Abstract
Background
Rhipicephalus sanguineus belongs to a complex of hard tick species with high veterinary-medical significance. Recently, new phylogenetic units have been discovered within R. sanguineus, which therefore needs taxonomic revision. The present study was initiated to provide new information on the phylogeography of relevant haplotypes from less studied regions of Europe and Africa. With this aim, molecular-phylogenetic analyses of two mitochondrial markers were performed on 50 ticks collected in Hungary, the Balkans, countries along the Mediterranean Sea, Kenya and Ivory Coast.
Results
In the “temperate lineage” of R. sanguineus, based on cytochrome c oxidase subunit 1 (cox1) and 16S rRNA genes, Rhipicephalus sp. I was only found in the eastern part of the Mediterranean Basin (with relatively homogenous haplotypes), whereas Rhipicephalus sp. II occurred in the middle-to-western part of this region (with phylogenetically dichotomous haplotypes). Ticks identified as R. leporis (based on morphology and cox1 gene) were found in Kenya and Ivory Coast. These clustered phylogenetically within R. sanguineus (s.l.) (“tropical lineage”).
Conclusions
In the Mediterranean Basin two mitochondrial lineages of R. sanguineus, i.e. Rhipicephalus sp. I and Rhipicephalus sp. II exist, which show different geographical distribution. Therefore, data from this study confirm limited gene flow between Rhipicephalus sp. I and Rhipicephalus sp. II, but more evidence (analyses of nuclear markers, extensive morphological and biological comparison etc.) are necessary to infer if they belong to different species or not. The phylogenetic relationships of eastern and western African ticks, which align with R. leporis, need to be studied further within R. sanguineus (s.l.) (“tropical lineage”).
Keywords: Phylogeography, cox1, 16S rRNA gene, Rhipicephalus sanguineus, Rhipicephalus leporis
Background
Rhipicephalus sanguineus (sensu lato) (Acari: Ixodidae) belongs to a complex of at least 17 hard tick species, some with high medical and veterinary importance [1]. The type-species of this group was formerly called R. sanguineus (sensu stricto) (the brown dog tick), with cosmopolitan distribution owing to its high adaptability (i.e. being able to utilize endophilic and exophilic habitats in both urban and rural environment; having year-round activity and up to four generations per year; exhibiting passive or active host-seeking behaviour) [2]. Although typically a dog parasite (in all three developmental stages), R. sanguineus may also infest a wide range of domestic and wild animal host species, even humans. There are several animal and human pathogens (including zoonotic ones) that are or may be transmitted by R. sanguineus (reviewed in [3]). On account of its preference of warmer climates, global warming was predicted to induce the expansion of the geographical range of R. sanguineus [2].
According to current knowledge, R. sanguineus is not a single species. Molecular phylogeographic studies have found high intraspecific divergence of mitochondrial DNA between R. sanguineus from Brazil and Argentina, while a strong genetic relationship was detected between its European and Argentinean populations [4]. The differences between these strains were also demonstrated with morphological comparisons under scanning electron microscopy [5] and crossbreeding experiments [4]. Similarly, R. sanguineus collected in the USA and Mexico were shown to be genetically different [6]. A latitude-linked geographical pattern of the two major R. sanguineus groups has been confirmed by further, global scale studies, which showed with molecular-phylogenetic methods [7–9] or crossbreeding experiments [10] that (at least) two species might exist under this name, and both occur in the New World and in the Old World. These two clades have been designated as “tropical species” or northern lineage and “temperate species” or southern lineage [8, 11]. Moreover, a comprehensive morphological and phylogenetic study drew the attention to the existence of further operational taxonomic units (Rhipicephalus sp. I-IV) in addition to the “tropical species” [1]. The geographical distribution of these groups has recently been shown to be associated with climate variables, such as temperature [12].
In the above studies, certain regions of the globe appear to be underrepresented, as exemplified by several countries in or close to the Mediterranean Basin. Accordingly, it has been stated that further morphological and genetic studies of ticks in the R. sanguineus complex are needed from the Old World [1]. Thus, the primary aim of the present study was to provide relevant data from less studied regions, i.e. to report and compare in a phylogeographical context two mitochondrial markers of R. sanguineus from Hungary (where its occasional emergence can be anticipated; [13]), the Balkans and in a broader sense the Mediterranean Basin, as well as western and eastern Africa. In this way, representatives of R. sanguineus from both the “temperate” and “tropical” lineages have been included. The nomenclature of these categories is used sensu Dantas-Torres et al. [1] throughout the text.
Methods
In this study, 68 ticks were collected (mainly from dogs) in 14 countries between 2010 and 2016. Eighteen ticks, morphologically identified as R. sanguineus and collected in France, Morocco, Algeria, Tunisia, Serbia and Turkey did not yield DNA, or their sequencing was not successful, therefore these samples were excluded from further study. Data of the remaining 50 ticks (collected in 11 countries) are shown in Table 1.
Table 1.
