Abstract
Intracellular protein delivery has potential biotechnological and therapeutic application, but remains technically challenging. In contrast, a plethora of nucleic acid carriers have been developed, with lipid-based nanoparticles (LNPs) among the most clinically advanced reagents for oligonucleotide delivery. Here, we validate the hypothesis that oligonucleotides can serve as packaging materials to facilitate protein entrapment within and intracellular delivery by LNPs. Using two distinct model proteins, horseradish peroxidase and NeutrAvidin, we demonstrate that LNPs can yield efficient intracellular protein delivery in vitro when one or more oligonucleotides have been conjugated to the protein cargo. Moreover, in experiments with NeutrAvidin in vivo, we show that oligonucleotide conjugation significantly enhances LNP-mediated protein uptake within various spleen cell populations, suggesting that this approach may be particularly suitable for improved delivery of protein-based vaccines to antigen-presenting cells.
1. Introduction
Protein therapeutics comprise a major class of pharmaceuticals, including enzyme replacement, monoclonal antibodies, hormones, and protein-based vaccines[1]; however, most protein therapeutics act extracellularly, at the cell surface, or if endocytosed, carry out their function within lysosomes. Effective intracellular protein delivery remains a challenge[2, 3], yet offers the potential to broaden the scope of diseases amenable to this class of drugs and circumvent the inherent risks of gene therapeutics[4].
Cell-penetrating peptides[5] represent one widely studied approach to achieving intracellular protein delivery through covalent or non-covalent attachment of protein transduction domains, which include TAT-derived peptides[6–8], arginine-rich peptides[9, 10], penetratin peptides[11], and amphiphilic peptide carriers such as Pep-1[12]. A related strategy involves fusion of a protein known or engineered to achieve intracellular localization, such as herpes simplex virus protein VP22[13, 14], or more recently, a supercharged variant of GFP[15]. Limitations with these methods include potential immunogenicity and the lack of protection from proteases.[16, 17].
A number of materials have been described for nanoparticulate encapsulation and intracellular delivery of proteins, including lipids and liposomes[18, 19], charge-conversional polyionic micelles[20], cationic amphiphiles[21], and various polymers[22–24]. Nonetheless, because protein loading depends on the strength of non-covalent interactions between the protein cargo and the material, these techniques may be useful only for a limited subset of proteins with suitable physicochemical properties. An alternative approach involves synthesis of a biodegradable polymeric shell directly from the protein itself, so that the protein core is encapsulated within a nanocarrier that facilitates intracellular delivery[25]. In general, these materials have not yet achieved clinical or even preclinical demonstration of their safety and efficacy for systemic delivery.
By comparison, a much wider array of materials has been explored for intracellular delivery of nucleic acids[26]. One example includes a group of lipid-like molecules termed lipidoids, which demonstrated potent delivery of RNAi therapeutics in preclinical studies involving mice, rats, and nonhuman primates[27–29]. We hypothesized that oligonucleotides could serve as packaging materials to facilitate protein entrapment within and intracellular delivery by these lipid-based nanoparticles (LNPs) (Figure 1a). In this report, we investigate this hypothesis by attempting to demonstrate effective LNP-mediated intracellular delivery of two model proteins, horseradish peroxidase (HRP) and NeutrAvidin (nAv).
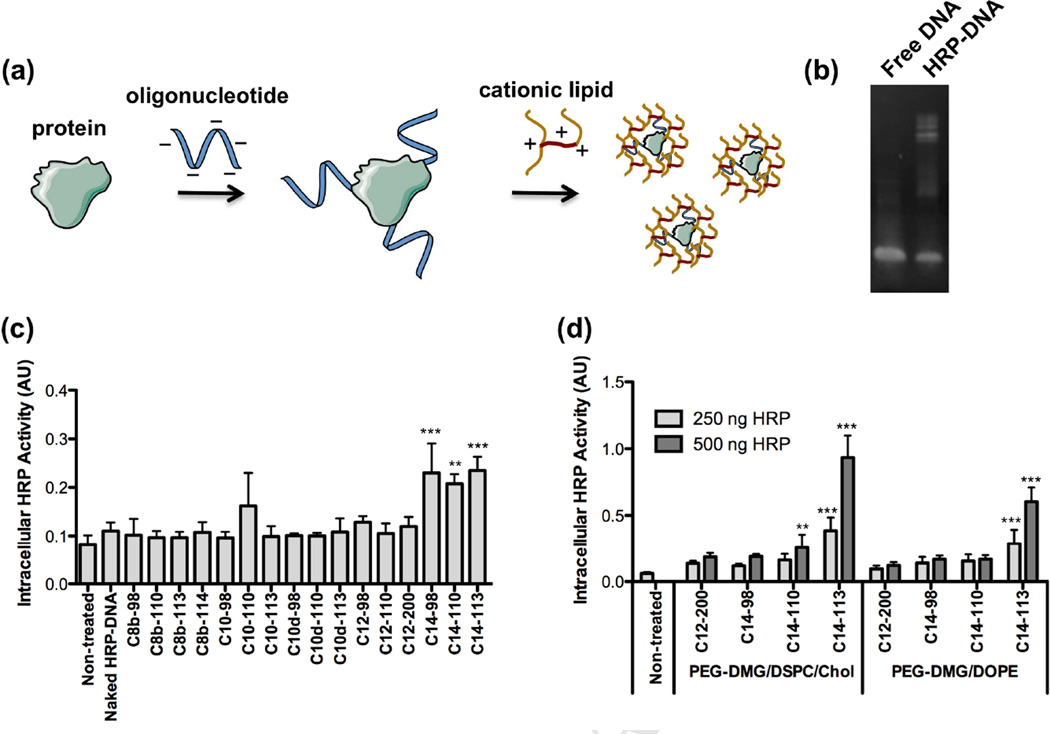
Figure 1.
(a) Schematic illustrating the use of oligonucleotides (oligos) as packaging materials to enhance encapsulation within lipid-based nanoparticles (LNPs). (b) Polyacrylamide gel electrophoresis (PAGE) of free DNA oligos and HRP-DNA oligo conjugates. (c) Screening of various cationic lipidoids for the intracellular delivery of active HRP-DNA conjugates to HeLa cells. An OPD assay was used to measure HRP activity (mean ± SD, n = 4) following a 3 h incubation of the LNPs (250 ng HRP/well). ** indicates p <0.01, *** indicates p < 0.001, compared to naked HRP-DNA control treatment. (d) Optimization of formulation composition to achieve effective intracellular HRP delivery (mean ± SD, n = 4) to HeLa cells using various cationic lipidoids. ** indicates p <0.01, *** indicates p < 0.001, compared to non-treated control.
