Abstract
Background
Polyploid hybrids represent a rich natural resource to study molecular evolution of plant genes and genomes. Here, we applied a combination of karyological and molecular methods to investigate chromosomal structure, molecular organization and evolution of ribosomal DNA (rDNA) in nightshade, Atropa belladonna (fam. Solanaceae), one of the oldest known allohexaploids among flowering plants. Because of their abundance and specific molecular organization (evolutionarily conserved coding regions linked to variable intergenic spacers, IGS), 45S and 5S rDNA are widely used in plant taxonomic and evolutionary studies.
Results
Molecular cloning and nucleotide sequencing of A. belladonna 45S rDNA repeats revealed a general structure characteristic of other Solanaceae species, and a very high sequence similarity of two length variants, with the only difference in number of short IGS subrepeats. These results combined with the detection of three pairs of 45S rDNA loci on separate chromosomes, presumably inherited from both tetraploid and diploid ancestor species, example intensive sequence homogenization that led to substitution/elimination of rDNA repeats of one parent. Chromosome silver-staining revealed that only four out of six 45S rDNA sites are frequently transcriptionally active, demonstrating nucleolar dominance. For 5S rDNA, three size variants of repeats were detected, with the major class represented by repeats containing all functional IGS elements required for transcription, the intermediate size repeats containing partially deleted IGS sequences, and the short 5S repeats containing severe defects both in the IGS and coding sequences. While shorter variants demonstrate increased rate of based substitution, probably in their transition into pseudogenes, the functional 5S rDNA variants are nearly identical at the sequence level, pointing to their origin from a single parental species. Localization of the 5S rDNA genes on two chromosome pairs further supports uniparental inheritance from the tetraploid progenitor.
Conclusions
The obtained molecular, cytogenetic and phylogenetic data demonstrate complex evolutionary dynamics of rDNA loci in allohexaploid species of Atropa belladonna. The high level of sequence unification revealed in 45S and 5S rDNA loci of this ancient hybrid species have been seemingly achieved by different molecular mechanisms.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-0978-6) contains supplementary material, which is available to authorized users.
Keywords: Ribosomal DNA, Concerted evolution, Homogenization, Polyploidy, Solanaceae
Background
The realization that a large number of plant species, including many important industrial crops, evolved through circles of hybridization and/or polyploidization [1] has attracted intensive studies on the different aspects of polyploidy including mechanisms of polyploidy genome evolution [2]. Recent advances in plant genome and genomics research clearly demonstrate that hybridization/polyploidization involves intensive genomic rearrangements including exchanges between genomes, and loss or variation of gene copies and expression. These molecular processes are fundamental for species adaptive evolution and performance.
Atropa belladonna is a member of a small genus of ancient allopolyploid plants from the Solanaceae family with a history of medical applications due to its alkaloids, atropine and scopolamine [3, 4]. For a long time, its origin and taxonomic position remained an enigma. However, recent comparative DNA analysis has suggested that the genus Atropa, represented by 2 to 5 closely related species [5, 6] originated about 10 to 15 Myr (Million years) ago due to hybridization between a tetraploid species of Hyoscyameae and a now-extinct diploid species sister to the tetraploid lineage [6–8]. The uncertainty about one of the founding parents further complicates the tracing of species evolution. To learn more about the origin and genome evolution of this ancient natural polyploid, we studied genomic and molecular organization of the Atropa belladonna ribosomal DNA (rDNA).
Tandemly arranged repeated rDNA units found in genomes of all eukaryotes contain evolutionarily conserved sequences coding for ribosomal rRNAs and more rapidly evolving intergenic spacer regions (IGS). Because of its high copy representation in the genome and special arrangement of conserved coding regions linked with variable IGS, rDNA became an attractive focus for investigations of molecular evolution of repeated sequences and phylogenetic studies in different taxonomic groups [9–12]. Genomic loci representing 5S rDNA (5S rRNA gene plus the IGS) and 45S rDNA (genes coding for 18S, 5.8S, and 25S rRNAs and the spacer regions) are mostly arranged in head-to-tail tandem repeats. In contrast to the majority of repeated sequences, whose functions mostly remain unclear, activities of 5S and 45S rDNA genes are vital for organisms, providing rRNA species necessary for assembly of functional ribosomes, which account for more than 90% of total cellular RNA. In eukaryotes, the copy number (CN) of rDNA repeats is higher than is required for rRNA synthesis, and the redundant copies of rDNA are transcriptionally silenced [10, 13–15]. Transcriptionally active 45S rDNA loci (also known as nucleolus organizer regions, NORs) can be recognized by cytological chromosome analysis. Active loci produce nucleoli in interphase and secondary constriction (SC) regions of satellite-bearing chromosomes in metaphase [10, 13, 14]. Vascular plants often possess only single loci for both 5S and 45S rDNA, although multiple loci were also observed [10, 16–18].
Although numerous copies of rDNA repeats co-exist in the same genome, they tend to be nearly identical in many diploid species due to the process of sequence homogenization [19–21], i.e. individual copies of the repeated elements evolve not independently, but in a concerted manner [22, 23]. However, recently accumulated data suggested that a number of rDNA repeat units with different levels of sequence similarity can be simultaneously present in the same genome [24, 25]. This is especially true for species of hybrid origin (for review see [10, 26]), where the inheritance and evolution of rDNA can follow various scenarios. Often in the first generation hybrids, the 45S rDNA loci inherited from both parents remain structurally intact while enduring differential transcriptional silencing [13, 15, 27–30]. In ancient allopolyploid species, a more complicated picture is usually observed with uniparental inheritance and/or structural rearrangements of parental 45S rDNA. For example, in 0.2 Myr old natural allotetraploid Nicotiana tabacum all parental 45S rDNA loci were detected on chromosomes of ancestor diploids, N. sylvestris and N. tomentosiformis. However, the 45S rDNA repeats specific for N. sylvestris were almost completely eliminated and replaced by rearranged repeats of N. tomentosiformis [19, 31]. On the other hand, both parental 5S rDNA variants remained conserved in N. tabacum [32]. In contrast, in 4.5 Myr old Nicotiana alloploids of sect. Repandae both 5S and 45S rDNA loci and corresponding repeat variants of one parental species were not detected [33], indicating that the age of alloploid genome could be an important factor determining the character of rDNA in the hybrids.
Here, we present our data on the chromosomal localization/activity and molecular structure of 45S and 5S rDNA genes in Atropa belladonna. Based on uncovered specific loci representation and DNA sequences of 45S and 5S rDNA repeats, presumptive factors and mechanisms determining evolutionary dynamics of rDNA in polyploids are discussed.
Methods
Plant material
Seeds of Atropa belladonna (accession nos. 986 and 987) were obtained from the collection of the Botanical Garden, University of Tübingen.
Chromosome analysis
Karyological analysis was performed as previously described [17]. Briefly, the primary root meristems of germinated seeds were pre-treated with 2 mM 8-hydroxyquinoline for 2 h at room temperature, fixed in ethanol-glacial acetic acid (3:1) and stored at −20 ° C. Excised roots were washed in 0.01 M citrate buffer (pH 4.8) prior to digestion in an enzyme mixture of 20% (v/v) pectinase (Sigma), 1% (w/v) cellulase (Calbiochem) and 1% (w/v) cellulase ‘Onozuka R-10’ (Serva) for 1.5–2 h at 37 ° C. Meristems were dissected out from root tips, squashed in drops of 45% acetic acid and frozen. After removal of coverslips, the preparations were post-fixed in 3:1 ethanol : glacial acetic acid, followed by dehydration in absolute ethanol and air-dried.
