Abstract
Objective:
To explore the effects of protein factor Oncostatin M (OSM), a member of the Interleukin-6 (IL-6) family on cell proliferation, osteogenic differentiation and mineralization.
Materials and methods:
Basal nutrient solutions of different concentrations of OSM (0, 5, 10, 20, 40, 80 ng/ml) were used. In order to divide embryonic origin between mesenchymal stem cells C3H10T1/2 of in vitro cultured mice, and the effects of in vitro proliferation efficiencies of C3H10T1/2 cells of different concentrations of OSM, the C3H10T1/2 cells were divided into four groups: (1) Basal nutrient solution group (negative control); (2) Osteogenesis induced liquid group (positive control); (3) OSM (20 ng/ml) group; (4) Experimental group (osteogenesis induced liquid + OSM (20 ng/ml)). The expressions levels of relevant osteogenesis and mineralization genes were detected.
Results:
OSM had several effects on promoting the proliferation of embryonic origin mesenchymal stem cells C3H10T1/2 with respect to time of exposure as well as concentrations. In the present study, it has been shown that when the concentration of OSM is 20 ng/ml, the effects of promoting proliferation are most obvious. OSM can induce osteogenic differentiation of C3H10T1/2, make the process of osteogenic differentiation in advance, and promote the formation of end-stage calcium deposits and mineralized nodule, and osteogenic differentiation of C3H10T1/2 is finally achieved.
Conclusion:
OSM can promote the proliferation of C3H10T1/2, and induce its osteogenic differentiation and end-stage mineralization.
Keywords: C3H10T1/2, Oncostatin, Proliferation, Osteogenic Differentiation, Bone Metabolic Marker
Introduction
Currently, the major plans of treating massive bone defects include: (i) Bone transplantation (autogenous bone transplantation, allograft bone transplantation and xenogenic bone transplantation), (ii) Artificial bone (bone cement, biological ceramics), (iii) Bone tissue engineered and bone transport operations. However, due to their limitations in clinical applications, there are huge gaps in clinically used bones. Currently, the construction of, appropriate bones with specific characteristics such as high biocompatibility, safety, economically practical and, good reliability is considered a major necessity in the field[1,2]. In recent years, autologous polyphyly stem cells have been widely used in various studies. In vitro culture and osteoinductive differentiation have provided more tools for new developments in this field[3].
Recently, a growing number of studies have reported that inflammatory factors might play an important role in bone damage repair process[4]. As a type of protein factor of IL-6 family, OSM is indispensable for the control and the adjustment of internal and external micro-environments in the osteogenic differentiation process[5]. Recent in vitro studies showed that OSM could induce the osteogenic differentiation in human-source mesenchymal stem cells and preosteoblasts of mice[6,7]. However there is no report on OSM acting on embryonic origin in murine C3H10T1/2. Murine C3H10T1/2 cells have many features of mesenchymal stem cells (MSCs).
The present work intends to discuss the effects of OSM on C3H10T1/2 cell proliferation and osteogenic differentiation. Also, it aims to explore the probable mechanisms involve in the process of inducing osteogenic differentiation. The ultimate goal is to provide a new cell model for osteogenic differentiation.
Materials and methods
Cells and regents
Murine C3H10T1/2 was purchased from Shanghai Model Cell Institute (Shanghai, China). Manufacturer information for other regents were detailed as follow: fetal calf serum (GIBCO BRL, Grand Island, NY, USA), TRIZOL agent (Bio-sharp, Hefei, China), RIPA lysate (Bio-sharp, Hefei, China), Metabolic activity test (MTS) detection kit (Beyotime, Shanghai, China), Oncostatin M protein (Bio-sharp, Hefei, China), Alizarin red dye solution (Jiancheng Biotech, Nanjing, China), BCA kit (BCA Protein Assay Kit for detection of protein concentration) (Bio-sharp, Hefei, China), Real-time-RT-PCR kit (Bio-sharp, Hefei, China), reverse transcription kit (Bio-sharp, Hefei, China), ALP buffer (Jiancheng Biotech, Nanjing, China), rabbit anti-mice OC, OPN, COL-1, BSP primary antibodies were purchased from Abcam (Cambridge, MA, USA).
Cell culture and osteogenesis induction
Cells at passage 3-5 were used to inoculate at 96-well plate (105/ml). The day after, they were swapped into substrates with the OSM concentrations of 0, 5, 10, 20, 40 and 80 ng/ml, and MTS detection method was used to evaluate the efficiency of cell proliferation after 48 and 72 hours. The cells were divided into four groups: (i) Basal nutrient solution group (CON); (ii) osteogenesis induced liquid group (OS); (iii) OSM (20 ng/ml) group and (iv) experimental group (OS+ 20 ng/ml OSM).