2. Materials and Methods
2.1. Materials
Lipidoids were synthesized as previously described[27]. Horseradish peroxidase (type VI, 250–330 units/mg), copper (II) sulfate, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), cholesterol, and O-phenylenediamine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Unlabeled NeutrAvidin, Oregon Green 488-labeled NeutrAvidin, and AlexaFluor 488 carboxylic acid, succinimidyl ester, were purchased from Life Technologies (Grand Island, NY, USA). Cy5.5 mono-reactive NHS ester was acquired from GE Healthcare (Pittsburgh, PA, USA). Three oligonucleotide variants (unmodified, 5’-hexynyl, and 5’-biotin-PEG4-modified) with the following sequence were custom synthesized and desalted by Integrated DNA Technologies (Coralville, IA, USA): 5’-CGGGCGCGACTAGTGTGAAATCTGAATAATTTTGTGTTACTC-3’. NHS-PEG4-Azide crosslinker, Slide-A-Lyzer G2 dialysis cassettes (20K MWCO), and Zeba Spin desalting columns (40K MWCO) were acquired from Pierce Biotechnology (Rockford, IL). mPEG2000-DMG was synthesized by Alnylam (Cambridge, MA, USA) as described[30]. 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were obtained from Avanti Polar Lipids (Alabaster, AL, USA). HeLa cells (ATCC, Manassas, VA, USA) were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Filamentous actin (F-actin) specific probe Alexa Fluor® 555 Phalloidin and Prolong gold anti-fade mounting medium were purchased from Life Technologies. Alexa Fluor® 647-AffiniPure Rabbit Anti-Horseradish Peroxidase antibody was obtained from Jackson ImmunoResearch (West Grove, PA, USA).
2.2. Protein-oligonucleotide conjugation
For DNA oligonucleotide conjugation to horseradish peroxidase (HRP), HRP was first azide-functionalized using NHS-PEG4-Azide crosslinker according to the manufacturer’s directions. Briefly, NHS-PEG4-Azide (2.27 mM, MW = 388.37 Da) in anhydrous DMSO was added to HRP (0.176 mM, MW = 44 kDa) in a total volume of 2.425 mL PBS (20 mM sodium phosphate, 0.15 M sodium chloride, pH 7.2) and then incubated for 30 min at RT. To quench the reaction, 1 M Tris-HCl (pH 8.0) was added to achieve a final Tris-HCl concentration of 0.1 M. The reaction was incubated for an additional 5 min at RT and then dialyzed against 3 L PBS overnight at 4°C using a Slide-A-Lyzer G2 cassette (20 K MWCO) with two buffer exchanges. Azide-functionalized HRP (72.5 µM) was then incubated with 5’-alkyne-modified oligonucleotide (109 µM, MW = 13.14 kDa), CuSO4 (1 mM), TCEP (4 mM), and TBTA (100 µM) in a final volume of 3.9 mL for 5 h at RT, with minor modification from a previous report[31]. The reaction was then purified by dialysis against 3 L PBS overnight at 4°C using a Slide-A-Lyzer G2 cassette (20 K MWCO) with two buffer exchanges. The HRP-oligo conjugate concentration was determined in two ways with roughly equivalent results: absorbance measurement at 403 nm (εHRP, 403 nm = 102 mM−1 cm−1) using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and a BCA Protein assay (Thermo Scientific).
For oligo conjugation to NeutrAvidin (nAv), 5’-biotin-PEG4-functionalized oligo (37.2 µM, MW = 13.46 kDa) was incubated with fluorescently labeled nAv (18.6 µM, MW = 60 kDa) in a final volume of 0.75 ml PBS for 1 h at RT and used without purification. nAv was pre-labeled with either Cy5.5 mono-reactive NHS ester (GE Healthcare) or AF 488 carboxylic acid, succinimidyl ester, (Life Technologies) using the manufacturers’ instructions and was purified prior to conjugation using a Zeba Spin desalting column (40 K MWCO) equilibrated with PBS. For both labeling reactions, the final fluorophore:protein labeling molar ratio was approximately 1:1 as determined with a spectrophotometer. The conjugate concentration was determined by using the absorbance measurement at 280 nm (εnAv, 280 nm= 99.6 mM−1 cm−1).
2.3. Microfluidic device formulation of lipid nanoparticles (LNPs)
Stock solutions of lipidoid, cholesterol (MW = 387 Da), DSPC (MW = 790 Da), and mPEG2000-DMG (MW = 2660 Da) were prepared in ethanol at concentrations of 100, 20, 20, and 20 mg/mL, respectively. In most experiments, the components were combined to yield molar fractions of 50⋮38.5:10⋮1.5; however, in one experiment, DSPC was substituted with DOPE (MW =744 Da) at the same molar fraction.
The HRP-DNA conjugate, or the free HRP control, was diluted in 10 mM citrate buffer (pH 3.0) at a concentration of 0.5 mg/mL DNA, corresponding to an HRP concentration of 1.13 mg/mL. Due to differences in stabilities between the free nAv protein and the nAv-DNA conjugate, the nAv-DNA conjugate was diluted in 25 mM citrate buffer (pH 5.2) at a concentration of 0.375 mg/mL DNA (equivalent protein concentration of 0.845 mg/mL), while the free nAv was diluted to 0.845 mg/mL in 10 mM citrate buffer (pH 3.0). The lipid solution was diluted as necessary to yield a lipidoid:DNA weight ratio of 10:1.
Microfluidic devices were fabricated as described previously[32]. To prepare LNPs, the protein solution and the lipid solution were injected into the microfluidic device at a relative volumetric flow rate of 3:1 (0.9 mL/min : 0.3 mL/min) using two syringes (Gastight syringes, Hamilton Company, NV, USA) that were controlled by two syringe pumps (PHD 2000, Harvard Apparatus, MA, USA). To remove ethanol, the freshly prepared LNPs were dialyzed for 2 h against 3 L PBS using Slide-A-Lyzer G2 cassettes (20K MWCO).
2.4. Gel electrophoresis
Oligos and protein-oligo conjugates were run on a pre-cast 4–20% polyacrylamide-TBE gel (Bio-Rad, Hercules, CA, USA) for 1 h at 100 V. The gel was then stained for 30 min with SYBR Gold (Invitrogen) and visualized with a Bio-Rad GelDoc XR+ imager.
LNPs formulated with nAv or nAv-oligo conjugates were run on a pre-cast Any kD mini-Protean TGX gel (Bio-Rad) for 30 min at 100 V. The gel was stained for 1 h with Bio-Safe Coomassie Stain (Bio-Rad), destained overnight, and visualized using a Bio-Rad GelDoc XR+ imager. The entrapment efficiency was determined as: (Sfree − SLNP)/Sfree, where Sfree is the total signal per lane for the free protein or conjugate and SLNP that for the LNP-encapsulated protein or conjugate.