Data of Rhipicephalus spp. ticks used in this study. The sex/stage of ticks and date of collection are not shown
| Speciesa | Country | Location | Host of origin | cox1 sequence identityb | cox1 sequence accession number | Number of identical sequences | 16S sequence accession number |
|---|---|---|---|---|---|---|---|
| “R. sanguineus” | Serbia | Kajtasovo | dog | 630/6302 | KX757879 | KX793717 | |
| Brnjica | dog | 630/6301 | KX757880 | KX793718 | |||
| Jajinci | dog | 629/6301 | KX757881 | KX793719 | |||
| 630/6301 | KX757882 | KX793719 | |||||
| Petnica | dog | 629/6302 | KX757883 | KX793720 | |||
| Lebane | dog | 629/6301 | KX757885 | KX793722 | |||
| Boka | dog | 628/6301 | KX757906 | KX793738 | |||
| 629/6301 | KX757907 | KX793738 | |||||
| 629/6302 | KX757905 | KX793739 | |||||
| Croatia | Rovinj | unknown | 630/6302 | KX757887 | 2 | KX793724 | |
| Pula | dog | 629/6302 | KX757888 | KX793725 | |||
| 630/6302 | KX757889 | KX793726 | |||||
| Zagreb | dog | 629/6302 | KX757890 | 1 | KX793727 | ||
| 630/6302 | KX757896 | 2 | KX793727 | ||||
| 628/6303 | KX757893 | 2 | KX793730 | ||||
| Zadar | dog | 629/6302 | KX757892 | 2 | KX793729 | ||
| Sibenik | dog | 629/6302 | KX757895 | KX793731 | |||
| Romania | Babadag | golden jackal | 630/6301 | KX757915 | KX793746 | ||
| Histria | dog | 628/6301 | KX757916 | KX793746 | |||
| Hungary | Szekszárdc | dog | 628/6302 | KX757901 | 2 | KX793734 | |
| Malta | Siggiewi | dog | 630/6302 | KX757902 | 6 | KX793735 | |
| Italy | Piacenza | dog | 630/6302 | KX757904 | KX793737 | ||
| 626/6303 | KX757903 | KX793736 | |||||
| Greece | Thessaloniki | dog | 629/6301 | KX757908 | 2 | KX793740 | |
| Algeria | Kehf Lagareb | bat (Myotis punicus) | 630/6302 | KX757910 | KX793742 | ||
| Morocco | Al-Hoceima | dog | 623/6303 | KX757909 | KX793741 | ||
| Ivory Coast | Abidjan | dog | 620/6204 | KX757914 | KX793745 | ||
| R. rossicus | Romania | Caraorman | dog | 624/6305 | KX757897 | KX793732 | |
| Lazuri | dog | 628/6305 | KX757898 | KX793733 | |||
| 630/6305 | KX757899 | KX793733 | |||||
| Grindul | dog | 629/6305 | KX757900 | – | |||
| R. leporis | Kenya | Turkana, Samburu | dog, cattle | 627/6306 | KX757911 | 4 | KX793743 |
| Ivory Coast | Bas-Sassandra | dog | 627/6306 | KX757912 | 1 | KX793744 | |
| 625/6306 | KX757913 | 1 | KX793744 | ||||
| 625/6306 | KX757917 | 2 | KX793744 |
aCurrently species delineation within R. sanguineus (s.l.) requires revision [1]: here species names designating reference sequences in GenBank are used
bNumber of nucleotides identical with reference sequence expressed as bp/bp. Superscript numbers indicate reference sequences (also shown on Figs. 2 and 3) as follows: 1KF219745; 2KU556745; 3AF081829; 4KF200084; 5JX394215; 6KM235720
c R. sanguineus is not regarded as indigenous to Hungary; these specimens (collected in 2012 from a dog that has never left the country) exemplify rare autochthonous cases
All ticks were stored in 96% ethanol. The morphology of ticks was preliminarily assessed using a stereo microscope (SMZ-2 T, Nikon Instruments, Japan, illuminated with model 5000–1, Intralux, Urdorf-Zürich, Switzerland) and standard keys (R. sanguineus: [14]; R. rossicus: [15]). In the category of R. sanguineus the adanal plate length to breadth ratio (which was reported to provide the only significant difference between Rhipicephalus sp. I-II: [1]) showed extreme variation even between conspecific males collected from the same dog (Fig. 1), therefore measurements were not taken. Identification of R. leporis males was based on the adanal and spiracular plates [16]. Pictures were made with a VHX-5000 (Keyence Co., Osaka, Japan) digital microscope.
Fig. 1.
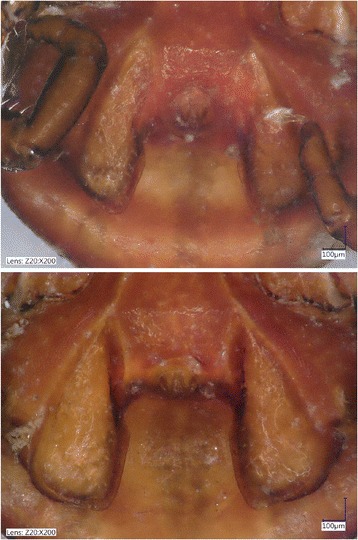
Adanal plates of two Rhipicephalus sp. I males (with only 1 bp difference in the amplified part of the cox1 gene; collected from the same dog in Jajinci, Serbia) showing similar shape (e.g. posteromedial corner), but highly different length-to-breadth ratio (upper: 2.7, lower: 2)
DNA was extracted from ticks using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instruction, and including an overnight digestion in tissue lysis buffer with proteinase K at 56 °C. The cytochrome c oxidase subunit 1 (cox1) gene was chosen as the first target for molecular analysis, because of its suitability as a DNA-barcode marker for tick species identification [17]. PCR was modified from Folmer et al. [18] and amplified approximately 710 bp using the primers HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) and LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) in a reaction volume of 25 μl, which contained 1 U (0.2 μl) HotStarTaq Plus DNA polymerase, 2.5 μl 10× CoralLoad Reaction buffer (including 15 mM MgCl2), 0.5 μl PCR nucleotide Mix (0.2 mM each), 0.5 μl (1 μM final concentration) of each primer, 15.8 μl ddH2O and 5 μl template DNA. For amplification, an initial denaturation step at 95 °C for 5 min was followed by 40 cycles of denaturation at 94 °C for 40 s, annealing at 48 °C for 1 min and extension at 72 °C for 1 min. Final extension was performed at 72 °C for 10 min.