Double fluorescent staining with CMA (chromomycin A3) and DAPI (4′,6-diamidino-2-phenylindole) was performed according to [34]. Preparations were stained with CMA solution (0.5 mg/mL, Serva) for 1 h in the dark, briefly rinsed in distilled water and air-dried. Then slides were stained with DAPI solution (2 μg/mL, Serva) for 30 min in the dark, briefly rinsed in distilled water and mounted in antifade buffer (Citifluor, Ted Pella Inc.). Transcriptional activity of 45S rRNA genes was determined using silver staining following the method of [35]. Slides were treated with a borate buffer (pH 9.2) and air-dried. Then a few drops of freshly prepared 50% silver nitrate were applied to each preparation. Slides were covered with a nylon mesh and incubated in a humid chamber at 42 °C for 20 min, washed in distilled water, and air-dried. The karyological analysis was conducted on at least 10 slides of both the A. belladonna accessions 986 and 987. In each slide 10 metaphase plates were analysed.
Fluorescence in situ hybridization (FISH)
For FISH, the following ribosomal DNA sequences were used as probes: 5S rDNA (pTa794) [36] labelled using PCR with tetramethyl-rhodamine-5-dUTP (Roche), and a 2.3-kb ClaI subclone of the 25S rDNA coding region of Arabidopsis thaliana [37] labelled by nick translation using digoxigenin-11-dUTP (Roche). The latter probe was used to determine the chromosomal localization of 45S rDNA. The following in situ hybridization of the probes and immunodetection of digoxigenated probe using FITC-conjugated anti-digoxigenin antibodies (Roche) were performed as described [17]. The fluorescence images were acquired using either an Olympus Camedia C-4040Z digital camera attached to a Leica DMRB epifluorescence microscope or a Hamamatsu C5810 CCD camera attached to an Olympus Provis AX epifluorescence microscope.
Cloning and sequence analysis of 45S rDNA intergenic spacer (IGS)
Genomic DNA was isolated from leaves of 3-month-old plants using DNeasy Plant kit (Qiagen, Valencia, CA).
Our early restriction mapping experiments revealed that the 45S rDNA of A. belladonna possesses EcoRI recognition sites in the 18S and 25S rRNA coding regions, whereas no EcoRI site is present in the IGS [38] (Additional file 1: Figure S1). Therefore, EcoRI can be used for cloning of the complete IGS. Accordingly, DNA of A. belladonna (acc. no. 986) was digested with EcoRI, ligated into pBluescript SK and transformed into E. coli strain XL-blue. The library was screened for 45S IGS using 32P labelled DNA probe specific for 3′ end of 25S rRNA as describer earlier [19], and three clones - Ab-IGS-1S, −2S, −1L - were identified. One of the clones (Ab-IGS-1S), containing the complete IGS of the shorter size class of two rDNA repeats [38] was selected for detailed restriction mapping, generation of subclones and sequencing. In order to evaluate molecular heterogeneity of the 45S rDNA, the transcribed part of the 45S IGS, i.e. the 5′ ETS (external transcribed spacer, extended from presumptive transcription initiation site (TIS) to 18S rRNA coding region) was amplified by PCR for both accessions of A. belladonna, cloned and sequenced (clones Ab-ETS-4, −5, −6, −7, −8, −9, −10, −11, −12, −14, −15, −16, −18, −19, −21) as described earlier [21].
Molecular analysis of 5S rDNA
The 5S rDNA units of A. belladonna were amplified by PCR using genomic DNA isolated from leaves of 3-month-old plants by DNeasy Plant kit (Qiagen, Valencia, CA), Pfu DNA polymerase (Thermo Fisher Scientific, Inc.) and primers Pr5S-L (5′-CAATGCGGCCGCGAGAGTAGTACTAGGATGCGTGAC-3′) + Pr5S-R (5′-CATTGCGGCCGCTTAACTTCGGAGTTCTGATGGGA-3′) complimentary to the 5S rRNA coding region [20] were used for amplification. For subsequent cloning, NotI recognition sites (GCGGCCGC, printed in bold above) were added at the 5′ ends of both primers.
The reaction was performed in 50 μl of reaction mixture containing the following components: 0.1 μg of the genomic DNA, 1.0 U of DNA polymerase, 1 × PCR buffer, 4 mM MgCl2, 0.4 mM of each dNTPs, and 1 μM of each primer. The amplification was carried out at “standard” or “soft” conditions applying the following programs: (1) initial DNA polymerase activation at 95 °C, 4 min; (2) DNA denaturation at 94 °C, 40 s; (3) primer annealing at 57 °C, 45 s (standard) or at 54 °C, 90 s (soft); (4) DNA synthesis at 72 °C, 50 s (standard) or 20 s (soft); (5) amplification completion at 72 °C for 8 min. The total number of amplification cycles was 35. Optimization of the PCR soft conditions, which favours amplification of shorter 5S rDNA repeats (see Results for details), was carried out in the preliminary experiments.
PCR amplification of 5S rDNA was performed in triplicates and pooled PCR products of each accession were used for cloning. The fragments of different length were cut out from the gel, purified with a Gel Band Purification Kit (Qiagen), digested with NotI (Fermentas, Lithuania), ligated into Eco52I site of pLitmus 38 and transformed into E. coli strain XL-blue. Plasmid DNA isolation, restriction mapping and other standard procedures were carried out according to [39]. Inserts of selected clones were sequenced using the Big Dye Terminator Cycle Sequencing Kit and ABI Prism 310 sequencer (PE Applied Biosystems, USA). Sequence alignment was performed by CLUSTAL W method [40].
Results
Chromosomal organization of 5S and 45S rDNA
The chromosome number for A. belladonna, 72 chromosomes per somatic cell, was estimated by DAPI staining of root meristems. For 45S rDNA six distinct hybridization signals specific for 45S rDNA cluster were detected on separate chromosomes (Fig. 1). Similarly, chromomycin A3 (CMA) staining produced six signals, two of which were relatively slight ones. Determination of 45S rDNA location was complemented by silver-staining, an indicator of transcriptional activity of these sites. Chromosome silver-staining resulted in four signals per cell, suggesting that only four 45S rDNA sites are transcriptionally active.
Fig. 1.
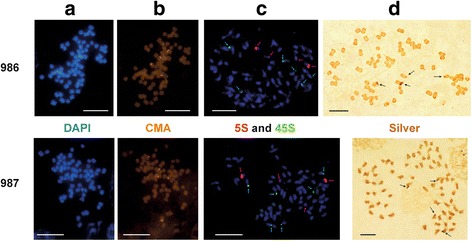
Karyological characterization of Atropa belladonna (accessions 986 and 987). a and b Double fluorescent staining with 4′,6-diamidino-2-phenylindole (DAPI) and chromomycin A3 (CMA), respectively; c Localization of 5S and 45S rDNA sequences on chromosomes; The chromosomes were stained by DAPI (blue fluorescence), hybridization signals of 5S (red) and 45S rDNA (green) are marked by arrows; d Active 45S rDNA (nucleolar organizing region, NOR) sites (arrows) in Atropa belladonna detected by silver staining; Scale bar, 10 μm
In contrast to 45S rDNA, only four 5S rDNA specific signals – two very strong and two weak – were detected at metaphase chromosomes of A. belladonna (Fig. 1). After double FISH with rDNA probes, hybridization signals specific for 5S rDNA and 45S rDNA were observed on separate chromosomes, indicating that there is no co-localization of 5S and 45S rDNA gene clusters.