2.5. In vitro protein uptake experiments
One day before nanoparticle treatment, HeLa cells (100 µL) were seeded in a clear 96-well tissue culture plate at 15,000 cells per well. LNPs and naked protein/conjugate control treatments were diluted in freshly warmed growth medium as necessary to achieve the indicated protein dose (generally 100 ng − 1 µg per 150 µL per well). The conditioned medium was aspirated just before treatment and replaced with the LNPs/proteins diluted in fresh medium. The nanoparticles were incubated with cells for 3 h at 37°C.
2.6. HRP activity assay
After incubation with the nanoparticles, the HeLa cells were washed 3 times with PBS (200 µL) and treated with 0.25% w/v trypsin-EDTA (25 µL) for 5 min at 37°C to detach cells as well as to digest any remaining membrane-bound HRP. Upon addition of growth medium (50 µL), the cells were pipet-mixed, transferred to a clear 96-well assay plate, pelleted using a centrifuge, re-suspended in 100 µL of substrate solution, and repeatedly pipet-mixed. Substrate solution was prepared by dissolving a 15 mg tablet of o-phenylenediamine (OPD) in 12.5 mL 100 mM sodium citrate, pH 4.5, and adding 3.5 µL of hydrogen peroxide solution (30% w/v) immediately before use. The assay plate was incubated for 10 min at RT with orbital shaking prior to addition of 50 µL 2.5 M H2SO4. The absorbance at 490 nm was then immediately measured using a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The activity assay was performed in an analogous manner on a series of dilutions of the free HRP protein and the conjugate to generate a standard curve for each.
2.7. FACS analysis
After aspirating conditioned medium and washing cells three times with PBS, cells were detached using 25 µl per well of 0.25% trypsin-EDTA (Invitrogen). Following a 5 min incubation at 37°C, fresh medium (50 µl) was added to the cells, which were mixed thoroughly and then transferred to a 96-well round-bottom plate. Cells were then pelleted, re-suspended in fixation buffer (4% v/v formaldehyde in PBS), incubated for 10 min at RT, pelleted again, and finally re-suspended in ice-cold FACS running buffer (2% v/v FBS in PBS). The cells were kept at 4°C until FACS analysis using a BD LSR II (Becton Dickinson, San Jose, CA, USA). Gating and analysis were performed using FlowJo v8.8 software (TreeStar, Ashland, OR, USA).
2.8. HRP immunocytochemistry
HeLa cells were treated on glass cover slips in 6-well plates for 3 h with an LNP dose corresponding to 10 µg of HRP per well. The cells were washed 3 times with PBS and then fixed using a 4% paraformaldehyde solution at room temperature for 30 min. The cells were then washed twice with PBS, permeablized for 30 min using a 0.1% Triton X100 solution, and subsequently blocked for 1 h using a 5% normal goat serum solution. Next, the cells were incubated for 1 h in an immunostaining cocktail solution consisting of DAPI (3 µM), Alexa Fluor® 555 Phalloidin (1:200 dilution), Alexa Fluor® 647-AffiniPure Rabbit Anti-Horseradish Peroxidase antibody (1:100 dilution), and 5% normal goat serum. After staining, the cover slips were washed three times with a 0.1% Tween 20 solution and mounted using Prolong gold anti-fade. Immunostained cells were imaged using an LSM 700 point scanning confocal microscope (Carl Zeiss Microscopy, Jena Germany) equipped with a 40 × oil immersion objective. Obtained images were adjusted linearly for presentation using Photoshop (Adobe Inc. Seattle, WA).
2.9. Animal experiments
Animal experiments were performed using 6–8 wk old, female C57BL/6 mice (Charles River, Wilmington, MA) in accordance with protocols approved by MIT’s Committee on Animal Care (CAC). Mice were injected intravenously via the tail-vein with 0.2 mL of LNP formulations, unformulated controls, or PBS at a nAv protein dose of 2.5 mg/kg. After 2 h, the animals were euthanized, and the livers, spleens, kidneys, lungs and heart were dissected and analyzed for fluorescence (ex = 675 nm, em = 720 nm) using an IVIS imaging system (Xenogen, Alameda, CA). Xenogen Living Image v. 4.2 acquisition and analysis software was used to quantify fluorescent radiant efficiency of each organ from the optical images. Spleens were snap frozen on dry ice and freshly embedded in optimal cutting temperature compound (OCT). Cryostat sections were cut and collected on superfrost plus treated slides. Prepared frozen sections were kept at −20 °C until needed. The livers and spleens were then frozen on dry ice using Tissue-Tek OCT (Sakura, Torrance, CA, USA) and sectioned with a cryotome. In accordance with a previously developed method[33], some frozen sections were thawed at room temperature for 30 min and then scanned for Cy5.5 fluorescence intensity using a Li-COR Odyssey fluorescence scanner (Lincoln, NE, USA) at a resolution of 21 µm.
The other frozen sections were fixed using a 4% paraformaldehyde solution at room temperature for 30 min. The cryosections were then washed twice with PBS, permeablized for 30 min using a 0.1% Triton X100 solution, and subsequently blocked for 1 h using a 5% normal goat serum solution. Next, the sections were incubated for 1 h in an immunostaining cocktail solution consisting of DAPI (3 µM), Alexa Fluor® 555 Phalloidin (1:200 dilution), and 5% normal goat serum. After staining slides were washed three times with a 0.1% Tween 20 solution and mounted using Prolong gold anti-fade. Immunostained sections were imaged using an LSM 700 point scanning confocal microscope (Carl Zeiss Microscopy, Jena Germany) equipped with a 40 × oil immersion objective. Obtained images were adjusted linearly for presentation using Photoshop (Adobe Inc. Seattle, WA).
For analysis of splenocyte uptake, spleens were harvested 2 h post-injection, and the spleen cell suspension was analyzed for different immune cell populations and for AF488-labeled nAv uptake by flow cytometry (CD11b+: cells of the macrophage/monocyte lineage; CD11b+GR1+: neutrophils; CD11c+: dendritic cells; GR1+: granulocytes; CD19+: B-cells; TCRb+: T cells).
2.10. Statistics
Data are expressed as mean ± S.D. for groups of at least 3 replicates. Data were analyzed for statistical significance by one-way ANOVA with Bonferroni multiple comparison correction, as implemented in GraphPad Prism 5 (*: p < 0.05, **: p < 0.01, and ***: p < 0.001).