To confirm the results obtained with cox1, another PCR was used to amplify approximately 460 bp of 16S rDNA of Ixodidae [19], using the primers 16S + 1 (5′-CTG CTC AAT GAT TTT TTA AAT TGC TGT GG-3′) and 16S-1 (5′-CCG GTC TGA ACT CAG ATC AAG T-3′). Other reaction components, as well as cycling conditions were the same as above, except for an annealing temperature of 51 °C.
PCR products were visualized in a 1.5% agarose gel. Purification and sequencing was done by Biomi Inc. (Gödöllő, Hungary). The sequences were submitted to the GenBank database under accession numbers KX757879–KX757917 (cox1) and KX793717–KX793746 (16S) (see Table 1). The MEGA model selection method was applied to choose the appropriate model for phylogenetic analyses. Phylogenetic analyses were conducted using the neighbour-joining method (p-distance model) and maximum likelihood method (Jukes-Cantor model) using MEGA version 6.0.
Results
Out of the 50 molecularly analysed ticks, 38 were identified as R. sanguineus based on the amplified parts of their cox1 and 16S rRNA genes (i.e. corresponding to groups Rhipicephalus sp. I and Rhipicephalus sp. II of the “temperate lineage”, and R. sanguineus (s.l.) “tropical lineage”).
The cox1 sequences from Rhipicephalus sp. I had one to three nucleotides different among them (627–630/630 bp; 99.5–100% sequence similarity). In the subgroup Rhipicephalus sp. IIa, one to three nucleotides were different from each other (627–630/630 bp; 99.5–100% sequence similarity), whereas subgroup Rhipicephalus sp. IIb was more heterogeneous, with up to seven bp differences (623/630 bp; 98.9% sequence similarity). Comparisons between the two subgroups (a + b) of Rhipicephalus sp. II showed 28–32 bp differences among haplotypes, resulting in 94.9–95.6% (598–602/630 bp) sequence similarity. The cox1 sequences differed by 56–60 nucleotides between Rhipicephalus sp. I and II (570–574/630 bp; 90.5–91.1% similarity).
Phylogenetic analyses of cox1 sequence data indicated that haplotypes of Rhipicephalus sp. I formed a single clade (Figs. 2 and 3). On the other hand, representative sequences from Rhipicephalus sp. II formed two subgroups (a + b), with strong support (bootstrap = 99–100%) (Figs. 2 and 3). These relationships were confirmed following phylogenetic analysis of 16S rDNA sequence data, i.e. 16S rRNA gene haplotypes formed two main clusters (Rhipicephalus sp. I and II) within the “temperate lineage” (Figs. 4 and 5), although the separation of Rhipicephalus sp. IIa and IIb subgroups (based on these shorter sequences) was poorly supported in the maximum likelihood tree (bootstrap = 53%) (Fig. 5).
Fig. 2.
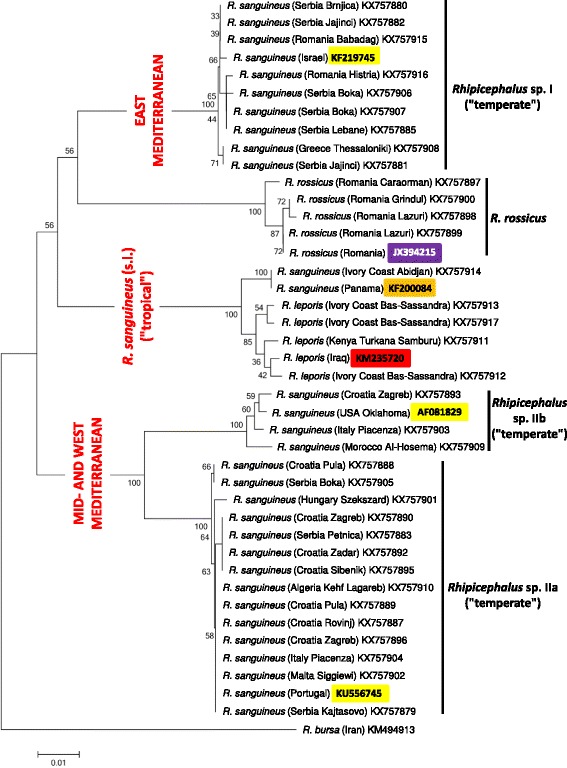
Phylogeny of Rhipicephalus spp. following neighbor-joining analysis of cox1 gene. For clarity, only one reference sequence (the closest Rhipicephalus haplotype available in GenBank from other studies) is included for each (sub)group. These reference sequences are indicated with coloured background of their accession numbers according to their taxonomic groups. Branch lengths represent the number of substitutions per site inferred according to the scale shown
Fig. 3.