Cell viability detection
MTS assay was performed to explore cell viability. We employed 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethophenyl) 2-(4-sulfophenyl)-2Htetrazolium (MTS) and electron coupling agent phenazine methosulphate (PMS). MTS is converted into a medium soluble formazan product by dehydrogenase enzymes found in metabolically active cells. 20 µl MTS solution was added to experiment culture to terminate the exposure of the tested agent. After 2 hours incubation, absorbance was measured at 490 nm using microtiter plate reader (VeraMax, Molecular Devices, USA). The quantity of formazan products is proportional to the number of viable cells in the culture.
Alizarin red staining and calcium quantitative analysis
After being fixed in 4% paraformaldehyde for 10 min, cells were stained with 2% ARS staining reagent (Jiancheng Biotech, Nanjing, China) for 15 min and then washed twice with deionized water. The red positions were recognized as calcium deposits. Calcium content were detected after 14 and 21 days Images were taken using microscope (Olympus, Tokyo, Japan) for further calcium quantitative analysis. Detailed protocols for calcium quantitative analysis was according to a previous report[8].
ALP activity detection
After being seeded at a density of 2´105 cells per well in 24-well plates, cells were and induced into osteoblast differentiation. At 7, 14 and 21 days after induction, cells were lyzed with a lysis buffer composed of 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1% Triton X-100. Intracellular alkaline phosphatase activity was determined by using an ALP activity kit (Jiancheng Biotech, Nanjing, China). The protein concentration of cell lysate was determined by using Bradford assay (Bio-Rad, USA) at 595 nm on a microplate spectrophotometer (TECAN, Australia). The specific ALP activity was normalized to the protein concentration. Each experiment was repeated in sextuple.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted after induction for 3, 7, 14 and 21 days using TRIzol reagent. Reverse transcription was performed using a PrimeScript RT reagent kit (TaKaRa, Dalian, China). Osteogenesis-related genes such as RUNX2, BMP2, SP7, ALP, DLX5, OC, OPN, ON, BSP and COL-1 was detected. The sequences of the primers used are shown in Table 1. Amplification and detection were performed using the SYBR Premix Ex Taq II kit (TaKaRa) and the Applied Biosystems ABI Prism 7500 HT sequence detection system (Applied Biosystems, Darmstadt, Germany), respectively. β-actin was used as internal control.
Table 1.
The sequences of common primers used in RT-PCR.
| GAPDH | Forward | GGAAGGTGAAGGTCGGAGTCAAC |
| Reverse | CTCGCTCCTGGAAGATGGTGATG | |
| COL-I | Forward | CCCGGGTTTCAGAGACAACTTC |
| Reverse | TCCACATGCTTTATTCCAGCAATC | |
| OC | Forward | CTATTGGCCCTGGCCGCACT |
| Reverse | AGCGCCGATAGGCCTCCTGA | |
| BSP | Forward | ACGGGGAACCTCGTGGGGAC |
| Reverse | AGGCTGGAGCTTCACTGGTGGT | |
| OPN | Forward | CCCACGGACCTGCCAGCAAC |
| Reverse | GGCAACGGGGATGGCCTTGT | |
| ALP | Forward | CTGGACGGACCCTCGCCAGT |
| Reverse | TTGCGCTTGGTCTCGCCAGT | |
| RUNX2 | Forward | CACTGGCGCTGCAACAAGA |
| Reverse | CATTCCGGAGCTCAGCAGCAGAATAA | |
| SP7 | Forward | ACTGAACCCCCAGCTGCCCA |
| Reverse | TGCTGCTCCCAGCCGCTCTA | |
| DLX5 | Forward | TCCTCCTCTCCTCCTCCTC |
| Reverse | GCGGTGGCTGCTCTTCTACT | |
| BMP2 | Forward | TGAGGATTAGCAGGTCTTTGC |
| Reverse | GCTGTTTGTGTTTGGCTTGA | |
| ON | Forward | CCGCCTACAAACGCATCTAT |
| Reverse | CAGGGCAGAGAGAGAGGACA |
Western blot
Cell lysates were extracted after induction of 14 and 21 days with lysis buffer and total protein concentration was determined with a BCA kit (Bio-sharp, Hefei, China) following the manufacturer’s instructions. A Total of 20 µg protein was boiled for 5 min in 1´ loading buffer, chilled on ice, and then separated on 10% SDS-PAGE and transferred to a PVDF membrane (Millipore, Bedford, MA, USA). Non-specific protein interactions were blocked by incubation with 5% fat-free milk in TBST buffer (50mM Tris–HCl, 150 mM NaCl, 0.05% Tween 20, pH 7.6) at 4°C for 1 h. The membranes were incubated overnight with primary antibodies in the blocking buffer at 4°C. Unbound antibody was removed by washing in TBST buffer three times (5 min/wash). Then, the membranes were incubated with horseradish peroxide-conjugated secondary antibody for 1 h at 20°C, followed by washing with TBST buffer for three times (5 min/wash). The membranes were developed using ECL according to the manufacturer’s protocols (Millipore, Billerica, MA, USA).