3. Results and discussion
To develop lipid-based nanoparticles (LNPs) delivery of protein-oligonucleotide conjugates, horseradish peroxidase (HRP), a 44 kDa enzyme with an isoelectric point of approximately 8.0–9.0, was selected as a model protein for delivery. We prepared HRP-oligonucleotide conjugates via an azide-alkyne click reaction. Briefly, HRP was azide-functionalized using an NHS-ester crosslinker with a short PEG spacer. The azide-modified HRP was then conjugated to a 42 base 5’-hexynyl-functionalized DNA oligonucleotide (oligo) with reaction conditions as reported previously[31] using a molar ratio of 1.5:1.0 oligo:HRP. A gel shift assay confirmed the synthesis of HRP conjugates bearing one or more DNA oligonucleotides (Figure 1b).
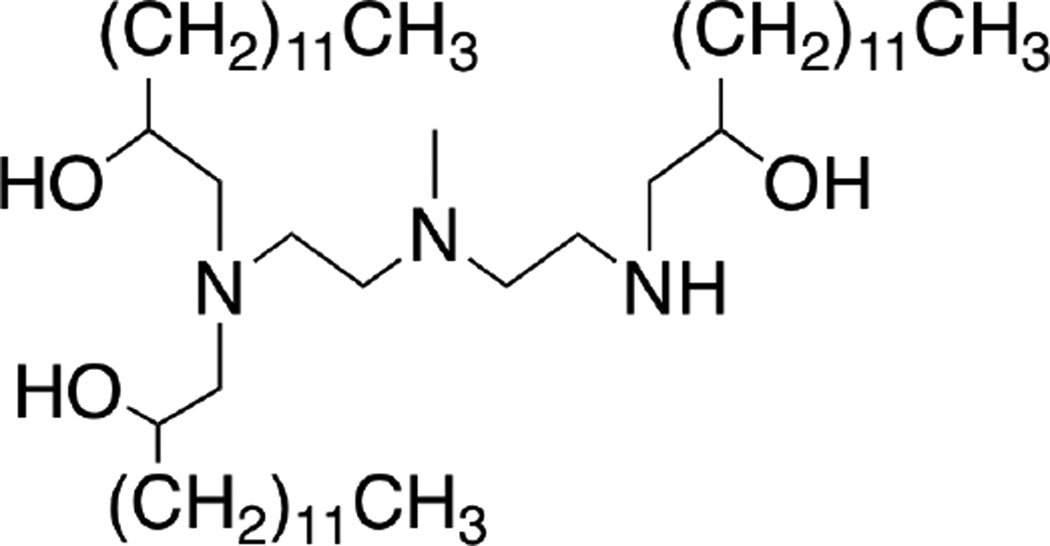
We first screened a panel of lipidoid formulations for in vitro delivery of the HRP-DNA conjugates. Sixteen lipidoids varying in their amine cores and alkyl tails as described[27] were mixed with DSPC, mPEG2000-DMG, and cholesterol and then formulated with HRP-oligonucleotide conjugates. The resulting nanoparticle formulations were incubated with HeLa cells for 3 h, after which the cells were washed 3 times with PBS and treated with trypsin to digest any remaining membrane-bound HRP. Subsequently, the cells were transferred to an assay plate, pelleted, and re-suspended in OPD substrate buffer for an HRP activity assay. As shown in Figure 1c, lipidoids bearing longer alkyl tails (C14) generally displayed more effective delivery of active HRP-DNA conjugates. Using the top four lipidoids from this screen, further optimization of formulation conditions indicated that formulations including DSPC and cholesterol delivered HRP conjugates more effectively than those containing DOPE (Figure 1d). In particular, the lipidoid C14-113 (Figure 2) outperformed the other lipidoids with respect to delivery activity.
Figure 2.
Structure of C14-113 lipidoid.
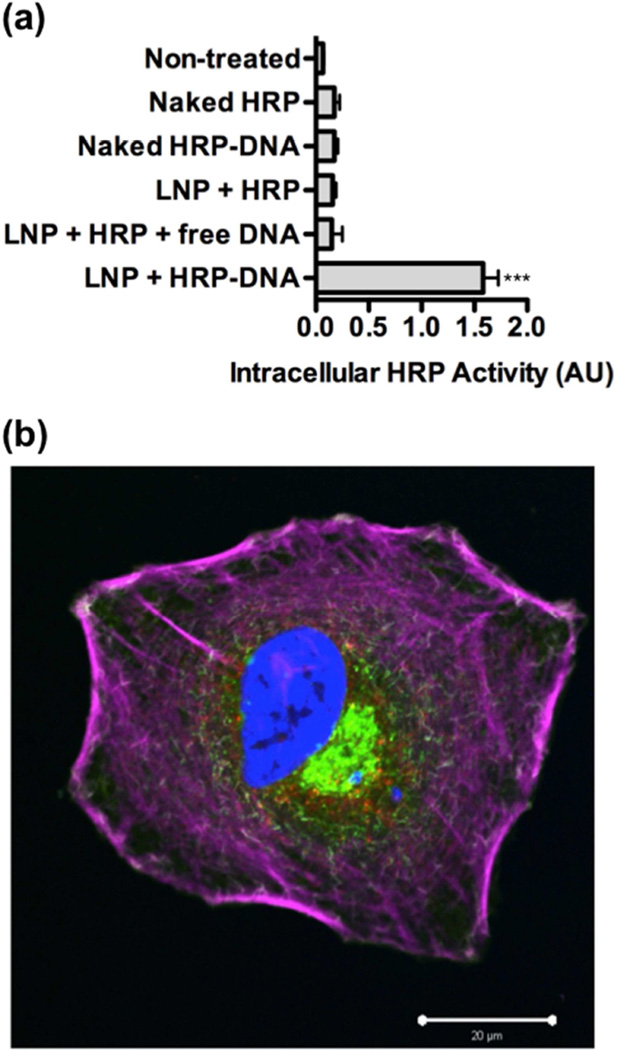
Having identified an LNP formulation mediating delivery of HRP-DNA conjugates, we then tested whether effective delivery depends on oligonucleotide conjugation. C14-113 LNPs were formulated in one of three ways: with HRP protein alone; with a mixture of HRP protein and free, unconjugated oligonucleotide; or with the HRP-oligonucleotide conjugates prepared by click chemistry. In addition to naked HRP protein and naked HRP-DNA conjugates as control treatments, the formulations were incubated for 3 h with HeLa cells. As seen in Figure 3a, only the C14-113 LNPs formulated with HRP-DNA conjugates yielded significant HRP activity after washing and trypsinizing the cells. This result suggests that HRP-oligonucleotide conjugation is required to achieve effective intracellular HRP delivery with these LNPs.
Figure 3.