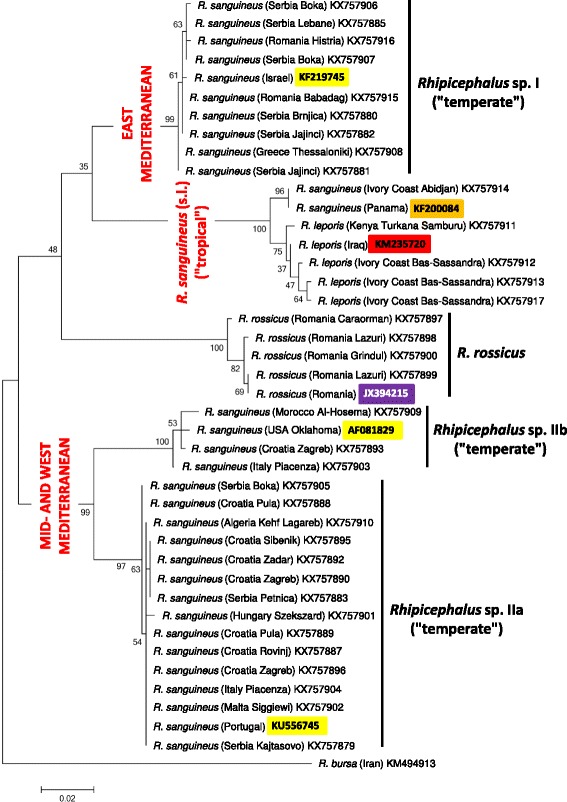
Phylogeny of Rhipicephalus spp. following maximum likelihood analysis of cox1 gene. For clarity, only one reference sequence (the closest Rhipicephalus haplotype available in GenBank from other studies) is included for each (sub)group. These reference sequences are indicated with coloured background of their accession numbers according to their taxonomic groups. Branch lengths represent the number of substitutions per site inferred according to the scale shown
Fig. 4.
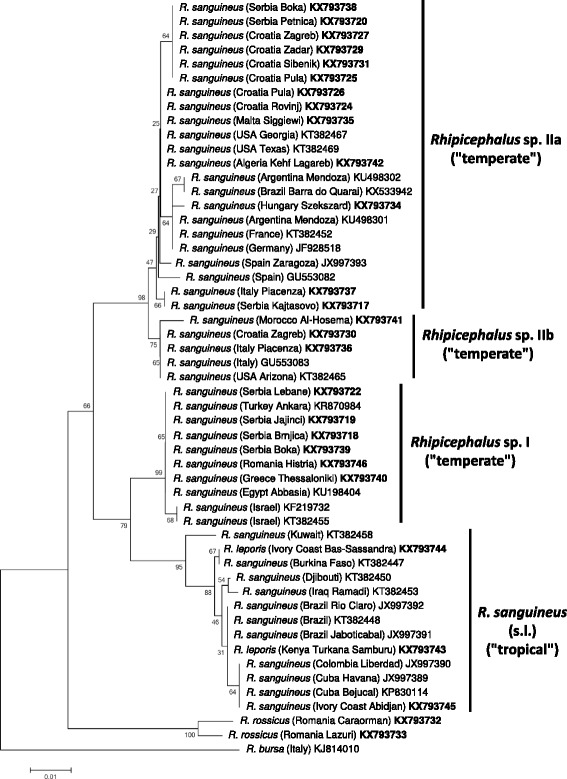
Phylogeny of Rhipicephalus spp. following neighbor-joining analysis of 16S rRNA gene. Sequences from this study are indicated with bold accession numbers. Branch lengths represent the number of substitutions per site inferred according to the scale shown
Fig. 5.
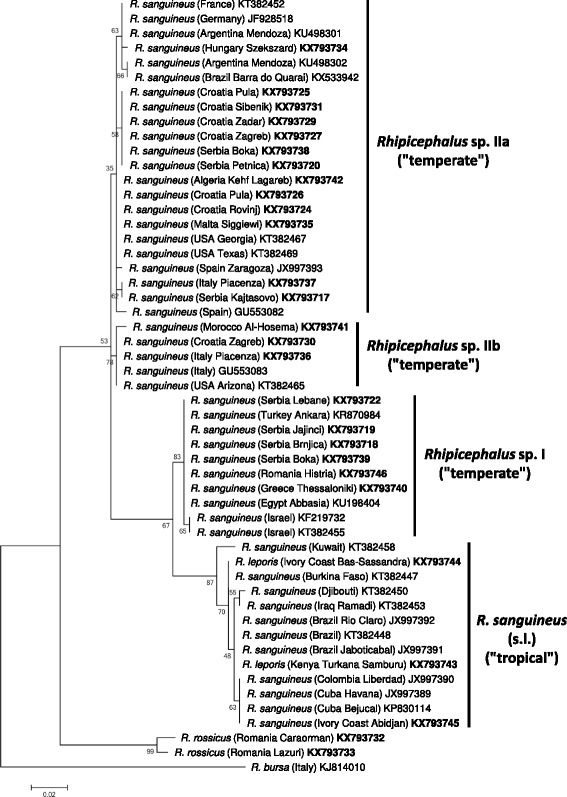
Phylogeny of Rhipicephalus spp. following maximum likelihood analysis of 16S rRNA gene. Sequences from this study are indicated with bold accession numbers. Branch lengths represent the number of substitutions per site inferred according to the scale shown
Geographically, samples of Rhipicephalus sp. I. have only been collected in the eastern part of the Mediterranean Basin, similarly to the origin of other sequences from this group available in GenBank (Fig. 6). Complementarily to this, samples of the subgroup Rhipicephalus sp. IIa have been collected in the middle and western part of the Mediterranean Basin, as well as in Hungary, showing a zone of overlap with Rhipicephalus sp. I in Serbia. The geographical occurrence of Rhipicephalus sp. IIb was focal within the range of Rhipicephalus sp. IIa (in northern Morocco, Italy and Croatia-Zagreb; Fig. 6).