Statistical analyses
SPSS 17.0 software was used for the statistical data analyses. One-way analysis of variance was used for comparisons among groups. Dunett, T3 test was adopted for the comparisons between any two means in each group. ANOVA was used for comparisons among multiple groups, and the results were represented by mean ± standard deviation (x̄ ± s); *P<0.05, **P<0.01, and P<0.05 was considered as of statistical significance. All experiments were performed in triplicates.
Results
The effects of OSM on the proliferation of murine C3H10T1/2 cells
After adding OSM in different concentrations to substrate containing 10% fetal calf serum, the C3H10T1/2 cells proliferation efficiency was verified at two time points of 48 h and 72 h. The results showed that OSM treatment at a concentration of 20 ng/ml had stimulating effects on C3H10T1/2 proliferation at both 48 and 72 h. We deducted that proliferation stimulatory effect of OSM on C3H10T1/2, correlated positively with time and concentration. The proliferation efficiency was significantly higher at 20 ng/ml (Figure 1).
Figure 1.

The effect of OSM on C3H10T1/2 cells proliferation at different time points and different concentrations. OSM treatment at 20 ng/ml significantly increased cell viability. *p<0.05, Compared to 0 ng/ml (OSM); **p<0.01, Compared to 0 ng/ml (OSM).
The effect of OSM on ALP activations in the process of murine C3H10T1/2 cells osteogenic differentiation
In the process of osteogenic differentiation, ALP staining method was used to detect ALP activity at different experimental time points after 7, 14 and 21 days. Results showed that ALP activity in the OS group had no significant difference with that in CON group after 7 days. Although we observed higher expression which peaked at 14 days and then declined slowly, the difference was not statistically significant for the CON group (P<0.01). Variation trends in the OSM group and the OS + OSM group were similar, which showed that they expressed higher ALP at 7 days (P<0.05), and then declined, however we found no significant differences compared with CON group after 21 days (P>0.05) (Figure 2). OSM and osteogenesis induced media can both stimulate ALP activity, but the effect of OSM was detected earlier than those of osteogenesis induced media. We deducted that OSM can induce the process of osteogenesis differentiation in advance.
Figure 2.

Expression of ALP activity in the process of osteogenic differentiation.
Calcium quantitative analysis and calcium staining in the process of osteogenic differentiation
Results of calcium deposition and formation of mineralized nodule showed that calcium deposition was enhanced after treatment with a combination of OS and OSM at 14 or 21 days. However, there were no statistically significant differences among other the 3 groups (CON, OS and OSM) both at 14 and 21 days. We also found that calcium deposition was even more markedly increased at 21 days (Figure 3). Calcium staining photos showed more perceptual and obvious results. Calcium staining photos of the experimental group had few calcium deposits and sparse mineralized nodule at 14 days, while at 21 days, abundant calcium deposits and the formation of mineralized nodule was observed under the microscope; while no obvious mineralized nodule was detected in the negative control group, the positive control group and the OSM group (Figure 4). These results suggested that in the process of inducing C3H10T1/2 cells osteogenic differentiation, OSM can promote calcium deposits formation in cellular matrix and the formation of mineralized nodules.
Figure 3.

Calcium quantitative analysis in the process of osteogenic differentiation.
Figure 4.

Alizarin red staining during the process of osteogenic differentiation.
Transcription levels of genes involved in osteogenesis and osteogenic differentiation in murine C3H10T1/2 cells
The expression levels of genes such as RUNX2, BMP2, SP7, ALP, DLX5, OC, OPN, ON, BSP and COL-1 in the process of osteogenic differentiation in each experimental groups at four time points of 3, 7, 14 and 21 days were evaluated. For mRNA expression of RUNX2, BMP2, SP7, DLX5, OC, OPN, ON, BSP and COL-1, only the combined treatment of OS + OSM significantly upregulated compared with CON group, especially at 21 days. For ALP mRNA, OS treatment alone also elevated its expression at 14 and 21 days (Figure 5).
Figure 5.