Sequence organization of 45S rDNA intergenic spacer region in A. belladonna
In our cloning experiments we have isolated two short and one long DNA fragments containing IGS regions of the short and long variants of 45S rDNA repeats of A. belladonna (Additional file 1: Figure S1). Sequencing of one of the short clones (Ab-IGS-1S) showed the IGS region of 3710 bp. The sequence can be subdivided into six structural regions (SR I to SR VI; Fig. 2) according to Harr-plot analysis (Fig. 3), GC-content calculations and comparison with 45S rDNA IGS of other Solanaceae (see below).
Fig. 2.
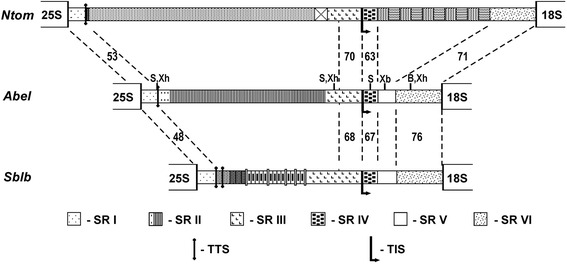
Organization and sequence similarity of the intergenic spacer (IGS) of 45S rDNA of Atropa belladonna (Abel - clone Ab-IGS-1S; Genbank Acc. No KF492694), Solanum bulbocastanum (Sblb – [27]) and Nicotiana tomentosiformis (Ntom – [19]). Percents of similarity for different structural regions (SR I – SR VI) of IGS are given. TIS: transcription initiation site; TTS: putative transcription termination site. Localization of restriction endonucleases recognition sites (B: Bam HI, EI: Eco RI, EV: Eco RV, S: Sph I, Xb: Xba I, Xh: Xho I) used for IGS mapping of A. belladonna rDNA is shown
Fig. 3.
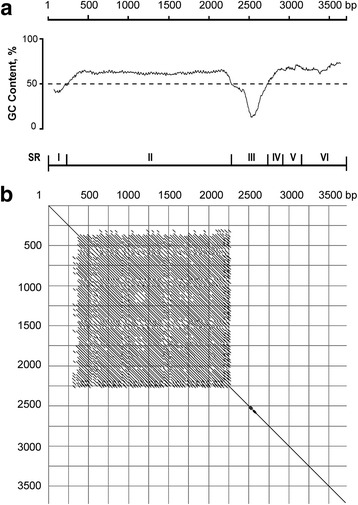
GC-content (a) and Harr-plot analysis (b) of nucleotide sequence of 45S IGS of Atropa belladonna (clone Ab-IGS-1S; Acc. No KF492694). A self-comparison of the IGS was made using the following parameters: window = 30, percentage = 70. Borders of structural regions (SR I – SR VI; see Fig. 2) are shown
The SR I (222 bp in length, 44.1% GC) consists of a unique sequence and exhibits moderate sequence similarity of 48 to 53% to representatives of distantly related Solanaceae species, Solanum bulbocastanum and Nicotiana tomentosiformis (Fig. 2). A pyrimidine-rich motif CCCTCCCCCTCC is present at the beginning of SR I (Additional file 2: Figure S2); similar motives were previously identified in the corresponding region of 45S rDNA in higher plants of different families [9, 41, 42]. At the 3′ end of SR I a GAGGTTTTT motif is located. From 1 to 4 copies of this sequence were found in representatives of distantly related genera of Solanaceae: Nicotiana, Solanum and Capsicum [19, 27, 31, 42, 43]. Obvious evolutionary conservation indicates functional importance of this motif, e.g. for transcription termination.
The next IGS region, SR II (2055 bp in length, 61.8% GC) contains subrepeats (Figs. 2 and 3). This region can be subdivided in two sub-regions, SR II-A (162 bp) and -B (1893 bp). The SR II-B is composed of numerous copies of short subrepeats, two variants of which – Z1 (32 bp long) and Z2 (33 bp long) – can be distinguished (Fig. 4). In contrast, no perfect repeated elements, but only short fragments demonstrating similarity to Z-sub-repeats, were found in SR II-A.
Fig. 4.
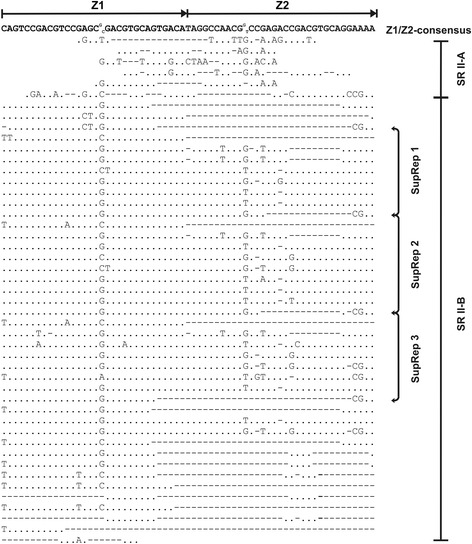
Molecular organization of structural region II (SR II) in the 45S IGS of Atropa belladonna. Alignment of individual Z-subrepeats and consensus sequences of Z1/Z2 subrepeats, borders of SR-IIA and -IIB and localization of Z-“super-repeats” (SupRep 1–3) are shown
All together, 37 perfect or partially deleted copies of Z1, 23 copies of Z2 and several short fragments of Z1/Z2-subrepeats were identified within SR II-B. The subrepeats are arranged as dimers Z1 + Z2 in the middle part of SR II-B, whereas the beginning and the end of SR II-B consist of Z1 subrepeats only. Further analysis revealed that some Z-subrepeats contain specific point mutations, which are periodically repeated within the SR II-B. Hence, in course of molecular evolution not only short motives and single Z-subrepeats, but also long arrays composed of several subrepeats were amplified. Such long blocks are shown as “super-repeats” in Fig. 4. A similar mode of amplification was described for C-subrepeats in SR II of Nicotiana [19]. Comparative restriction mapping of the cloned short and long IGS fragments demonstrated that two short clones appear to be identical whereas the long one differs by the length of the SR II by 0.75 kb (see Additional file 1: Figure S1 and Fig. 2). This difference is probably attributable to different numbers of Z-subrepeats.
The following SR III (464 bp long) is represented by a unique AT-rich (33% GC) sequence. It contains the putative promoter region including the transcription initiation site (TIS) at the 3′ end. A similar AT-rich region preceding the TIS has been found in Solanum [11, 27, 42], Nicotiana [19, 31], Capsicum [43] and other plant species [9, 41, 44].
The SR III can be further subdivided in two parts, A and B. The 185 bp-long SR III-A of A. belladonna exhibits a low similarity to 45S IGS of other Solanaceae, whereas the following 279 bp-long SR III-B and especially the region around the putative TIS are more conserved (Fig. 2 and Additional file 3: Figure S3). The two parts of SR III display a difference in GC-content that amounts to 44.9% for SR III-A vs. 25.1% for SR III-B. The difference is attributable to nine short GC-rich motives “imbedded” in the AT-rich sequence of SR III-A.
In the 45S IGS of A. belladonna no subrepeats are present down-stream of TIS. According to the comparison with the 45S IGS of other Solanaceae species, three regions – SR IV, V and VI – can be distinguished (Fig. 2).