Fig. 6.
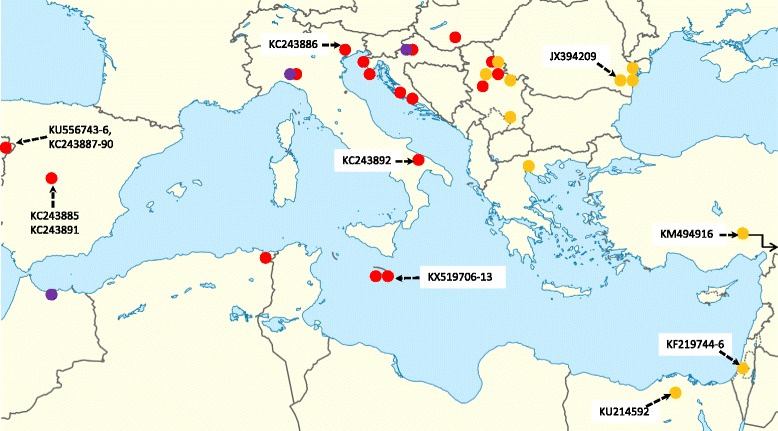
Geographical distribution of Rhipicephalus sanguineus cox1 haplotypes in and near the Mediterranean Basin. Coloured circles without accession number indicate samples from this study (yellow, Rhipicephalus sp. I; red, Rhipicephalus sp. IIa; purple, Rhipicephalus sp. IIb). Among these, multiple sequences for the same location are not shown. Haplotypes from other studies (which had 99–100% similarity with those in this study) are marked with GenBank accession numbers, connected with dash line arrows to the relevant location. Overlapping circles indicate the same location (where different haplotypes were found); the zigzag arrow marks the direction of location (outside this map) for the sample from Iran (Tehran). The location within a country is accurately indicated for samples of this study, as well as for Egypt, Italy and Romania from other studies, but for the rest of the samples only the country was known
There was only one specimen from the Ivory Coast which was molecularly identified as R. sanguineus (s.l.) (“tropical lineage”). The cox1 sequence from this specimen was 100% identical to one sequence in GenBank, from Central America (Panama: KF200084; [20]).
Based on cox1 gene, all remaining (eight) ticks from Kenya and the Ivory Coast (Table 1) clustered phylogenetically with R. leporis (Figs. 2 and 3). Males of R. leporis identified morphologically in the present study had very long and narrow dorsal prolongation of spiracular plates, and tear-drop shaped adanal plates rounded posteriorly (Fig. 7). Comparison of the cox1 sequences of relevant specimens with a voucher sequence in GenBank (Iraq: KM235720) confirmed them as R. leporis (624–626/629 bp; 99.2–99.5% similarity). The cox1 haplotypes of R. leporis and R. sanguineus (s.l.) (“tropical lineage”) showed ten bp differences (620/630 bp; 98.4% similarity) within the same country (Ivory Coast), and their separation was phylogenetically well supported (100%: Figs. 2 and 3).
Fig. 7.
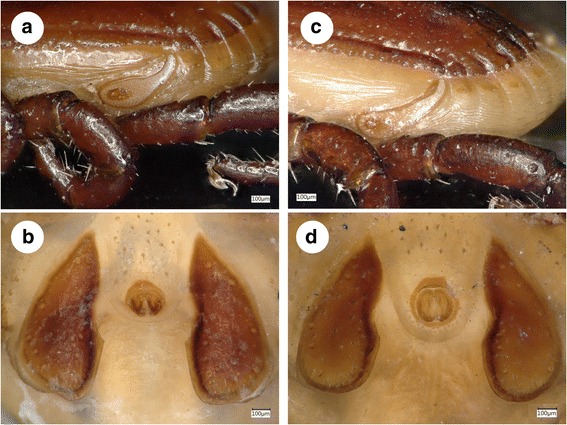
Diagnostically important structures of R. sanguineus and R. leporis males. a Left spiracular plate of Rhipicephalus sp. I collected in Histria, Romania. b Adanal plates of Rhipicephalus sp. II collected in Pula, Croatia. c Long and narrow dorsal prolongation of spiracular plates in R. leporis. d Tear-drop shaped, posteriorly rounded adanal plates in R. leporis
In the 16S rRNA gene sequence analysis, the difference between R. leporis and R. sanguineus (s.l.) (“tropical lineage”) was not so evident. For instance, there was only one nucleotide difference between R. leporis isolates from east and west Africa (Kenya vs Ivory Coast). Rhipicephalus leporis from the Ivory Coast was not different from a R. sanguineus haplotype (KT382447) collected in neighbouring Burkina Faso, but both had five bp differences from R. sanguineus (s.l.) collected in Kuwait (394/399 bp; 98.7% similarity). Phylogenetically, R. leporis 16S haplotypes clustered separately from the latter (KT382458), but together with R. sanguineus (s.l.) isolates from the Old and New Worlds (Figs. 4 and 5).