(a) Protein-oligonucleotide conjugation is required for effective intracellular delivery of HRP by C14-113 LNPs. HeLa cells were treated as indicated for 3 h (500 ng HRP/well) and then subjected to an HRP activity assay (mean ± SD, n = 4). *** indicates p < 0.001 comparing LNP + HRP-DNA treatment to all other control treatments. (b) Following treatment of HeLa cells with C14-113 LNPs encapsulating HRP-oligo conjugates, immunocytochemical staining and confocal fluorescence microscopy confirmed intracellular HRP localization. Cells were treated in 6-well plates for 3 h with an LNP dose corresponding to 10 µg of HRP per well. Blue: nuclei; green: membrane; purple: F-actin; red: HRP.
To confirm that the HRP-oligonucleotide conjugates localized within the cells, we performed immunocytochemistry (ICC) and confocal microscopy (Figure 3b, Figure S1). The imaging data revealed substantial intracellular HRP staining within cells treated with LNPs containing HRP-DNA conjugates, but no such staining in cells treated with naked HRP-DNA conjugates (Figure S1). These data substantiate the conclusions from the enzymatic activity assay that the C14-113 LNPs mediate intracellular uptake of horseradish peroxidase-oligonucleotide conjugates.
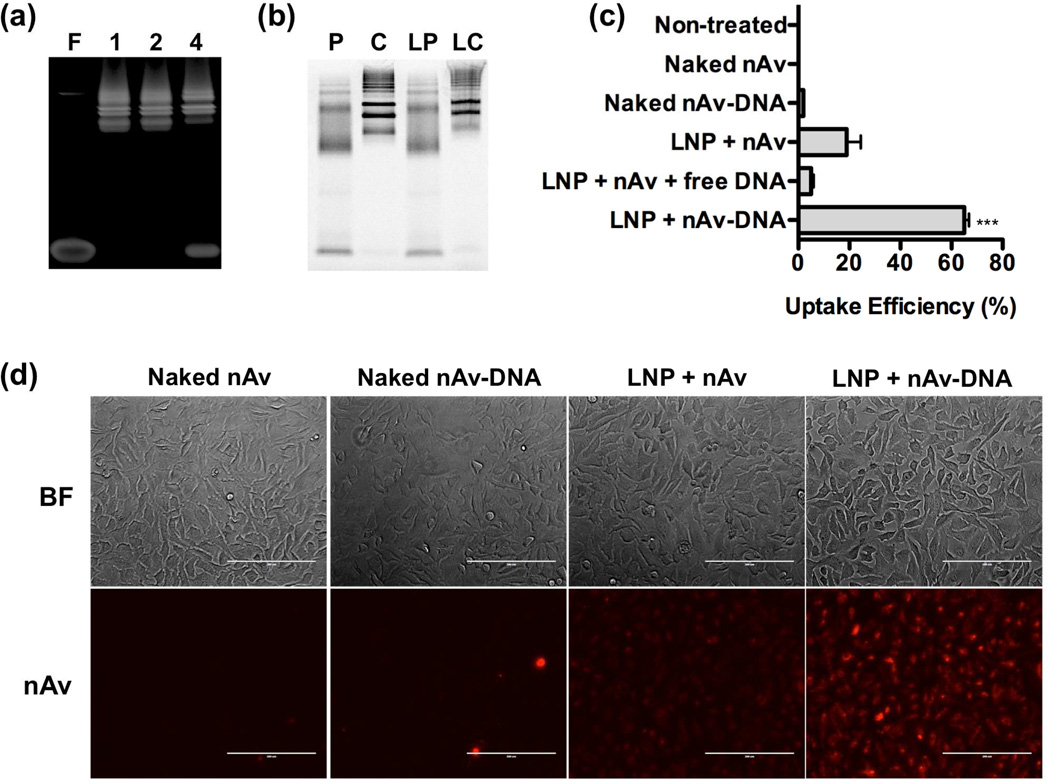
The same principle was illustrated for intracellular delivery of another model protein, NeutrAvidin (nAv), a deglycosylated variant of avidin with a molecular weight of 60 kDa and an isoelectric point of 6.3. Fluorescently labeled nAv-oligonucleotide conjugates were prepared by incubating Oregon Green 488-labeled nAv, which has four biotin binding sites, with various stoichiometric ratios of 5’-biotinylated oligonucleotides. The resulting conjugates were analyzed with a gel shift assay, which showed efficient conjugation at the 1:1 and 2:1 molar ratios of oligo:nAv (Figure 4a). Excess free oligo was detected in the mixture prepared at a 4:1 oligo:nAv ratio, most likely due to interference of the fluorescent label with one or more of nAv’s four potential biotin-binding sites. For ensuing experiments, the oligo:nAv molar ratio was maintained at 2:1.
Figure 4.
(a) Polyacrylamide gel electrophoresis (PAGE) of free (F) DNA oligos and NeutrAvidin-DNA oligo conjugates prepared at various molar ratios of oligo:DNA (1:1, 2:1, and 4:1). (b) SDS-PAGE of naked nAv protein (P), naked nAv-oligo conjugate (C), C14-113 LNPs encapsulating nAv protein (LP), and C14-113 LNPs encapsulating nAv-oligo conjugates (LC). (c) Protein-oligonucleotide conjugation is required for efficient intracellular delivery of nAv by C14-113 LNPs. HeLa cells were treated as indicated for 3 h (100 ng Oregon Green 488-labeled nAv/well), and the uptake efficiency was assessed by FACS analysis (mean ± SD, n = 4). *** indicates p < 0.001 comparing LNP + nAv-DNA treatment to all other control treatments. (d) Fluorescence microscopy images of HeLa cells treated for 3 h with the indicated Cy5.5-labeled nAv formulations (250 ng nAv/well).
To determine whether oligonucleotide conjugation affected the encapsulation efficiency of nAv within LNPs, C14-113 LNPs were formulated either with free nAv or with nAv-DNA conjugates and characterized by SDS-PAGE, alongside naked nAv and naked nAv-DNA conjugates (Figure 4b). Quantification of the gel results indicated that the encapsulation efficiency of nAv alone within C14-113 LNPs was ~11.0%, in comparison with ~30.8% for nAv-DNA conjugates (Table 1). These data suggest that oligonucleotide conjugation enhances the encapsulation efficiency of nAv within C14-113 LNPs approximately threefold.
Table 1.