The SR IV (185 bp long, 64.3% GC) of A. belladonna exhibits a moderate similarity with the corresponding IGS regions of Solanum and Nicotiana (Fig. 2 and Additional file 4: Figure S4). In the central part of this region a short, conserved element (CE: 41 bp) occurs, which demonstrates significant similarity – 76-80% – with other Solanaceae. Previously it was shown that CE is duplicated in the 45S IGS of potato S. tuberosum [21], and multiplicated in tomato S. lycopersicum and closely related species [11]. It was proposed that CE could be involved in transcription regulation, because differential transcription/silencing of parental 45S rDNA in interspecific hybrids of Solanum correlates with the number of CE [27]. In contrast to SR IV the following SR V (234 bp long, 68.0% GC) has no essential similarity with the 45S IGS of Solanum and Nicotiana; the level of sequence identity amounts to 58 and 41%, respectively.
Region SR VI adjacent to the 18S rRNA gene is 550 bp long (68.2% GC). The region exhibits comparatively high sequence similarity (71–76%) with the distantly related Solanaceae species (Fig. 2 and Additional file 5: Figure S5). Several segments of particularly high sequence identity were found in SR VI. These segments may be involved in regulation of transcription and/or processing of 45S rRNA.
In order to evaluate the level of intragenomic heterogeneity of individual repeats of 45S rDNA of A. belladonna we have amplified by PCR, cloned and sequenced the transcribed portion of 45S IGS (i.e., 5′ETS from presumptive TIS to 18S rRNA coding region). In total, 20 5′ETS clones were obtained and subjected to restriction mapping. For all clones, identical fragment patterns were obtained (data not shown). Afterwards, ten 5′ETS clones were randomly selected, sequenced and compared with the sequence of the complete 45S IGS described above. The results (Additional file 6: Figure S6) demonstrate that the level of sequence similarity between these eleven individual clones ranges from 98.2 to 100%. In the majority of clones, deviations from the consensus sequence were presented by 1 to 3 base substitutions, excepting clones Ab-ETS-9 and −10, which contain 11 and 10 substitutions, respectively. Also, 1- and 2-bp-long deletions and 1-bp-long insertion were found in the clones Ab-ETS-14, −16 and −12, respectively.
Molecular organization of the 5S rDNA repeats
Agarose gel separation of PCR products demonstrated that the main class of 5S rDNA repeats in A. belladonna has a length of about 260 bp (Fig. 5a). An additional shorter DNA fragment was detected when a large excess of sample was used for electrophoretic analysis (see Fig. 5a, right panel). The data show that the second minor class of 5S rDNA repeats, which has a length of about 180 bp, is present in the genome of A. belladonna. Evaluation of relative intensity of bands by the image analyzer showed that in accessions 986 and 987, respectively, from 5 to 7% and less than 2% of 5S rDNA repeats belong to the second minor class.
Fig. 5.
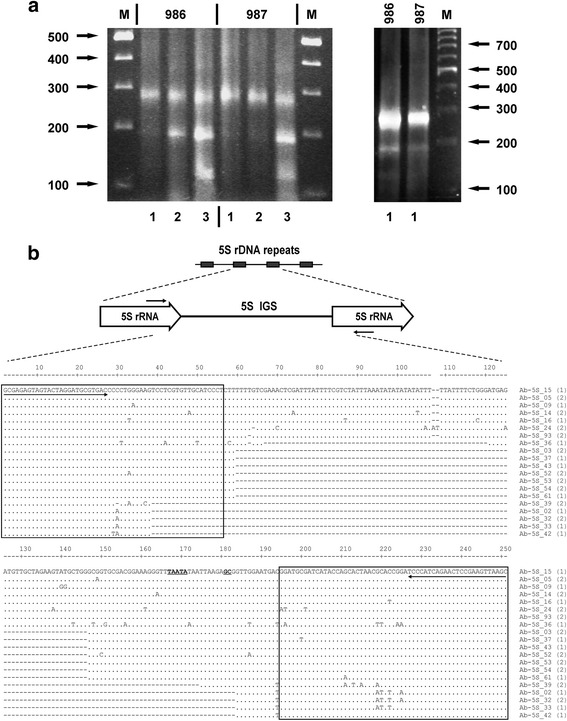
Molecular organization and polymorphisms of 5S rDNA repeats of Atropa belladonna. a Electrophoretic separation of 5S rDNA PCR products obtained for Acc. Nos 986 and 987; PCR amplification was performed (1) at standard conditions (see Methods), (2) at soft conditions, or (3) at soft conditions using increased (4 μM) concentration of primers; M, molecular weight marker; a ten-fold excess of the PCR product was loaded on gel in the right panel in comparison to the left panel. b Sequence comparison of 5S rDNA variants; Sequences of primers used for PCR amplification are marked by arrows, coding regions are shown as boxes and elements of presumptive external promoter are printed in bold underlined text; Numbers 1 and 2 shown in brackets are referred to Acc. Nos 986 and 987, respectively
In order to clone different length variants of 5S rDNA, we performed optimization of PCR conditions (shortened elongation time, prolonged primer annealing under lower temperature, increased concentration of primers) to improve amplification of the shorter minor 180 bp-fragments. This resulted in preferential amplification of underrepresented 5S repeat variants. In particular, higher primer concentration led not only to improved generation of the 180 bp fragments, but also to amplification of a third class of 5S rDNA repeats, which have a length of about 120 bp (Fig. 5a, left panel, variant 3). This class remained undetectable under standard PCR amplification conditions. Hence, the 120 bp-repeats appear to be represented in the genome by very low CN.
Applying agarose gel electrophoretic fractionation, we have cloned PCR products representing all three size classes of 5S rDNA repeats. In total, for the two studied accessions of A. belladonna, 32 recombinant clones were isolated and, after determination of the insert lengths by restriction mapping, 20 of them were selected for sequencing. Comparison of the obtained sequences showed that the 5S repeats of A. belladonna can be classified into 3 groups: long (257–259 bp; clones pAb-5S-05, -09, −14, −15, −16, −24, −93), intermediate (171–203 bp; clones pAb-5S-03, −36, −37, -43, -52, -53, -54, -61), and short (113–121 bp; clones pAb-5S-02, −32, −33, −39, -42) repeats (Fig. 5b).
The long repeats are composed of the region coding for 5S rRNA and an IGS. Taking into account location of the primers used for PCR we calculated that as in other eukaryotes the length of the rRNA coding region is 120 bp. The length of 5S IGS of long repeats ranges from 137 to 139 bp, which is shorter compared to other representatives of Solanaceae, e.g. 165–229 bp in Solanum [20, 45] and 310–560 bp in Nicotiana [32, 46] species. In contrast to Solanum, no subrepeats were found in the central non-transcribed part of A. belladonna 5S IGS.
Sequence comparison showed that the long 5S rDNA repeats of both accessions of A. belladonna are very similar (96.4-99.6% of similarity, except one clone, pAb-5S_24 - see Fig. 5b). The minor differences are mainly due to occasional base substitutions and a few single nucleotide indels in the IGS. The 5S rRNA coding region of A. belladonna is identical to that of tomato Solanum lycopersicum and very similar to other Solanaceae (Additional file 7: Figure S7).
Similar to other plant species [20, 32, 47], an oligo-dT motif downstream of the coding region was found in the long 5S rDNA repeats of A. belladonna. These motives have been shown to function in Pol III transcription termination of the eukaryotic 5S rRNA genes [48]. Sequence comparison also revealed that in the long 5S IGS variant of A. belladonna, similar to other plant species [20, 32, 47], a TATA-like motif and a GC dinucleotide are located, respectively, at the positions −28 to −24 bp and −14 bp upstream of the coding region. These motives – together with the internal promoter elements – were proposed to form the Pol III transcription initiation site [48]. Thus, the long 5S rDNA repeats of A. belladonna contain the structurally normal 5S rRNA coding region and all known signals required for transcription initiation and termination. Therefore, they appear to be functionally active.