Rhipicephalus rossicus was only identified in Romania (represented by four samples: Table 1). In the neighbor-joining analysis of cox1 sequences (Fig. 2) R. rossicus clustered as the sister group to eastern Mediterranean isolates (Rhipicephalus sp. I). However, based on 16S rRNA gene, R. rossicus formed a sister group to all R. sanguineus isolates (Figs. 4 and 5).
Discussion
In a previous comprehensive study on the morphological and genetic diversity of R. sanguineus [1] it was shown that two groups under the “temperate lineage” (i.e. Rhipicephalus sp. I and Rhipicephalus sp. II) diverge molecularly to the point where they may be considered separate species. However, except for the adanal plate length-to-breadth ratio, no consistent morphological differences were observed between them [1]. Furthermore, in the present study the adanal plate length-to-breadth ratio was shown to vary even between almost identical haplotypes of Rhipicephalus sp. I. Therefore, for the investigation of their geographical distribution, haplotypes were here assigned to specimens of R. sanguineus based on molecular data, involving the analysis of two mitochondrial genetic markers. However, it also should be considered that analysis of relatively short sequences may cause an over-resolution of phylogenetic trees based on mitochondrial markers.
Phylogenetic analysis of cox1 sequences from new isolates of “R. sanguineus” in the present study confirmed the existence of paraphyletic groups previously referred to under the same species name [1]. Here it was also shown that in contrast to Rhipicephalus sp. I (which is a homogenous group), Rhipicephalus sp. II is rather heterogenous, consisting of two, phylogenetically well-defined clades (a + b). Phylogenetic analysis of 16S rDNA sequence data verified that these categories are also represented by formerly reported sequences from the New World: the clade composed of sequences classified as Rhipicephalus sp. IIa, based on phylogenetic analysis, included samples from Argentina, southernmost Brazil and the USA (Georgia, Texas), while another 16S rDNA sequence from the USA (Arizona) (as well as a cox1 sequence from Oklahoma) belonged to Rhipicephalus sp. IIb (Figs. 2, 3, 4 and 5).
The “tropical” and “temperate” lineages of “R. sanguineus” are reported to have a latitude related geographical pattern [1, 7]. Adding to this, it was demonstrated here that within the “temperate lineage” the distribution of Rhipicephalus sp. I and sp. II reflect longitudinal separation within and close to the Mediterranean Basin. Based on cox1 sequences, R. rossicus was demonstrated to phylogenetically cluster with the eastern Mediterranean Rhipicephalus sp. I, the former having an eastern distribution in Europe, emerging towards the west [15].
In this study the great majority of ticks from Rhipicephalus sp. I and Rhipicephalus sp. II mitochondrial lineages were collected in countries with a uniform Mediterranean climate (in Serbia haplotypes of these two categories even originated in the same location), and the phylogenetically separate subgroups Rhipicephalus sp. IIa and IIb occurred simultaneously in certain sampling sites (in Piacenza/Italy, Zagreb/Croatia). Therefore, the geographical patterns of Rhipicephalus sp. I, Rhipicephalus sp. IIa and Rhipicephalus sp. IIb in the Mediterranean Basin appear to be independent of current climatic conditions (unlike the geographical distribution of the “tropical” and “temperate” lineages of R. sanguineus: [12]).
If not current climatic conditions, then other factors influencing the tick life-cycle may provide a plausible explanation for the parapatric separation of Rhipicephalus sp. I and Rhipicephalus sp. II lineages in the Mediterranean Basin. Molecular evidence from a broad range of invertebrate and vertebrate taxa (i.e. potentially encompassing ticks and their hosts) indicate that southern peninsulas of Europe acted as major refugia during ice age(s), from which genetically distinct clades emerged [21]. While recolonization events to northern parts of Europe may have resulted in secondary sympatry for these clades, their genetic differences are still maintained and demonstrable. Thus, several (potential) host species of R. sanguineus had also been affected by glacial isolation in the same way. For example, wolf haplotype lineages and hedgehog species differ between Italy and the Balkans [21, 22], and genetically distinct populations of bank voles exist in the western and eastern Balkans [23]. These geographical patterns are similar to the one observed for Rhipicephalus sp. I and Rhipicephalus sp. II in the present study, suggesting that during ice age(s) the Mediterranean range of R. sanguineus (in sympatry with the above hosts) was not confluent, but inhabited by reproductively isolated tick populations. Nevertheless, successful interbreeding between ticks from Rhipicephalus sp. I and Rhipicephalus sp. IIb populations (listed as reference sequences from Israel and USA, Oklahoma on Figs. 2 and 3) had already been demonstrated [10].