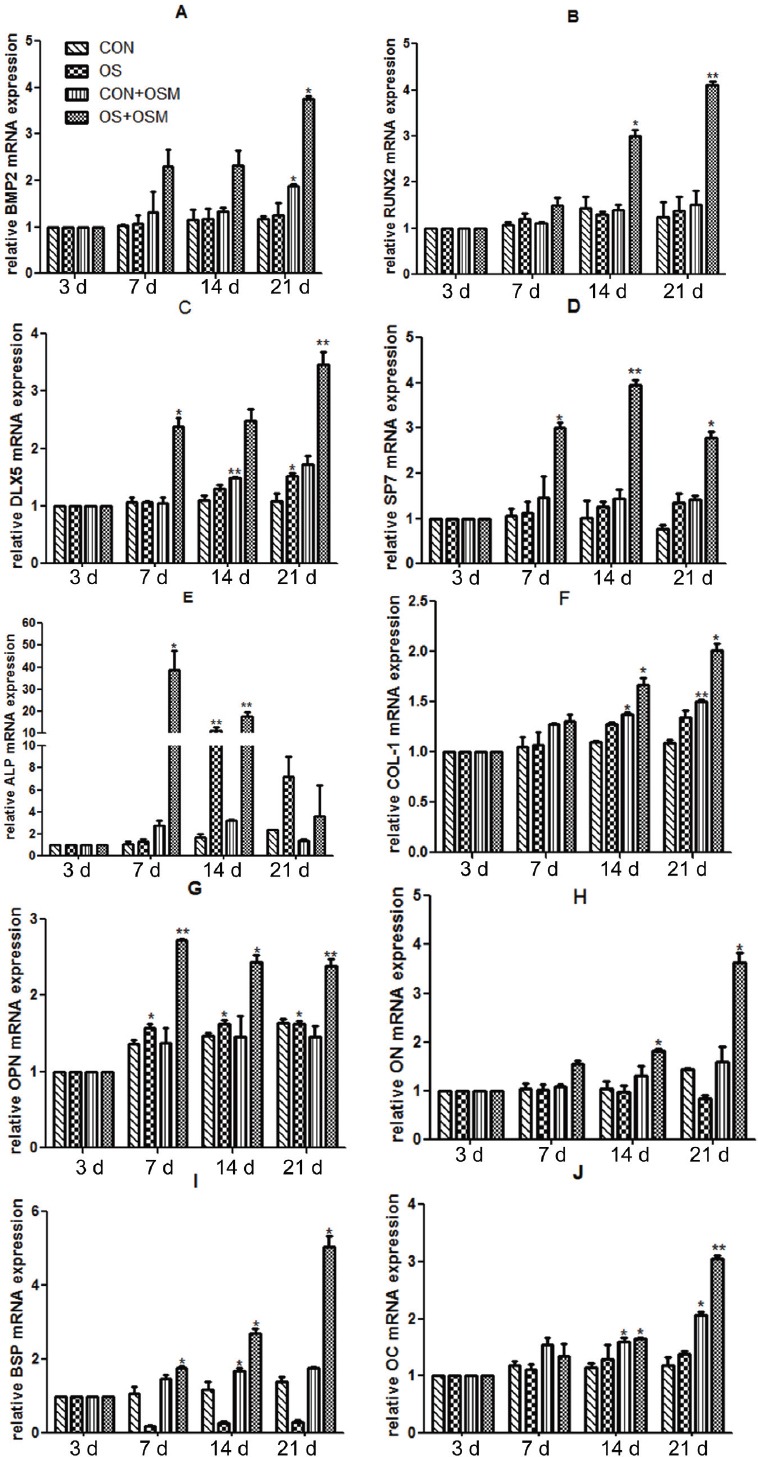
Expression levels of genes involved in osteogenesis in the process of osteogenic differentiation detected by RT-PCR.
Expression levels for proteins involved in osteogenesis and osteogenic differentiation in murine C3H10T1/2 cells
Western-Blot and double antibodies hybrid immune method were performed on protein extractions in order to analyze the relative expression levels of COL-1, OPN, OC and BSP proteins in osteogenic differentiation. Results showed the presence of these proteins at 14 and 21 days, and the variance regulations were similar to the results of PCR (Figure 6).
Figure 6.

(A) Zebra Imaging of OC Western-blot; (B) Zebra Imagings of COL-1, OPN and BSP Western-blot.
Discussion
In the present study, we found that OSM can not only promote proliferation of C3H10T1/2 cells, but also its osteogenic differentiation, evidenced by significantly increased ALP activity, up-regulated osteogenesis-associated mRNAs and proteins, enhanced calcium deposition and increased Alizarin red staining.
Previous evidence has demonstrated that combinations of OSM and gp130/OSMR and gp130/LIFR activates JAK family signal transducer factors[9] through the channel of winding phosphorylated amino acid, further activates STAT signal factors and selectively combines with MAPK or PI3K/AKT[10,11], which play biological roles such as adjusting cell differentiation and regeneration[12-16]. Currently, more reports about the influences of OSM on osteogenic differentiation in various cell lines are available; however there are no reports about the effects of OSM on C3H10T1/2 cell lines. Our study intended to evaluate the effects of OSM on osteogenic differentiation adjustments of C3H10T1/2 cell lines under different conditions.