Biophysical characterization of LNPs formulated with nAv or nAv-DNA conjugates
| Encapsulation Efficiency (%) |
Size (nm) | PDI | ζ-potential (mV) | |
|---|---|---|---|---|
| LNP + nAv | 11.0 | 79.4 ± 1.9 | 0.115 | +0.18 |
| LNP + nAv-DNA | 30.8 | 234.3 ± 13.4 | 0.151 | +0.65 |
To characterize the effect of oligo conjugation on intracellular nAv delivery in vitro, HeLa cells were incubated with one of three C14-113 LNP formulations: those encapsulating nAv protein alone, those encapsulating a mixture of nAv protein and unconjugated, unmodified DNA oligos; and those encapsulating the nAv-DNA conjugates. HeLa cells were treated for 3 h with these formulations, along with the naked protein and the naked conjugate treatments. The cells were then washed several times, trypsinized, and analyzed by fluorescence-activating cell sorting (FACS) for Oregon Green 488 signal. As displayed in Figure 4c, neither the naked nAv nor the naked nAv-DNA conjugates yielded significant cellular internalization. At the same nAv dose, the C14-113 LNPs encapsulating nAv-DNA conjugates achieved uptake in ~65% of HeLa cells, in comparison with ~19% uptake efficiency for LNPs encapsulating the nAv protein alone, and ~5% uptake efficiency for LNPs encapsulating a mixture of nAv and unconjugated oligo. These results demonstrate that oligo conjugation improves intracellular delivery of nAv mediated by C14-113 LNPs. Meanwhile, the reduction in uptake observed for LNPs formulated with a mixture of nAv and unconjugated oligo compared with those formulated with nAv alone suggests that the unconjugated oligo may compete with nAv for loading into the C14-113 LNPs. The extent of protein uptake for these treatments was also evaluated by fluorescence microscopy, which confirmed that only the HeLa cells treated with C14-113 LNPs encapsulating nAv-DNA conjugates showed significant uptake (Figure 4d).
To examine whether oligo conjugation also affects LNP-mediated protein uptake in vivo, we investigated the biodistribution of these LNPs 2 h after intravenous (IV) administration to mice. As shown by fluorescence imaging of the dissected organs, both the LNPs encapsulating the nAv-DNA conjugates (LNP + nAv-DNA) and the LNPs encapsulating the free nAv protein (LNP + nAv) were associated with greatly increased nAv-associated Cy5.5 signal in the liver as compared with naked nAv-oligo conjugates (Figure S2). However, the C14-113/nAv-DNA LNP-treated mice showed higher Cy5.5 signal within the spleens and lower signal within the kidneys than those that were treated with C14-113/nAv LNPs, suggesting altered biodistribution of the protein as a result of protein-oligo conjugation.
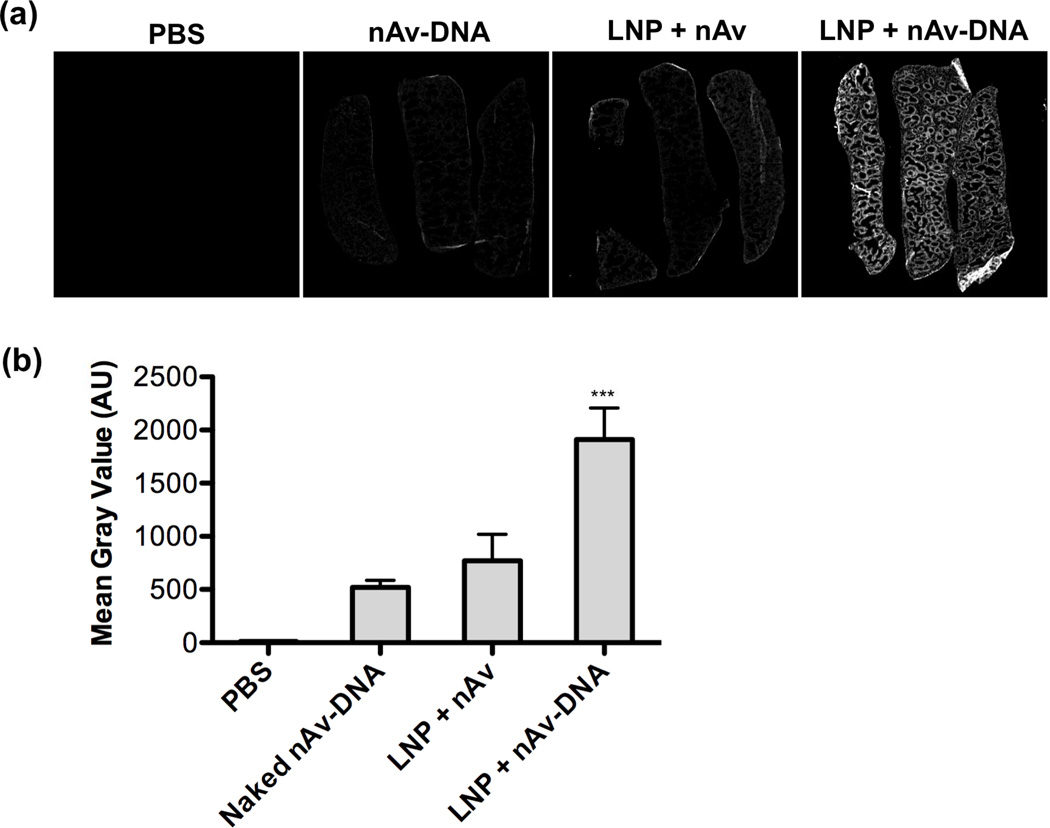
To quantify the difference in protein localization observed in the spleens, we imaged the organ sections using a Li-COR Odyssey near-infrared fluorescence scanner (Figure 5a). The results indicated approximately 2.5-fold higher uptake of nAv in the spleens of mice treated with LNPs encapsulating the nAv-oligo conjugates compared with those treated with LNPs containing the nAv protein alone (Figure 5b). Furthermore, comparing treatment with C14-113/nAv-DNA LNPs and treatment with naked nAv-DNA conjugates, encapsulation within LNPs increased spleen protein uptake approximately 3.5-fold. Immunohistochemical staining and confocal microscopy of spleen sections confirmed increased accumulation of Cy5.5-labeled nAv in those mice treated with C14-113/nAv-DNA LNPs (Figure 6a). By comparison, much less accumulation was observed for those mice treated with either naked nAv-DNA conjugates or with C14-113/nAv LNPs.
Figure 5.
(a) Near-infrared fluorescence scans of Cy5.5 signal in spleen sections harvested from mice 2 h after IV injection of the indicated Cy5.5-labeled nAv treatments. (b) Quantification of mean gray values of spleens for each group (n = 3). *** indicates p < 0.001 comparing LNP + nAv-DNA treatment to all other control treatments.
