatment of Helicobacter pylori infection. Helicobacter 2009; 14: 46–51. [DOI] [PubMed] [Google Scholar]
- 17. Rüssmann H, Kempf VAJ, Koletzko S, et al Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimens. J Clin Microbiol 2001; 39: 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 2001; 45: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malfertheiner P, Megraud F, O'Morain CA, et al Management of Helicobacter pylori infection–the Maastricht IV/Florence Consensus Report. Gut 2012; 61: 646–664. [DOI] [PubMed] [Google Scholar]
- 20. Forbes GM, Glaser ME, Cullen DJE, et al Duodenal ulcer treated with Helicobacter pylori eradication: seven‐year follow‐up. Lancet 1994; 343: 258–260. [DOI] [PubMed] [Google Scholar]
- 21. Xia HX, Fan XG, Talley NJ. Clarithromycin resistance in Helicobacter pylori and its clinical relevance. World J Gastroenterol 1999; 5: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yılmaz Ö, Demiray E, Tümer S, et al Detection of Helicobacter pylori and determination of clarithromycin susceptibility using formalin‐fixed paraffin‐embedded gastric biopsy specimens by fluorescence in situ hybridization. Helicobacter 2007; 12: 136–141. [DOI] [PubMed] [Google Scholar]
- 23. Taylor DE. Pathophysiology of antibiotic resistance: clarithromycin. Can J Gastroenterol 2000; 14: 891–894. [DOI] [PubMed] [Google Scholar]
- 24. Şimşek H, Balaban YH, Güneş DD, et al Alarming clarithromycin resistance of Helicobacter pylori in Turkish population. Helicobacter 2005; 10: 360–361. [DOI] [PubMed] [Google Scholar]
- 25. Cambau E, Allerheiligen V, Coulon C, et al Evaluation of a new test, GenoType HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori . J Clin Microbiol 2009; 47: 3600–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaynor M, Mankin AS. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr Top Med Chem 2003; 3: 949–961. [DOI] [PubMed] [Google Scholar]
- 27. Kusters JG, Kuipers EJ. Antibiotic resistance of Helicobacter pylori . J Appl Microbiol 2001; 90: 134S–144S. [DOI] [PubMed] [Google Scholar]
- 28. Toracchio S, Aceto GM, Mariani‐Costantini R, et al Identification of a novel mutation affecting domain V of the 23S rRNA gene in Helicobacter pylori . Helicobacter 2004; 9: 396–399. [DOI] [PubMed] [Google Scholar]
- 29. Chen S, Li Y, Yu C. Oligonucleotide microarray: a new rapid method for screening the 23S rRNA gene of Helicobacter pylori for single nucleotide polymorphisms associated with clarithromycin resistance. J Gastroenterol Hepatol 2008; 23: 126–131. [DOI] [PubMed] [Google Scholar]
- 30. De Francesco V, Margiotta M, Zullo A, et al Clarithromycin‐resistant genotypes and eradication of Helicobacter pylori . Ann Intern Med 2006; 144: 94–100. [DOI] [PubMed] [Google Scholar]
- 31. Fontana C, Favaro M, Minelli S, et al New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother 2002; 46: 3765–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrido L, Toledo H. Novel genotypes in Helicobacter pylori involving domain V of the 23S rRNA gene. Helicobacter 2007; 12: 505–509. [DOI] [PubMed] [Google Scholar]
- 33. Monteiro L, Oleastro M, Lehours P, et al Diagnosis of Helicobacter pylori infection. Helicobacter 2009; 14: 8–14. [DOI] [PubMed] [Google Scholar]
- 34. Megraud F. Macrolide resistance in H. pylori In Macrolide Antibiotics, Schönfeld W, Kirst HA. (eds). Birkhauser: Basel, 2002;243–260, Deutsche Bibliothek Cataloging‐in‐Publication Data. [Google Scholar]
- 35. Megraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev 2007; 20: 280–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lehours P, Megraud F. Helicobacter pylori molecular diagnosis. Expert Rev Mol Diagn 2011; 11: 351–355. [DOI] [PubMed] [Google Scholar]
- 37. Malfertheiner P. Helicobacter pylori infection‐ Management from a European Perspective. Dig Dis 2014; 32: 275–280. [DOI] [PubMed] [Google Scholar]
- 38. Gerrits MM, van Vliet AHM, Kuipers E, et al Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis 2006; 6: 699–709. [DOI] [PubMed] [Google Scholar]
- 39. Rüssmann H, Adler K, Haas R, et al Rapid and accurate determination of genotypic clarithromycin resistance in cultured Helicobacter pylori by fluorescent in situ hybridization. J Clin Microbiol 2001; 39: 4142–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Francesco V, Giorgio F, Ierardi E, et al Primary clarithromycin resistance in Helicobacter pylori: the Multicentric Italian Clarithromycin Resistance Observational (MICRO) study. J Gastrointestin Liver Dis 2011; 20: 235–239. [PubMed] [Google Scholar]
- 41. Dixon MF, Genta RM, Yardley JH, et al Classification and grading of gastritis: the updated Sydney system. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol 1996; 20: 1161–1181. [DOI] [PubMed] [Google Scholar]
- 42. Clinical and Laboratory Standards Institute . Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; Approved Guideline (2nd edn), 2010; 30(18): M45–MA2.
- 43. Panayotopoulou EG, Sgouras DN, Papadakos K, et al Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J Clin Microbiol 2007; 45: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Şen N, Yılmaz Ö, Simsek İ, et al Detection of Helicobacter pylori DNA by a simple stool PCR method in adult dyspeptic patients. Helicobacter 2005; 10: 353–359. [DOI] [PubMed] [Google Scholar]
- 45. Brandi G, Biavati B, Calabrese C, et al Urease‐positive bacteria other than Helicobacter pylori in human gastric juice and mucosa. Am J Gastroenterol 2006; 101: 1756–1761. [DOI] [PubMed] [Google Scholar]
- 46. Osaki T, Mabe K, Hanawa T, et al Urease‐positive bacteria in the stomach induce a false‐positive reaction in a urea breath test for diagnosis of Helicobacter pylori infection. J Med Microbiol 2008; 57: 814–819. [DOI] [PubMed] [Google Scholar]
- 47. Binh TT, Shiota S, Suzuki R, et al Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next‐generation sequencing. J Antimicrob Chemother 2014; 69: 1796–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sheu SM, Sheu BS, Chiang WC, et al H. pylori clinical isolates have diverse babAB genotype distributions over different topographic sites of stomach with correlation to clinical disease outcomes. BMC Microbiol 2012; 12: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rolig AS, Shanks J, Carter JE, et al Helicobacter pylori requires TlpD‐Driven chemotaxis to proliferate in the antrum. Infect Immun 2012; 80: 3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cerqueira L, Fernandes RN, Ferreira RM, et al Validation of a Fluorescence In Situ Hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J Clin Mıcrobiol 2013; 51: 1887–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Erzin Y, Altun S, Dobrucali A, et al Analysis of serum antibody profile against H. pylori VacA and CagA antigens in Turkish patients with duodenal ulcer. World J Gastroenterol 2006; 12: 6869–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]