Using osteosarcoma cells and human bone marrow-derived mesenchymal stem cells (hBMMSCs) in vitro, it was shown that OSM regulated the expression of osteogenetic genes, promoted the proliferation of bone cells and inhibited the proliferation of Osteosarcoma cells through PKC, ERK1/2 and STAT3 pathways[17]. Scaffidi et al[18] revealed that OSM can enhance cell mitosis and promote the proliferation of lung fibroblast by activating MAPK signal pathway in vitro. The intensity of OSM effects on promoting cell proliferation was found to be related to the concentration and culturing time of OSM. In stimulating the proliferation of lung fibroblast, the most effective concentration was 2 ng/ml and the best time was 48 h[18]. Sims Na et al[19] found that OSM, as a multifunctional transcriptional regulator, promoted the proliferation of bone cells by regulating the function of osteoblast and osteoclast cells under physiological and pathological conditions. In this experiment, after C3H10T1/2 cells were stimulated by OSM at different concentration, higher levels of cell proliferation was observed after 48 h and 72 h, with higher cell proliferation rates at 72 h. Our results showed that the most effective concentration of OSM for promoting the proliferation of C3H10T1/2 cells was 20 ng/ml and the highest proliferation rate took place at 72 h. Comparing our results to those reported by Scaffidi et al[18], we obtained different conclusions. Those differences might be attributed to the fact that these two studies used different cell lines and these cells reacted differently under OSM induction. OSM in vitro promoted C3H10T1/2 cell proliferation and maturation thus established the foundation for osteogenic differentiation.
Inflammatory condition was once regarded as the important factor inhibiting the process of bone formation and bone resorption during bone injury repair. Nevertheless, new findings revealed that inflammatory cytokines played an important role in the process of promoting bone formation and bone resorption. There is a complex relation between the immune system and the bone tissues, in which inflammatory cytokines promote bone formation. In chronic inflammation, activated T cells release soluble inflammatory factors that increase the ALP activity and induce over-expression of RUNX2, SP7, BSP and OC mRNAs in marrow stroma cells. This increases the level inflammatory cytokines originated from T cells and promote osteogenic differentiation and bone cell maturation by regulating the interaction between osteoblast and osteoclast cells, which accelerates the process of bone formation[20]. Bedi et al believe that activated T cells can promote osteogenic differentiation by regulating the signal pathway directed by Wnt[21]. According to this, inflammatory factors have a huge promotion effect on osteogenic differentiation.
Nicolaidou et al. prepared cultures with hMSC and macrophages or monocytes cells and reported that the secreted OSM increased ALP expression on the 7th day and the formation of mineralized nodules observed on the 21st day. Their results proved that OSM induced hMSC osteogenic differentiation and promoted mineralization[22,23]. OPN genes showed almost the same changing rules as ALP, and OPN appeared in the early period of osteogenic differentiation and induced bone cell adhesion. RUNX2 and SP7 genes are the key transcription factors[24] in regulating early-stage osteogenic differentiation and final-stage matrix mineralization. Zhou et al[25] proved that SP7 gene in newly-born mice, was one of the necessary in vivo factors in bone formation and bone balance. The result of the experiment showed that the expression of RUNX2 mRNA in the experiment group became higher starting the 1st week and continued until reaching its peak at the 3rd week. Expression of SP7 mRNA increased from the 1st week and reached its peak at the 2nd week, but still had higher edge compared to comparison group at the 3rd week. This proved that OSM regulated the continuous high expression of RUNX2 and SP7 in the process of osteogenic differentiation and induced stem cells to differentiate directionally towards bone cells and promoted bone cell maturation. Rattanasopha et al[26] knocked out neonatal mouse homologous transcription factor DLX5, and found that all the mice showed deformity on their fore or hind limbs, which proved that DLX5 was indispensable in the process of bone development. There are many kinds of matrix proteins in the extracellular microenvironment of osteoblasts, and as an extracellular matrix protein adhesion scaffold, COL-1 is mainly regulated by vitamin C and ALP activity. We know that ON combines with COL-1 to form a compound that induces calcium and phosphorus deposition in the extracellular matrix and forming hydroxyapatite (HA). Subsequently, BSP and OC show up in the medium and we have the final term of bone formation related to final mineralization. This experiment, through RT-PCR, detected the expression of extracellular matrix during the stem cell differentiation process and the result showed that in the process of OSM stimulating stem cells or bone differentiation, OC, ON, BSP and COL-1 mRNA in the experiment group showed stable growth trends and reached peaks in the final stage of osteogenic differentiation. Interestingly, finding that OSM can promote the expression of BMP-2 to increase in the process of experiment was in conformity with the report produced by Alfons et al[27]. In that experiment, Western Blot results suggested a steady expression of extracellular matrix proteins including OC, OPN, BSP and COL-1 in osteogenic differentiation. Additionally, calcium quantity analysis suggested that the level of calcium deposition in extracellular matrix in the experiment group at the 2nd and 3rd week were meaningfully higher than other groups. Mineralized nodule staining imaging results directly reflect the formation of a big quantity of extracellular matrix calcium nodule in vitro. Our result showed that OSM had the capability of inducing C3H10T1/2 osteogenic differentiation in vitro and promoted final formation of mineralized nodules.