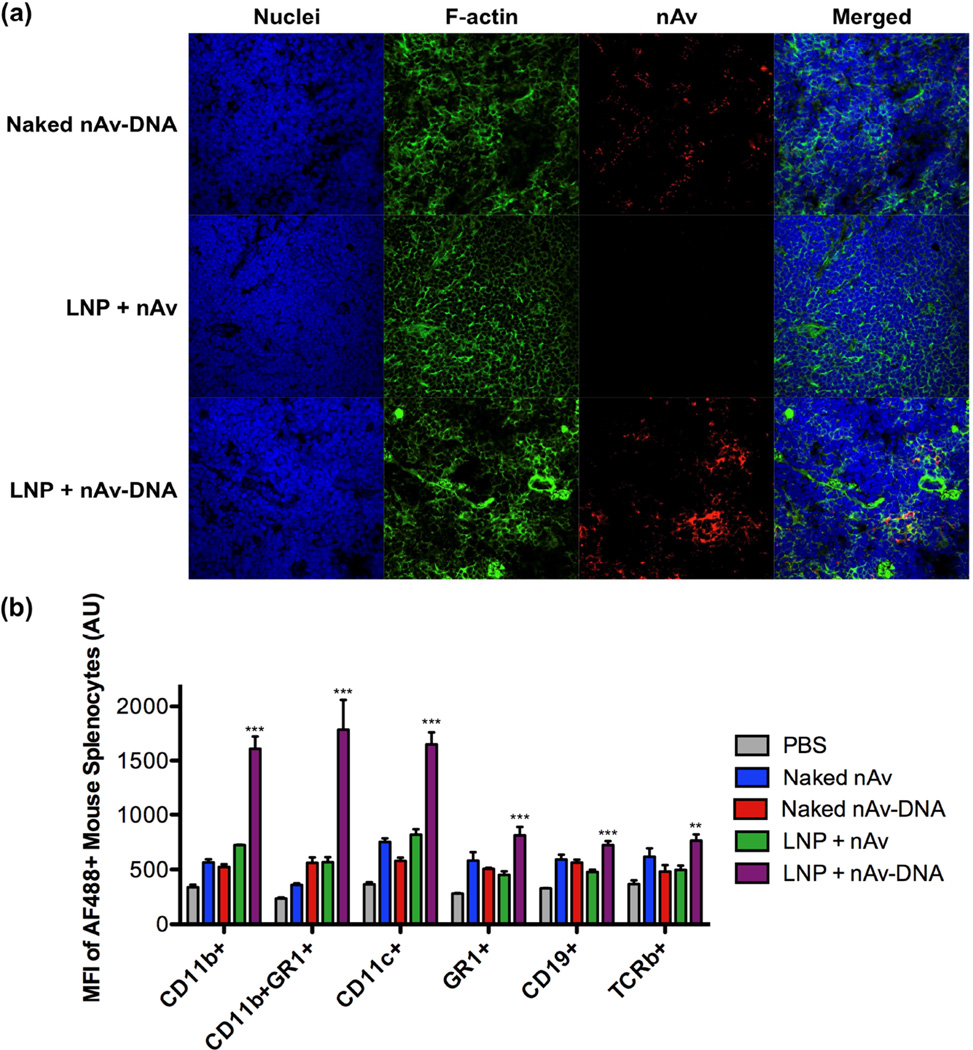
Figure 6.
(a) Immunohistochemical staining revealed accumulation of Cy5.5-labeled nAv in spleen sections harvested from mice 2 h after IV injection of C14-113 / nAv-DNA LNPs. (b) Mice were injected IV with the indicated AF488-labeled nAv treatments, and the spleens were harvested 2 h post-injection. The spleen cell suspension was analyzed by FACS for different immune cell populations as well as for the geometric mean fluorescent intensity (MFI) of positive (AF488+) cells. CD11b+: cells of the macrophage/monocyte lineage; CD11b+GR1+: neutrophils; CD11c+: dendritic cells; GR1+: granulocytes; CD19+: B-cells; TCRb+: T cells. ** indicates p <0.01, *** indicates p < 0.001, comparing LNP + nAV-DNA group to LNP + nAv group.
To determine more quantitatively which immune cell populations within the spleen were positive for protein uptake, mice were injected IV with LNPs encapsulating Alexa Fluor 488 (AF488)-labeled nAv protein or nAv-oligo conjugates, along with their naked counterparts. Two hours after injection, the spleens were harvested and the spleen cell suspension was analyzed by FACS for various immune cell markers as well as for AF488-labeled nAv uptake. Relative to naked nAv or nAv-DNA conjugates, both LNP treatments showed increased nAv uptake efficiency in dendritic cells (CD11c+) and cells of the macrophage/monocyte lineage (CD11b+) (Figure S3). Interestingly, comparing the two LNP treatments, the FACS data showed only modestly higher or equivalent nAv uptake efficiency within dendritic cells or macrophages/monocytes for the LNPs encapsulating nAv-oligo conjugates. For example, the C14-113/nAv-DNA LNPs yielded uptake of nAv in ~37% of splenic dendritic cells, compared with ~29% uptake efficiency for C14-113/nAv LNPs. While this difference was small but significant (p < 0.001), analysis of the AF488 geometric mean fluorescent intensity (MFI) of nAv+ macrophages/monocytes and dendritic cells indicated a substantial 2-to-3-fold enhancement in nAv uptake for the LNPs encapsulating the nAv-oligo conjugates (Figure 6b). This enhancement is noteworthy considering that there were no major differences in the uptake levels of nAv+ macrophage/monocyte cells or dendritic cells among the naked control treatment groups and the group treated with LNPs encapsulating free nAv. These results indicate that protein-oligo conjugation increases the level of protein uptake in those immune cells within the spleen susceptible to intracellular delivery by LNPs.
Recombinant protein-based vaccines are considered less toxic and easier to produce than traditional vaccines based on whole organisms; however, the comparatively lower immunogenicity of protein-based vaccines requires the development of methods for improved delivery to and activation of antigen-presenting cells including dendritic cells and macrophages[34, 35]. In this regard, our observation that oligonucleotide conjugation results in enhanced LNP-mediated protein uptake within splenic dendritic cells and macrophage/monocytes may indicate potential utility in the generation of vaccine responses to proteins. Further experimentation is necessary to determine whether this enhanced protein uptake is also associated with the induction of a more robust immune response characterized by increases in relevant cytokine levels and stimulation of specific T-cell and B-cell responses. Because immunostimulatory oligos such as those enriched in unmethylated CpG motifs are known to be vaccine adjuvants[36–38], the approach outlined here may be suited for the delivery of a vaccine comprising a protein antigen conjugated to a functional, immunostimulatory oligo adjuvant[39, 40]. These delivery studies with HRP and nAv suggest the potential for the use of this oligonucleotide conjugation approach for LNP-mediated delivery of other protein cargo.