The intermediate 5S rDNA repeats contain a deletion (53–85 bp) in the central part of the IGS, compared to the long repeats. Nevertheless, they still possess all external promoter elements and the conserved 5S rDNA coding region (except the clone pAb-5S-36, which contains seven base substitutions in the sequenced fragments of the coding region). However, the intermediate repeats completely or partially (clone pAb-5S-36) lack the oligo-dT sequence required for transcription termination. These structural defects indicate that the intermediate repeats are non-functional, or that there is an alternative transcription termination option.
In the short 5S rDNA repeats, nearly the entire IGS is missing, as is 18 bp at the 3′ end of the coding region. Additionally, the short repeats accumulated several nucleotide substitutions in the rudimental fragment of 5S coding region, and a cytosine residue in position −1, which is required for transcription initiation [48], is changed into thymidine in all short clones sequenced. Accordingly, it looks probable that the short repeats represent pseudogenes.
We have calculated the number of base substitutions in individual 5S rDNA repeats compared to the consensus sequence and found that the three groups of repeats significantly differ by this parameter (Table 1 Specifically, the frequency of base substitutions is 1.43, 2.26 and 9.74 per 100 bp in long, intermediate and short repeats, respectively. Hence, the frequency of base substitutions appears to be about 1.6 and 6.8 times higher in intermediate and short repeats, respectively.
Table 1.
Number of base substitutions in 5S rDNA of Atropa belladonna
| Clone | Length | Transversions | Transitions | Total number of base substitutions | |
|---|---|---|---|---|---|
| C → T, G → A | T → C, A → G | ||||
| Group I: long repeats | |||||
| Ab-5S-05 | 199 | 0 | 1 | 0 | 1 |
| Ab-5S-09 | 199 | 1 | 1 | 1 | 3 |
| Ab-5S-14 | 199 | 2 | 2 | 0 | 4 |
| Ab-5S-15 | 199 | 0 | 0 | 0 | 0 |
| Ab-5S-16 | 199 | 2 | 2 | 0 | 4 |
| Ab-5S-24 | 200 | 5 | 3 | 0 | 8 |
| Ab-5S-93 | 198 | 0 | 0 | 0 | 0 |
| Total for group I | 1393 | 10 (50.0%) | 9 (45.0%) | 1 (5.0%) | 20 (100%) |
| Group II: intermediate repeats | |||||
| Ab-5S-03 | 112 | 0 | 0 | 0 | 0 |
| Ab-5S-36 | 144 | 6 | 9 | 1 | 16 |
| Ab-5S-37 | 112 | 0 | 1 | 0 | 1 |
| Ab-5S-43 | 112 | 0 | 0 | 0 | 0 |
| Ab-5S-53 | 112 | 0 | 0 | 0 | 0 |
| Ab-5S-52 | 112 | 1 | 2 | 0 | 3 |
| Ab-5S-54 | 112 | 0 | 0 | 0 | 0 |
| Ab-5S-61 | 112 | 0 | 1 | 0 | 1 |
| Total for group II | 928 | 7 (33.3%) | 13 (61.9%) | 1 (4.8%) | 21 (100%) |
| Group III: short repeats | |||||
| Ab-5S-02 | 54 | 1 | 5 | 0 | 6 |
| Ab-5S-32 | 53 | 1 | 5 | 0 | 6 |
| Ab-5S-33 | 54 | 1 | 4 | 0 | 5 |
| Ab-5S-39 | 62 | 3 | 4 | 0 | 7 |
| Ab-5S-42 | 54 | 1 | 2 | 0 | 3 |
| Total for group III | 277 | 7 (25.9%) | 20 (74.1%) | 0 | 27 (100%) |
Note: The number of base substitutions was calculated comparing sequences of individual clones and the consensus sequence. Lengths of clones are presented without primers used for PCR
Additionally, we have compared the frequency of different types of mutations and found that transitions amount to 50.0, 66.7 and 74.1% of all base substitutions in long, intermediate and short repeats, respectively (see Table 1). Remarkably, among 44 transitions detected, 42 were represented by C → T and G → A, which could be related to 5-methyl-cytosine deamination. Accordingly, it looks probable that the intermediate and short repeats were highly methylated for a long time, which resulted in preferential accumulation of respective transitions. Taken together, these data strongly support our proposition that the intermediate and short subrepeats represent pseudogenes. Thus, in ancient hexaploid A. belladonna, redundant 5S rDNA repeats did not evolve in a concerted manner. They appear to be gradually changed into pseudogenes and partially eliminated from the genome.
Discussion
Chromosome analysis and origin of A. belladonna
The small Old World polyploid genus Atropa possesses unique morphological traits and occupies an isolated taxonomic position within Solanaceae [5]. While the exact taxonomic position of Atropa is still debated, the majority of available data [8, 49] place this group within the tribe Hyascyameae, in spite of a marked difference in fruit morphology (fleshy berry-like fruits of Atropa versus dry capsules of other Hyascyameae). Within the tribe, Atropa is a sister group to the other six genera of Hyascyameae (Anisodus, Atropanthe, Hyoscyamus, Physochlaina, Przewalskia, and Scopolia). It is generally believed that Atropa species originated about 10 to 15 Myr ago through hybridization between a tetraploid species of Hyoscyameae and an extinct diploid progenitor related to the tetraploid lineage [6–8]. Therefore, genomic constitution of Atropa could be presented as EEH1H1H2H2, where E and H represent the genomes of Extinct diploid and Hyascyameae tetraploid parents, respectively.
The majority of karyology studies showed that Atropa species possess a karyotype of 2n = 72, although 2n = 50, 60 and 74 were also reported (see references presented in the Index to plant chromosome numbers at TROPICOS database - http://www.tropicos.org/Project/IPCN). Our results of chromosome analysis in A. belladonna are clearly consistent with the counts 2n = 72 (see Fig. 1).
The available cytogenetic data for the tribe Hyascyameae were differently interpreted in the available publications. Yuan et al. [7] propose for the section a base chromosome number x = 12 that is concordant with the well-supported taxonomic position of Hyascyameae within Solanoideae, which together with Nicotianoideae belong to the strongly supported monophyletic “x = 12” clade [49]. Accordingly, Anisodus, Atropanthe and Scopolia (2n = 48), Przewalskia (2n = 44), and Physochlaina (2n = 42) are considered as tetraploids, whereas Hyoscyamus possesses various chromosome numbers and ploidy levels [7, 50, 51]. In contrast, Tu et al. [52] believe that the basic chromosome number in the section is 6 (x = 6). According to this view, A. belladonna would be considered a dodecaploid. Chromosome staining with CMA and FISH experiments conducted in our study demonstrated that three loci (three pairs of sites) of 45S rDNA are present on six different chromosomes. This observation further supports the hexaploid constitution of Atropa as proposed by Yuan et al. [7] and revealed that Atropa possesses one 45S rDNA locus per chromosomal set.
Generally, the presence of a single chromosome pair with satellites is common in the family Solanaceae, especially in “x = 12” clade, and for several representatives of the clade the existence of single 5S and 45S rDNA loci was demonstrated by FISH ([27, 53–55] and references therein). Accordingly, multiple rDNA loci appear to be rare in “x = 12” clade and were found only in a few terminal clades that demonstrate intensive chromosomal evolution [50, 53, 55]. Thus, the available data show that Atropa most probably originated from parental species possessing single 5S and 45S rDNA locus per chromosomal set. Accordingly, six sites each of 5S and 45S rDNA could be anticipated in the modern allohexaploid A. belladonna. Our data reveal that the six sites of 45S rDNA are really present in A. belladonna, but only four 5S rDNA sites were found, demonstrating that two 5S rDNA sites were lost since the polyploid formation.