Rhipicephalus leporis was hitherto known to occur in the Middle East and Central Asia [16], but here its specimens (identified both morphologically and genetically) are reported from Africa. Apart from a broad range of wild animals, dogs and goats are among the preferred hosts of this tick species [16]. Consequently, R. leporis could have been unknowingly transported on these hosts to regions outside its formerly known range, and not necessarily recently (considering the genetic divergence between its isolates from Iraq vs Kenya and the Ivory Coast). If confirmed, a likely explanation for R. leporis not being discovered in Africa until now is its morphological similarity to R. sanguineus (s.l.).
In the present study R. leporis and R. sanguineus (s.l.) (“tropical lineage”) clustered close to each other phylogenetically, with their cox1 sequences differing by 1.6%. This sequence divergence is within the range (i.e. 0.2–3%) of reported intraspecific nucleotide variation for the cox1 gene of R. sanguineus (s.l.) [1]. In addition, the cox1 sequence/phylogenetic difference between R. leporis and R. sanguineus (s.l.) (“tropical lineage”) was not reproducible with the analysis of 16S rRNA gene. When comparing 16S rDNA sequences of Rhipicephalus spp. it should be taken into account that the amplified part of the 16S rRNA gene was considerably shorter than cox1, and the average interspecific distance was reported to be lower for this gene than for either cox1 or 12S genes [17]. Therefore, the resolution of analysing these shorter 16S gene fragments may not suffice to distinguish closely related species. In addition, the sequence divergence between 12S gene sequences of R. leporis and R. sanguineus (s.l.) (“tropical lineage”) was reported to be of similar magnitude than between isolates of R. leporis or R. sanguineus (s.l.) themselves. For instance, sequences of the 12S gene of ticks from Kuwait, morphologically identified as R. leporis, were 99% similar to sequences of R. leporis from Iraq and R. sanguineus (s.l.) from South America [12]. Based on this ambiguity, further and larger scale studies may be needed to ultimately verify that R. leporis from east and west Africa are not intraspecific morphological variants of R. sanguineus (s.l.) within the “tropical lineage”.
Conclusions
Two mitochondrial lineages within the “temperate species” of R. sanguineus (i.e. Rhipicephalus sp. I and Rhipicephalus sp. II) show different geographical distribution in the region of Mediterranean Basin, confirming limited gene flow between them. However, more evidence (analyses of nuclear markers, extensive morphological and biological comparison etc.) are necessary to infer if they belong to different species or not. Similarly, the phylogenetic relationships of eastern and western African ticks, which align with R. leporis, need to be studied further within R. sanguineus (s.l.) (“tropical species”).
Acknowledgements
The survey was organized in the framework of the EurNegVec COST Action TD1303. The authors thank Elias Papadopoulos for providing ticks from Greece.
Funding
Molecular analyses were funded by OTKA 115854 (Hungary). Tick collection was supported by grants OI173006 (Serbia), PN-II-RU-TE-2014-4-1389 (Romania), GENOTICKTRECK-1957 (Croatia). GF receives János Bolyai Research Scholarship of the Hungarian Academy of Sciences. The publication of this research was supported by the 11475-4/2016/FEKUT grant of the Hungarian Ministry of Human Resources.
Availability of data and materials
The sequences obtained and/or analysed during the current study are deposited in GenBank under accession numbers KX757879–KX757917 (cox1) and KX793717–KX793746 (16S). All other relevant data are included in the manuscript.
Authors’ contributions
SH initiated and supervised the study, extracted the DNA, did part of the morphological and genetic comparisons, wrote the manuscript. ADS, ST, RB, JDA and RF provided samples from multiple locations for the study. JK made the microscopic pictures and performed phylogenetic analyses. NT performed the PCRs. TG collected the important sample from Hungary, and MLB those from Algeria. GF organized part of the sample collection. All authors contributed to, read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Ticks were collected from animals during regular veterinary care, therefore no permission was needed.
Contributor Information
Sándor Hornok, Email: hornok.sandor@univet.hu.
Attila D. Sándor, Email: adsandor@gmail.com
Snežana Tomanović, Email: snezanat@imi.bg.ac.rs.
Relja Beck, Email: relja.beck@gmail.com.
Gianluca D’Amico, Email: gianluca.damico@usamvcluj.ro.
Jenő Kontschán, Email: jkontschan@gmail.com.
Nóra Takács, Email: takacs.nora@aotk.szie.hu.
Tamás Görföl, Email: gorfol.tamas@gmail.com.
Mohammed Lamine Bendjeddou, Email: amine_bendjeddou@yahoo.fr.
Gábor Földvári, Email: foldvarigabor@gmx.de.
Róbert Farkas, Email: farkas.robert@univet.hu.