There is evidence showing that OSM realizes its biological regulating function by regulating JAK/STAT and MAPK signal pathways. In osteogenic differentiation, OSM activates these signal transcription pathways, regulating the expression of particular osteogenic genes that are linked to cell functions and may induce the obvious cell shape changes in osteogenic cell lines[28]. Also, mesenchymal stromal cells have multi-directional differentiation potentials in different culturing environments and OSM has effects on human adipose tissue derived stem cells in vitro, which can promote osteogenic differentiation and mineralization by upregulating ALP and inhibiting adipogenic differentiation[29]. Pierre et al reported that OSM induced the osteogenic directed differentiation in human bone marrow mesenchymal stem cells in vitro[30]. It can be inferred that OSM played a significant part in the process of stem cell directed differentiation. In the current study, we set 4 different groups and established different extracellular environment and our results revealed that OSM had exact effects on inducing stem cell osteogenic differentiation and matrix mineralization. OSM could promote bone formation in the process of osteogenic differentiation in each stage of osteogenic cells. Early in 2002, in vivo studies conducted on mice suggested that injecting Ad-mOSM into knee joint cavity can induce subperiosteal bone deposition and it was found at the same time that OSM had special inhibitory effect on bone sclerosis protein and disturbed the bone resorption process in osteoclast mediation[27]. Song et al pointed out that OSM promoted the high expression of ALP and OC while affecting the BMSCs. Mineralized nodules started to appear at the 2nd week and starting 3rd week, the expression of some relevant osteogenic genes declined, which was in accord with the physical changing trends seen in osteogenic differentiation[29].
In 2010 Sims et al[31] demonstrated the importance of gp130 family of transcription factors in bone repair phenomenon and proposed the OSM application in hospitals. Walker et al found out that some mice suffered from osteoporosis after knocking out their receptor gene[32]. Sato et al[33] proposed that OSM adjusted the oriented differentiation of mesenchymal bone cells in the bone marrow hematopoietic microenvironment. In 2012, Bolin et al reported that injecting OSM to Balb/c mice tibia could find obvious bone deposition and bone mineralization[34]. In 2015 Guihard et al successfully realized the bone injury repairing induced by membranous ossification bones under the periosteum in the mice tibia damage model by OSM injection[35]. Using calcium alizarin red staining, this trial discovered that calcium nodules deposits in the extracellular matrix both at 2 and 21 days, showing precisely the occurrence of the bone deposition during the culture process. Besides, OSM has close relation with VEGF, PTH, HIF1 and MIR200 in cellular microenvironment and those biomolecules play important roles during the process of osteogenic differentiation. To sum up, the function of OSM in the induction of osteogenic differentiation and repairing of bone defect is affirmed.
OSM is a multifunctional adjustable proteomics, meditating the whole process of osteogenic differentiation through different methods in extracellular microenvironment. This study established that OSM could promote the proliferation of murine C3H10T1/2 stem cells and induce its osteogenic differentiation and matrix mineralization. We also showed that OSM might control the process of osteogenic differentiation by BMP-2 or Wnt signal pathway, however this needs further verification. OSM can be developed and used in clinical nonunion and for its role in promoting the osteogenesis. It can also be useful for treating large segmental bone defects.
Footnotes
The authors have no conflict of interest.
Edited by: G. Lyritis
References
- 1.Case ND, Duty AO, Ratcliffe A, Muller R, Guldberg RE. Bone formation on tissue-engineered cartilage constructs in vivo: Effects of chondrocyte viability and mechanical loading. Tissue Eng. 2003;9:587–596. doi: 10.1089/107632703768247296. [DOI] [PubMed] [Google Scholar]