At the chromosomal level, contrasting evolutionary dynamics of plant 5S and 45S rDNA is a well-documented phenomenon. As demonstrated for several genera, 45S rDNA loci are more variable then 5S rDNA between closely related species, varieties and even individuals in terms of differences in size, number and loci locations [56–58]. Generally, in many plant species – both diploids and polyploids – the number of 5S loci is lower compared to 45S loci [16, 18], which could be used as another argument supporting different patterns of their molecular evolution.
Molecular organization and evolution of 45S rDNA in A. belladonna
Considering a high similarity of the plant rDNA sequences coding for the 18S-5.8S-25S ribosomal RNAs [10], we concentrated our efforts on the analysis of evolutionary variable IGS. Based on restriction analysis and sequence characterization, the cloned A. belladonna IGS sequences represent two rDNA length variants of 9.4 and 10.2 kb revealed earlier by rDNA mapping experiments based on Southern blotting [38]. Sequencing of the short clone Ab-IGS-1S resulted in a 3710 bp long IGS fragment. Combining this IGS sequence with the 18S and 25S rRNA coding sequences of tomato and potato available in Genbank (Acc. Nos X51576, X67238, X13557) and ITS1-5.8S-ITS2 region of Atropa described by Uhink and Kadereit [6] we calculated that the length of Atropa rDNA unit is about 9.45 kb, which is very close to 9.4 kb, estimated earlier by Southern analysis. The larger cloned IGS fragment with a length of 6.51 kb, as estimated by restriction analysis, differs by the size of a region that contains Z-subrepeats up-stream of TIS (Additional file 1: Figure S1 and Fig. 2) and obviously corresponds to the longer 10.2 kb rDNA repeat. Detailed sequence characterization of the A. belladonna IGS revealed a structural organization similar to rDNA spacers of other Solanaceae species (Fig. 2), with specific functional subdivision into (i) a region of rRNA transcription termination, (ii) long block of sub-repeats, (iii) AT-rich region up-stream of TIS and (iv) the 5′ ETS adjacent to the 18S rRNA coding sequence.
Of special interest regarding the genome evolution in A. belladonna is a high level of IGS sequence homogenization between the two types of 45S rDNA length repeats. The short and long rDNA variants are situated at different sites according to their clear segregation [38] in somatic hybrids between A. belladonna and tobacco with incomplete chromosomal sets of both parents [59]. Comparison of 15 individual genomic DNA fragments of PCR amplified ETS clones demonstrated 98.2 to 100% sequence similarity. This high level of similarity was observed even in the central part of 5′ ETS, i.e. in SRV (Fig. 2), which is known to be more variable compared to other 45S rDNA regions. These data are also in a good agreement with the results on high sequence homogeneity in the ITS region of Atropa 45S rDNA [6] and comparable to intragenomic similarity of 45S rDNA repeats in diploid (98.4 to 99.9%) and polyploid (93.5 to 99.6%) species from the related genus Solanum. For comparison, the interspecific sequence similarities vary from 81 to 88% for representatives of distantly related Solanum species within sect. Petota [11, 22, 28].
The revealed high sequence similarity of 45S rDNA individual copies in allohexaploid A. belladonna leads to the suggestion that they originated from a single parent despite the location on all three sets of chromosome pairs. Previous studies showed, that in some cases supposedly uniparental rDNA inheritance has actually resulted from the intergenomic homogenization of parental 45S rDNA repeats [25, 26]. Indeed, we demonstrated that 45S rDNA repeats of allotetraploid Nicotiana tabacum structurally differ from the rDNA variants of parental diploid species, N. sylvestris and N. tomentosiformis [19, 31]. However, the 45S rDNA loci have the same chromosomal location in N. tabacum and in the parental species [60, 61]. We have also detected that in N. tabacum, 45S rDNA repeats originating from the maternal diploid species N. sylvestris were nearly completely substituted by structurally rearranged rDNA of the paternal diploid N. tomentosiformis [19, 27]. Later partial or complete conversion of parental 45S rDNA was reported for other alloploids of Nicotiana [33, 62]. Based on these considerations, we suggest that in A. belladonna, similar to N. tabacum [19], the repeats of one parent were overwritten or substituted by rDNA of the second parent following the process of sequence conversion.
The molecular conversion of 45S rDNA sequences in alloploids appears to be time dependent. As a rule, in the first generation of symmetric hybrids rDNA arrays of both parental forms are present [26, 27, 30]. However, the appearance of novel variants of 45S rDNA in first generation somatic hybrid between N. tabacum and A. belladonna [38] and in synthetic tobacco [62] shows that rearrangement of 45S rDNA units may start immediately after hybridization/polyploidization.
In young natural allotetraploid hybrids Tragopogon mirus and T. miscellus, which have been formed repeatedly during the past 90 years, partial loss of 45S rDNA of one parental diploid was observed, although the chromosome complements of parental diploids are additive and no chromosomal rearrangements were found [63]. In Arabidopsis suecica, a natural allotetraploid derived from A. thaliana and A. arenosa, one pair of A. thaliana NORs is missing. Similarly, in artificially obtained A. suecica-like allotetraploids, pairs of A. thaliana NORs are gained de novo, lost, and/or transposed to A. arenosa chromosomes [64]. Therefore, it seems likely that the loss of one pair of A. thaliana NORs observed in natural A. suecica was attributed to rapid chromosomal rearrangements during the next few generations after polyploidization. Similarly, in synthetic Triticum/Aegilops allopolyploids certain parental NOR loci were completely eliminated within several generations [30].
However, selective elimination of parental NORs and/or 45S rDNA variants may require much more time. In 0.2 Myr old N. tabacum, residual parental sylvestris-like rDNA ranges from 2 to 10% of total nuclear rDNA in different cultivars and amounts to 25% in feral tobacco [62], but three pairs of 45S rDNA loci are retained on the sylvestris-donated chromosomes. In 4.5 Myr old Nicotiana alloploids of sect. Repandae, only maternal 45S rDNA was found, and this completed diploidization was attributed to locus loss [33]. To the contrary, more ancient Atropa, whose age seems to be at least 10 Myr [6, 8], still possess six homogenized 45S rDNA loci of both parents, suggesting that not only time, but also some other factors could be responsible for the elimination or homogenization of redundant 45S rDNA loci.
IGS subrepeats have been proposed as one of the factors influencing homogenization of rDNA in plants. Comparing 45S rDNA molecular organization in Brassica and Nicotiana, Kovarik et al. [62] suggested that the presence of IGS subrepeats up-stream and down-stream of TIS in the Nicotiana species may be responsible for enhanced recombination potential of the rDNA, resulting in effective homogenization of the parental 45S rDNA in allopolyploids. In the IGS of Atropa, subrepeats were found only up-stream, but not down-stream of TIS (Fig. 2), nevertheless, the parental 45S rDNA is effectively homogenized. Thus, this process probably does not require the presence of two subrepeat regions in the IGS.
Differential functional activity of the 45S rDNA loci in A. belladonna
In our karyological experiments we have found that mostly only two out of three 45S rDNA loci seem to be functionally active in A. belladonna. Accordingly, the question arises, which molecular mechanisms could be responsible for the differential activity of the 45S rDNA sites?