References
- 1.Dantas-Torres F, Latrofa MS, Annoscia G, Giannelli A, Parisi A, Otranto D. Morphological and genetic diversity of Rhipicephalus sanguineus sensu latofrom the New and Old Worlds. Parasit Vectors. 2013;6:213. [DOI] [PMC free article] [PubMed]
- 2.Dantas-Torres F. Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasit Vectors. 2010;3:26. [DOI] [PMC free article] [PubMed]
- 3.Dantas-Torres F. The brown dog tick, Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae): from taxonomy to control. Vet Parasitol. 2008;152:173–85. doi: 10.1016/j.vetpar.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 4.Szabó MP, Mangold AJ, João CF, Bechara GH, Guglielmone AA. Biological and DNA evidence of two dissimilar populations of the Rhipicephalus sanguineus tick group (Acari: Ixodidae) in South America. Vet Parasitol. 2005;130:131–40. doi: 10.1016/j.vetpar.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira PR, Bechara GH, Denardi SE, Saito KC, Nunes ET, Szabó MP, Mathias MI. Comparison of the external morphology of Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae) ticks from Brazil and Argentina. Vet Parasitol. 2005;129:139–47. doi: 10.1016/j.vetpar.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Eremeeva ME, Zambrano ML, Anaya L, Beati L, Karpathy SE, Santos-Silva MM, et al. Rickettsia rickettsii in Rhipicephalus ticks, Mexicali, Mexico. J Med Entomol. 2011;48:418–21. doi: 10.1603/ME10181. [DOI] [PubMed] [Google Scholar]
- 7.Burlini L, Teixeira KR, Szabó MP, Famadas KM. Molecular dissimilarities of Rhipicephalus sanguineus (Acari: Ixodidae) in Brazil and its relation with samples throughout the world: is there a geographical pattern? Exp Appl Acarol. 2010;50:361–74. doi: 10.1007/s10493-009-9321-8. [DOI] [PubMed] [Google Scholar]
- 8.Moraes-Filho J, Marcili A, Nieri-Bastos FA, Richtzenhain LJ, Labruna MB. Genetic analysis of ticks belonging to the Rhipicephalus sanguineus group in Latin America. Acta Trop. 2011;117:51–5. doi: 10.1016/j.actatropica.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Liu GH, Chen F, Chen YZ, Song HQ, Lin RQ, Zhou DH, Zhu XQ. Complete mitochondrial genome sequence data provides genetic evidence that the brown dog tick Rhipicephalus sanguineus (Acari: Ixodidae) represents a species complex. Int J Biol Sci. 2007;9:361–9. doi: 10.7150/ijbs.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin ML, Studer E, Killmaster L, Zemtsova G, Mumcuoglu KY. Crossbreeding between different geographical populations of the brown dog tick, Rhipicephalus sanguineus (Acari: Ixodidae) Exp Appl Acarol. 2012;58:51–68. doi: 10.1007/s10493-012-9561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nava S, Mastropaolo M, Venzal JM, Mangold AJ, Guglielmone AA. Mitochondrial DNA analysis of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) in the Southern Cone of South America. Vet Parasitol. 2012;190:547–55. [DOI] [PubMed]
- 12.Zemtsova GE, Apanaskevich DA, Reeves WK, Hahn M, Snellgrove A, Levin ML. Phylogeography of Rhipicephalus sanguineus sensu lato and its relationships with climatic factors. Exp Appl Acarol. 2016;69:191–203. doi: 10.1007/s10493-016-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornok S, Farkas R. [First autochthonous infestation of dogs with Rhipicephalus sanguineus (Acari: Ixodidae) in Hungary: case report and review of current knowledge on this tick species.] Magyar Állatorvos. 2005;127:623–9 (In Hungarian with English abstract).
- 14.Estrada-Peña A, Bouattour A, Camicas J-L, Walker AR. Ticks of domestic animals in the Mediterranean region: a guide to identification of species. Zaragoza: University of Zaragoza Publishing House; 2004. [Google Scholar]
- 15.Mihalca AD, Kalmár Z, Dumitrache MO. Rhipicephalus rossicus, a neglected tick at the margin of Europe: a review of its distribution, ecology and medical importance. Med Vet Entomol. 2015;29:215–24. doi: 10.1111/mve.12112. [DOI] [PubMed] [Google Scholar]
- 16.Walker JB, Keirans JE, Horak IG. The genus Rhipicephalus (Acari, Ixodidae). A guide to the brown ticks of the world. Cambridge: Cambridge University Press; 2000.
- 17.Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida) Parasit Vectors. 2014;7:93. doi: 10.1186/1756-3305-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mel Marine Biol Biot. 1994;3:294–9. [PubMed] [Google Scholar]
- 19.Black WC, Piesman J. Phylogeny of hard and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Natl Acad Sci USA. 1994;91:10034–8. [DOI] [PMC free article] [PubMed]
- 20.Miller MJ, Esser HJ, Loaiza JR, Herre EA, Aguilar C, Quintero D, et al. Molecular ecological insights into Neotropical bird-tick interactions. PLoS One. 2016;11:e0155989. doi: 10.1371/journal.pone.0155989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linnean Soc. 1999;68:87–112. doi: 10.1111/j.1095-8312.1999.tb01160.x. [DOI] [Google Scholar]
- 22.Verginelli F, Capelli C, Coia V, Musiani M, Falchetti M, Ottini L, et al. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and South-East European wolves. Mol Biol Evol. 2005;22:2541–51. doi: 10.1093/molbev/msi248. [DOI] [PubMed] [Google Scholar]
- 23.Kotlík P, Deffontaine V, Mascheretti S, Zima J, Michaux JR, Searle JB. A northern glacial refugium for bank voles (Clethrionomys glareolus). Proc Natl Acad Sci USA. 2006;103:14860–4. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences obtained and/or analysed during the current study are deposited in GenBank under accession numbers KX757879–KX757917 (cox1) and KX793717–KX793746 (16S). All other relevant data are included in the manuscript.