- 2.Khosla S, Westendorf JJ, Modder UI. Concise review: Insights from normal bone remodeling and stem cell-based therapies for bone repair. Stem cells. 2010;28:2124–2128. doi: 10.1002/stem.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruder SP, Kraus KH, Goldberg VM, Kadiyala S. The effect of implants loaded with autologous mesenchymal stem cells on the healing of canine segmental bone defects. J Bone Joint Surg Am. 1998;80:985–996. doi: 10.2106/00004623-199807000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Giannoudis PV, Hak D, Sanders D, Donohoe E, Tosounidis T, Bahney C. Inflammation, bone healing, and Anti-Inflammatory drugs: An update. J Orthop Trauma. 2015;29 Suppl(12):S6–S9. doi: 10.1097/BOT.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 5.Moxham JP. Oncostatin-M enhances osteoinduction in a rabbit critical calvarial defect model. Laryngoscope. 2007;117:1790–1797. doi: 10.1097/MLG.0b013e3180ed451b. [DOI] [PubMed] [Google Scholar]
- 6.Lee MJ, Heo SC, Shin SH, Kwon YW, Do EK, Suh DS, Yoon MS, Kim JH. Oncostatin M promotes mesenchymal stem cell-stimulated tumor growth through a paracrine mechanism involving periostin and TGFBI. Int J Biochem Cell Biol. 2013;45:1869–1877. doi: 10.1016/j.biocel.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Song HY, Jeon ES, Kim JI, Jung JS, Kim JH. Oncostatin M promotes osteogenesis and suppresses adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells. J Cell Biochem. 2007;101:1238–1251. doi: 10.1002/jcb.21245. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi C, Myoui A, Hashimoto N, Kuriyama K, Yoshioka K, Yoshikawa H, Itoh K. Continuous inhibition of MAPK signaling promotes the early osteoblastic differentiation and mineralization of the extracellular matrix. J Bone Miner Res. 2002;17:1785–1794. doi: 10.1359/jbmr.2002.17.10.1785. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland NG, Gilbert DJ, Jenkins NA, Hara T, Miyajima A. Mouse oncostatin M: An immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]
- 10.Ehlting C, Bohmer O, Hahnel MJ, Thomas M, Zanger UM, Gaestel M, Knoefel WT, Schulte AEJ, Haussinger D, Bode JG. Oncostatin M regulates SOCS3 mRNA stability via the MEK-ERK1/2-pathway independent of p38(MAPK)/MK2. Cell signal. 2015;27:555–567. doi: 10.1016/j.cellsig.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Smith DA, Kiba A, Zong Y, Witte ON. Interleukin-6 and oncostatin-M synergize with the PI3K/AKT pathway to promote aggressive prostate malignancy in mouse and human tissues. Mol Cancer Res. 2013;11:1159–1165. doi: 10.1158/1541-7786.MCR-13-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer LK, Wang S, Schaefer TS. Oncostatin M activates stat DNA binding and transcriptional activity in primary human fetal astrocytes: low- and high-passage cells have distinct patterns of stat activation. Cytokine. 2000;12:1647–1655. doi: 10.1006/cyto.2000.0774. [DOI] [PubMed] [Google Scholar]
- 13.Van Wagoner NJ, Choi C, Repovic P, Benveniste EN. Oncostatin M regulation of interleukin-6 expression in astrocytes: biphasic regulation involving the mitogen-activated protein kinases ERK1/2 and p38. J Neurochem. 2000;75:563–575. doi: 10.1046/j.1471-4159.2000.0750563.x. [DOI] [PubMed] [Google Scholar]
- 14.Arita K, South AP, Hans-Filho G, Sakuma TH, Lai-Cheong J, Clements S, Odashiro M, Odashiro DN, Hans-Neto G, Hans NR, Holder MV, Bhogal BS, Hartshorne ST, Akiyama M, Shimizu H, McGrath JA. Oncostatin M receptor-beta mutations underlie familial primary localized cutaneous amyloidosis. Am J Hum Genet. 2008;82:73–80. doi: 10.1016/j.ajhg.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka M, Miyajima A. Oncostatin M. A multifunctional cytokine. Rev Physiol Biochem Pharmacol. 2003;149:39–52. doi: 10.1007/s10254-003-0013-1. [DOI] [PubMed] [Google Scholar]
- 16.Savarese TM, Campbell CL, McQuain C, Mitchell K, Guardiani R, Quesenberry PJ, Nelson BE. Co-expression of oncostatin M and its receptors and evidence for STAT3 activation in human ovarian carcinomas. Cytokine. 2002;17:324–334. doi: 10.1006/cyto.2002.1022. [DOI] [PubMed] [Google Scholar]
- 17.Chipoy C, Berreur M, Couillaud S, Pradal G, Vallette F, Colombeix C, Rédini F, Heymann D, Blanchard F. Downregulation of osteoblast markers and induction of the glial fibrillary acidic protein by oncostatin M in osteosarcoma cells require PKCdelta and STAT3. J Bone Miner Res. 2004;19:1850–1861. doi: 10.1359/JBMR.040817. [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi AK, Mutsaers SE, Moodley YP, McAnulty RJ, Laurent GJ, Thompson PJ, Knight DA. Oncostatin M stimulates proliferation, induces collagen production and inhibits apoptosis of human lung fibroblasts. Br J Pharmacol. 2002;136:793–801. doi: 10.1038/sj.bjp.0704769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sims NA, Quinn JM. Osteoimmunology: oncostatin M as a pleiotropic regulator of bone formation and resorption in health and disease. Bonekey Rep. 2014;3:527. doi: 10.1038/bonekey.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rifas L, Arackal S, Weitzmann MN. Inflammatory T cells rapidly induce differentiation of human bone marrow stromal cells into mature osteoblasts. J Cell Biochem. 2003;88:650–659. doi: 10.1002/jcb.10436. [DOI] [PubMed] [Google Scholar]