It is well known that in interspecific hybrids/allopolyploids 45S rDNA loci of one parent species are usually functionally active. This issue was originally described as nucleolar dominance (ND) by Navashin [65], and it was demonstrated later that at the molecular level ND means transcriptional silencing of 45S rDNA due to siRNA mediated differential methylation of cytosine, post-transcriptional modification of histone proteins and chromatin remodelling [13, 15, 27, 28, 30, 66]. It was also argued that differential activity of 45S rDNA in hybrids can be controlled by structural peculiarities of parental rDNA repeats (especially by that of IGS). Especially, it was found that in the IGS of Xenopus and Arabidopsis, the upstream subrepeats represent transcriptional enhancers [67, 68]. Later, it was also shown that in Vigna [69] and Solanum [27] the 45S rDNA repeats bearing more subrepeats down-stream of TIS appear to be more transcriptionally active. If no down-stream subrepeats are present in 45S rDNA, the variants possessing more subrepeats up-stream of TIS seem to be dominant (for review see [10, 13, 15, 26]). Considering these data, it is possible to suppose that four highly active sites of Atropa contain the long variant of 45S rDNA repeats, whereas the other two loci harbour the short one. Also, the differential transcription of 45S rDNA may be due to inactivation of complete NOR caused by local chromosomal context [70, 71], which may differ for 45S rDNA sites on Atropa chromosomes inherited from distantly related ancestors, i.e. for two E- versus four H-subgenomes.
For allotetraploids of Nicotiana, [29] it was found that the 45S rDNA loci participating in intergenomic conversion should be transcriptionally active, whereas transcriptionally silenced, highly methylated rDNA units escape homogenization, resulting in long-term persistence of both parental rDNA variants in the allopolyploid genome. Accordingly, it is possible to speculate that immediately after formation of the allohexaploid genome of Atropa the 45S rDNA loci of both parents were active, allowing intergenomic conversion. As a result, the 45S rDNA units of one parent were removed and substituted by rDNA of the second parent and later two loci (probably containing the short 45S rDNA variant and/or located on E-chromosomes) became less or not active. Presently, the mechanisms of presumptive differential activity of 45S rDNA variants in modern A. belladonna remain not yet understood and will be the focus of future research.
Molecular evolution of A. belladonna 5S rDNA
In our karyological experiments four 5S rDNA specific signals – two very strong and two weak – were detected on A. belladonna chromosomes. Further molecular studies revealed that the 5S rDNA is represented by three length classes. The 5S rDNA repeats of the long class contain all structural elements required for transcription and, therefore, appear to be functionally active. In contrast, repeats of the intermediate and especially of the short class show structural defects like loss of signals involved in transcription initiation and even partial deletions of coding region. Also, they exhibit increased rates of base substitutions, i.e. typical features of non-functional pseudogenes. Remarkably, the increased rate of mutations seems to be preferentially attributed to the long-term increase of cytosine methylation.
The short and intermediate 5S rDNA repeats clearly differ in the character of structural defects and in the rate of base substitutions and, therefore, can be considered as early and late stages of functional rDNA transformation into pseudogenes.
Regarding the allohexaploid origin of Atropa, it was tempting to propose that the three structural classes of 5S rDNA repeats may represent three parental genomes and have to be located separately on three pairs of chromosomes. In order to clarify the origin of three 5S rDNA classes in A. belladonna, we have compared their IGS sequences, because it is well known that the region displays a very high rate of molecular evolution and may differ even between closely related species, e.g., in such genera as Solanum [20] and Nicotiana [32]. We have found (see Fig. 5) that three 5S rDNA classes present in the A. belladonna genome show high similarity in IGS and probably originate from the same parent. Hence, the 5S rDNA of the second parent was lost.
In order to assess the relative CN of 5S rDNA repeats of different length we have quantified the intensity of respective PCR products after electrophoretic separation and found that the long repeats are the most frequent in the 5S rDNA of A. belladonna. Respectively, comparing our karyological and molecular data it is possible to propose that two very large 5S rDNA sites contain potentially functional long repeats, whereas two weak sites contain intermediate repeats, i.e. slightly damaged pseudogenes. This proposition agrees well with earlier data that in other Solanacea 5S rDNA repeats are highly similar within an array [20, 32], and in other plants the similarity within an array is higher than between arrays [25].
The short repeats (i.e. hardly damaged pseudogenes) most likely exist as orphans outside of 5S rDNA loci and remained undetectable in karyological experiments due to their low CN. Alternatively, they could be interspersed with other repeats within 5S rDNA loci, but in this case it would be difficult to explain the high molecular evolution rate of short repeats. Similar to A. belladonna, 5S rDNA-related pseudogenes represented by minor sequence classes were found earlier in several species of different taxonomic groups, e.g., in Solanum [20], Vigna [12], Rosa [47] and Thinopyrum [25].
The origin of 5S rDNA was studied in several alloploids/hybrids; either bi- or uniparental inheritance was found [12, 20, 24, 32, 33, 64]. In the case of uniparental inheritance, elimination of 5S rDNA repeats of one parent was attributed to the loss of respective chromosomal loci, i.e. in contrast to 45S rDNA, no replacement of one parental 5S rDNA was observed. For instance, unchanged parental variants of 5S rDNA are still present in the genome of N. tabacum [32]. For Sanquisorba, it was found that duplicated 5S rDNA loci tend to have been lost after polyploidization, whereas 45S rDNA sites tend to have been conserved [72]. Similarly, Clarkson et al. [33] demonstrated that in recently formed (0.2 Myr old) alloploids of Nicotiana the number of loci equal the sum of those of their parents whereas in ancient (4.5 Myr old) alloploids the loss of 5S rDNA loci of one parent occurred. Regarding these data it could be anticipated that in 10 Myr old A. belladonna the 5S rDNA loci of one parent may have been eliminated. Supporting this assumption, our studies revealed that two chromosomal sets – probably originated from E-diploid – bear no 5S rDNA loci. Hence, all 5S rDNA repeats currently present in the Atropa genome seem to be derived from the H-tetraploid parent.
As mentioned above, the genomic constitution of allohexaploid Atropa can be presented as EEH1H1H2H2. Regarding the very high sequence similarity of GBSSI gene copies inherited from the Hyascyameae parent [7], it looks probable that two copies of the H-genome (i.e., H1 and H2) were very closely related. Accordingly, it could be expected that 5S rDNA repeats located on H1 and H2 chromosomes have to be very similar if not identical, and our sequencing data confirm this. However, in spite of the high similarity the evolutionary fate of the two sets of 5S rDNA repeats derived from the H-tetraploid parent seems to be different: one set remained functionally active, whereas the second one was transformed into pseudogenes.
Thus, elimination of redundant 5S rDNA repeats in Atropa occurred in two rounds: the loss of repeats donated by E-diploid is already completed whereas the removal of one set of H-tetraploid donated repeats is still in progress.
Distinct patterns of molecular evolution of 5S and 45S rDNA during Atropa diploidization
Taken together, our results demonstrate that in an ancient allohexaploid A. belladonna parental chromosomal loci of 45S rDNA were conserved, but at the sequence level the 45S rDNA endured interchromosomal conversion resulting in elimination of rDNA repeats of one parental species. In contrast, 5S rDNA sequences of one parent were either simply deleted from the genome, or firstly converted into pseudogenes and then lost. Hence, homogenization of 5S and 45S rDNA was achieved by different means: partial deletion of 5S rDNA repeats versus interchromosomal conversion of 45S rDNA. These rearrangements resulted in marked difference at chromosomal level: the number of 5S loci was reduced whereas that of 45S rDNA was preserved in evolution.