4. Conclusion
Using two distinct model proteins, we demonstrated that intracellular protein delivery with lipid-based nanoparticles is dramatically enhanced through conjugation of oligonucleotides to the protein cargo. Using click chemistry, we synthesized conjugates of horseradish peroxidase (HRP) and a DNA oligonucleotide, and we show that one particular lipidoid, C14-113, mediates effective intracellular delivery to HeLa cells of HRP-oligo conjugates but not free HRP protein. Similarly, with NeutrAvidin (nAv), a variant of avidin, we show that binding to a biotinylated oligo significantly enhances intracellular nAv delivery by C14-113 in HeLa cells. When mice were injected intravenously with C14-113 LNPs encapsulating either free nAv or nAv-oligo conjugates, we observed that oligonucleotide conjugation significantly improved intracellular nAv uptake in macrophage/monocytes and dendritic cells within the spleen. These in vivo results suggest that this approach may be suitable for enhanced delivery of protein-based vaccines.
Supplementary Material
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), as a Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C, as well as by the NIH Grants R01-EB000244-27 and 5-R01-CA132091-04. A.A.E. acknowledges graduate research fellowship support from the NSF. D.C. acknowledges the Juvenile Diabetes Research Foundation grant 17–2007-1063. O.V. acknowledges support through the Congressionally Directed Medical Research Programs (CDMRP), Department of Defense (DOD), postdoctoral fellowship award (Grant#W81XWH-13-1-0215).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: A summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Gu Z, Biswas A, Zhao M, Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin V. Intracellular delivery of protein and peptide therapeutics. Drug Discov Today Technol. 2008;5:e95–e103. doi: 10.1016/j.ddtec.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Ford KG, Souberbielle BE, Darling D, Farzaneh F. Protein transduction: An alternative to genetic intervention? Gene Ther. 2001;8:1–4. doi: 10.1038/sj.gt.3301383. [DOI] [PubMed] [Google Scholar]
- 5.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: From molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapoport M, Lorberboum-Galski H. Tat-based drug delivery system--new directions in protein delivery for new hopes? Expert Opin Drug Deliv. 2009;6:453–463. doi: 10.1517/17425240902887029. [DOI] [PubMed] [Google Scholar]
- 7.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, et al. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 9.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, et al. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 11.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 12.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 13.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 14.Phelan A, Elliott G, O'Hare P. Intercellular delivery of functional p53 by the herpesvirus protein vp22. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 15.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent delivery of functional proteins into mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Sayed A, Futaki S, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: Ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel LN, Zaro JL, Shen WC. Cell penetrating peptides: Intracellular pathways and pharmaceutical perspectives. Pharm Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 18.Debs RJ, Freedman LP, Edmunds S, Gaensler KL, Duzgunes N, Yamamoto KR. Regulation of gene expression in vivo by liposome-mediated delivery of a purified transcription factor. J Biol Chem. 1990;265:10189–10192. [PubMed] [Google Scholar]
- 19.Zelphati O, Wang Y, Kitada S, Reed JC, Felgner PL, Corbeil J. Intracellular delivery of proteins with a new lipid-mediated delivery system. J Biol Chem. 2001;276:35103–35110. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Ishii T, Cabral H, Kim HJ, Seo JH, Nishiyama N, et al. Charge-conversional polyionic complex micelles-efficient nanocarriers for protein delivery into cytoplasm. Angew Chem Int Ed Engl. 2009;48:5309–5312. doi: 10.1002/anie.200900064. [DOI] [PubMed] [Google Scholar]
- 21.Weill CO, Biri S, Adib A, Erbacher P. A practical approach for intracellular protein delivery. Cytotechnology. 2008;56:41–48. doi: 10.1007/s10616-007-9102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tinsley JH, Hawker J, Yuan Y. Efficient protein transfection of cultured coronary venular endothelial cells. Am J Physiol. 1998;275:H1873–H1878. doi: 10.1152/ajpheart.1998.275.5.H1873. [DOI] [PubMed] [Google Scholar]
- 23.Futami J, Kitazoe M, Maeda T, Nukui E, Sakaguchi M, Kosaka J, et al. Intracellular delivery of proteins into mammalian living cells by polyethylenimine-cationization. J Biosci Bioeng. 2005;99:95–103. doi: 10.1263/jbb.99.95. [DOI] [PubMed] [Google Scholar]
- 24.Coue G, Engbersen JF. Functionalized linear poly(amidoamine)s are efficient vectors for intracellular protein delivery. J Control Release. 2011;152:90–98. doi: 10.1016/j.jconrel.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 25.Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 26.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 27.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of rnai therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novobrantseva TI, Borodovsky A, Wong J, Klebanov B, Zafari M, Yucius K, et al. Systemic rnai-mediated gene silencing in nonhuman primate and rodent myeloid cells. Mol Ther Nucleic Acids. 2012;1:e4. doi: 10.1038/mtna.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of rnai therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, et al. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem Int Ed Engl. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, et al. Rapid discovery of potent sirna-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 33.Lee MJ-E, Veiseh O, Bhattarai N, Sun C, Hansen SJ, Ditzler S, et al. Rapid pharmacokinetic and biodistribution studies using cholorotoxin-conjugated iron oxide nanoparticles: A novel non-radioactive method. PLoS One. 2010;5:e9536. doi: 10.1371/journal.pone.0009536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peek LJ, Middaugh CR, Berkland C. Nanotechnology in vaccine delivery. Adv Drug Deliv Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh M, Chakrapani A, O'Hagan D. Nanoparticles and microparticles as vaccine-delivery systems. Expert Rev Vaccines. 2007;6:797–808. doi: 10.1586/14760584.6.5.797. [DOI] [PubMed] [Google Scholar]
- 36.Klinman DM. Cpg DNA as a vaccine adjuvant. Expert Rev Vaccines. 2003;2:305–315. doi: 10.1586/14760584.2.2.305. [DOI] [PubMed] [Google Scholar]
- 37.Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV. Cpg oligodeoxynucleotides act as adjuvants that switch on t helper 1 (th1) immunity. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weeratna RD, McCluskie MJ, Xu Y, Davis HL. Cpg DNA induces stronger immune responses with less toxicity than other adjuvants. Vaccine. 2000;18:1755–1762. doi: 10.1016/s0264-410x(99)00526-5. [DOI] [PubMed] [Google Scholar]
- 39.Datta SK, Cho HJ, Takabayashi K, Horner AA, Raz E. Antigen–immunostimulatory oligonucleotide conjugates: Mechanisms and applications. Immunol Rev. 2004;199:217–226. doi: 10.1111/j.0105-2896.2004.00149.x. [DOI] [PubMed] [Google Scholar]
- 40.Wilson JT, Keller S, Manganiello MJ, Cheng C, Lee CC, Opara C, et al. Ph-responsive nanoparticle vaccines for dual-delivery of antigens and immunostimulatory oligonucleotides. ACS Nano. 2013;7:3912–3925. doi: 10.1021/nn305466z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.