- 21.Bedi B, Li JY, Tawfeek H, Baek KH, Adams J, Vangara SS, Chang MK, Kneissel M, Weitzmann MN, Pacifici R.l. Silencing of parathyroid hormone (PTH) receptor 1 in T cells blunts the bone anabolic activity of PTH. Proc Natl Acad Sci USA. 2012;109:E725–E733. doi: 10.1073/pnas.1120735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golub EE, Boesze-Battaglia K. The role of alkaline phosphatase in mineralization. Current Opinion in Orthopaedics. 2007;18:444–448. [Google Scholar]
- 23.Nicolaidou V, Wong MM, Redpath AN, Ersek A, Baban DF, Williams LM, Cope AP, Horwood NJ. Monocytes Induce STAT3 Activation in Human Mesenchymal Stem Cells to Promote Osteoblast Formation. PLoS One. 2012;7:e39871. doi: 10.1371/journal.pone.0039871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai MT, Lin YS, Chen WC, Ho CH. Runx2 and Osterix Gene Expression in Human Bone Marrow Stromal Cells Are Mediated by Far-Infrared Radiation. WCE. 2011 Jul;:6–8. [Google Scholar]
- 25.Zhou X, Zhang Z, Feng JQ, et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. PNAS. 2010;107:12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rattanasopha S, Tongkobpetch S, Srichomthong C, Kitidumrongsook P, Suphapeetiporn K, Shotelersuk V. Absent expression of the osteoblast-specific maternally imprinted genes, DLX5 and DLX6, causes split hand/split foot malformation type I. J Med Genet. 2014;51:817–823. doi: 10.1136/jmedgenet-2014-102576. [DOI] [PubMed] [Google Scholar]
- 27.De Hooge ASK, Van De Loo FAJ, Bennink MB, Jong DS, Arntz OJ, Lubberts E, Richards CD, Wim B. Adenoviral transfer of murine oncostatin M elicits periosteal bone apposition in knee joints of mice, despite synovial inflammation and up-regulated expression of interleukin-6 and receptor activator of nuclear factor-κB ligand. American Journal of Pathology. 2002;160:1733–1743. doi: 10.1016/s0002-9440(10)61120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy JB, Schindler C, Raz R, Levy DE, Baron R, Horowitz MC. Activation of the JAK-STAT signal transduction pathway by OSM in cultured human and mouse osteoblastic cell. Endocrinology. 1996;137:1159–1165. doi: 10.1210/endo.137.4.8625884. [DOI] [PubMed] [Google Scholar]
- 29.Song HY, Jeon ES, Kim JI, Jung JS, Kim JH. Oncostatin M promotes osteogenesis and suppresses adipogenic differentiation of human adipose tissue derived mesenchymal stem cells. J Cell Biochem. 2007;101:1238–1251. doi: 10.1002/jcb.21245. [DOI] [PubMed] [Google Scholar]
- 30.Guihard P, Dangerc Y, Brounaisa B, Davida E, Briona R, Delecrind J, Richardse C, Chevalierc S, Redinia F, Heymanna D, Gascanc H, Blancharda F. Induction of osteogenesis in mesenchymal stem cells by activated monocytes/macrophages depends on oncostatin M signaling. Stem Cells. 2012;30:762–772. doi: 10.1002/stem.1040. [DOI] [PubMed] [Google Scholar]
- 31.Sims NA, Walsh NC. GP130 cytokines and bone remodeling in health and disease. BMB Rep. 2010;43:513–523. doi: 10.5483/bmbrep.2010.43.8.513. [DOI] [PubMed] [Google Scholar]
- 32.Walker EC, McGregor NE, Poulton IJ, Solano M, Pompolo S, Fernandes TJ, Constable MJ, Nicholson GC, Zhang JG, Nicola NA, Gillespie MT, Martin TJ, Sims NA. Oncostatin M promotes bone formation independently of resorption when signaling through leukemia inhibitory factor receptor in mice. J Clin Invest. 2010;120:582–592. doi: 10.1172/JCI40568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minehata K, Takeuchi M, Hirabayashi Y, Inoue T, Donovan PJ, Tanaka M, Miyajima A. Oncostatin M Maintains the Hematopoietic Microenvironment in the Bone Marrow by Modulating Adipogenesis and Osteogenesis. PLoS One. 2014;9:e116209. doi: 10.1371/journal.pone.0116209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolin C, Tawara K, Sutherland C, Redshaw J, Aranda P, Moselhy J, Anderson R, Jorcyk CL. Oncostatin m promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes Cancer. 2012;3:117–130. doi: 10.1177/1947601912458284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guihard P, Boutet MA, Brounais-Le Royer B, Gamblin AL, Amiaud J, Renaud A, Berreur M, Rédini F, Heymann D, Layrolle P, Blanchard F. Oncostatin M, an Inflammatory Cytokine Produced by Macrophages, Supports Intramembranous Bone Healing in a Mouse Model of Tibia Injury. Am J Pathol. 2015;185:765–75. doi: 10.1016/j.ajpath.2014.11.008. [DOI] [PubMed] [Google Scholar]