The partial loss of 5S and 45S rDNA repeats in Atropa represents an example of genomic diploidization, a well-known phenomenon occurring during the evolution of polyploid organisms. This phenomenon embraces numerous functional and structural changes at different levels like loss of redundant gene expression, rearrangements of repeated genomic sequences, loss of chromosomes, etc. [73–75].
Earlier, several modes of rDNA molecular evolution were proposed [22, 23, 76]. The first of them is a concerted evolution, where rDNA arrays evolve in a concerted manner, resulting in high similarity of individual repeats. The mechanisms of sequence homogenization are not fully understood. Yet, unequal crossing-over, gene conversion and purifying selection were proposed as molecular mechanisms of concerted evolution [76–78]. This mode of evolution can explain intragenomic variation of 45S rDNA ITS2 in 66% of plant species [24]. In Atropa, concerted evolution seems to have occurred for 45S rDNA, resulting in high similarity of repeated units separately located at the loci of three chromosomal pairs.
The second mode is birth-and-death evolution, whereby repeated units endure cycles of duplications, divergence and partial loss. Birth-and-death evolution is the best model to explain ITS2 variation in 27% of plant species [24]. In Atropa, such a scenario appears to be realised for the 5S rDNA.
Conclusions
The combined molecular, cytogenetic and phylogenetic data obtained in this study demonstrate complex evolutionary dynamics or rDNA loci in allohexaploid species of Atropa belladonna. Our study revealed the high level of rDNA sequence unification in this ancient hybrid species and showed different molecular mechanisms underlying sequence homogenization at the 45S and 5S rDNA loci. The 45S rDNA endured interchromosomal conversion resulting in elimination of rDNA repeats of one parental species. In contrast, 5S rDNA sequences of one parent were either eliminated from the genome, or converted into pseudogenes and then lost. These molecular events led to reduction of the 5S rDNA loci number on A. bellabonna chromosomes while the 45S loci were preserved. The presented data further contribute to better understanding of molecular processes underlying formation and evolution of nuclear genomes in polyploid plant species.
Acknowledgements
This work was supported by Alexander von Humboldt Foundation (Germany) research fellowships granted to RAV and NVB. We thank Taj Arndell (University of Adelaide, Australia) for careful reading of the manuscript and correction of English.
Availability of data and materials
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos: KF492694, KF496929-KF496945, KX196202-KX196207 and KY126357-KY126368.
Authors’ contributions
The experimental design was conceived by RAV, JM and VH. The experiments were performed by RAV, IIP, NVB and MHB. The data were analysed by RAV with assistance from NVB and IIP. This paper was written by RAV, NVB and VH. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Localization of restriction endonucleases recognition sites (B: Bam HI, EI: Eco RI, EV: Eco RV, S: Sph I, Xb: Xba I, Xh: Xho I) in the coding region (a) and IGS (b) of Atropa belladonna 45S rDNA. (JPG 2102 kb)
Nucleotide sequence comparison of 45S IGS structural region I (SR I) of Atropa belladonna (Abel), Solanum bulbocastanum (Sblb - Komarova et al. 2004) and Nicotiana tomentosiformis (Ntom - Volkov et al. 1999). Pyrimidine-rich motives present at the beginning of SRI in different Solanaceae species are boxed. Presumptive transcription termination signal GAGGTTTTTT at the end of SRI is shown in bold. (PDF 69 kb)
Nucleotide sequence comparison of 45S IGS structural region III-B (SR III-B) of Atropa belladonna (Abel) and corresponding regions of Solanum bulbocastanum (Sblb) and Nicotiana tomentosiformis (Ntom). (PDF 40 kb)
Nucleotide sequence comparison of 45S IGS structural region IV (SR IV) of Atropa belladonna (Abel) and corresponding regions of Solanum bulbocastanum (Sblb) and Nicotiana tomentosiformis (Ntom). (PDF 39 kb)
Nucleotide sequence comparison of 45S IGS structural region VI (SR VI) of Atropa belladonna (Abel) and corresponding regions of Solanum bulbocastanum (Sblb) and Nicotiana tomentosiformis (Ntom). (PDF 43 kb)
Nucleotide sequence comparison of individual copies of 5′ETS region of Atropa belladonna 45S rDNA. Sequence of presumptive TIS is presented in bold underlined. (PDF 152 kb)
Nucleotide sequence comparison of 5′ (a) and 3′ (b) portions of 5S rDNA coding region of Atropa belladonna (Abel: this study, consensus sequence – see Fig. 5), Solanum lycopersicum (Slyc: Genbank Acc. No X55697), S. tuberosum (Stub: X82780), S. pinnatisectum (Spnt: X82779) and Petunia hybrida (Phyb-A: X07930, Phyb-B: X07929, Phyb-C: X07928). Sequences of primers Pr5S-R (A) and Pr5S-L (B) used for A. belladonna 5S rDNA amplification are marked as arrows. (JPG 1600 kb)
Contributor Information
Roman A. Volkov, Phone: +380-372-584793, Email: r.volkov@chnu.edu.ua
Irina I. Panchuk, Email: i.panchuk@chnu.edu.ua
Nikolai V. Borisjuk, Email: nborisjuk@yahoo.com
Marta Hosiawa-Baranska, Email: marta.hosiawa@us.edu.pl.
Jolanta Maluszynska, Email: jolanta.maluszynska@us.edu.pl.
Vera Hemleben, Email: vera.hemleben@uni-tuebingen.de.
References
- 1.Sattler MC, Carvalho CR, Clarindo WR. The polyploidy and its key role in plant breeding. Planta. 2016;243:281–296. doi: 10.1007/s00425-015-2450-x. [DOI] [PubMed] [Google Scholar]
- 2.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Facchini PJ. Alkaloids biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:29–66. doi: 10.1146/annurev.arplant.52.1.29. [DOI] [PubMed] [Google Scholar]
- 4.Raskin I, Ribnicky DM, Komarnytsky S, Ilic N, Poulev A, Borisjuk N, et al. Plants and human health in the twenty-first century. Trends Biotech. 2002;20:522–531. doi: 10.1016/S0167-7799(02)02080-2. [DOI] [PubMed] [Google Scholar]
- 5.Hunziker A. Genera solanacearum: the genera of solanaceae illustrated, arranged according to a new system. Ruggell: ARG Gantner Verlag KG; 2001. [Google Scholar]
- 6.Uhink CH, Kadereit JW. Phylogeny and biogeography of the Hyoscyameae (Solanaceae): European – East Asian disjunctions and the origin of European mountain plant taxa. In: Uhink CH, editor. Biogeographische Beziehungen zwischen den Alpen, dem Kaukasus und den asiatischen Hochgebirgen: Ph. D. Thesis. Mainz: Der Johannes Gutenberg-Universitat Mainz; 2009.
- 7.Yuan Y, Zhang Z, Chen Z, Olmstead RG. Tracking ancient polyploids: a retroposon insertion reveals an extinct diploid ancestor in the polyploid origin of Belladonna. Mol Biol Evol. 2006;23:2263–2267. doi: 10.1093/molbev/msl099. [DOI] [PubMed] [Google Scholar]
- 8.Tu T, Volis S, Dillon MO, Sun H, Wen J. Dispersals of Hyoscyameae and Mandragoreae (Solanaceae) from the New world to Eurasia in the early Miocene and their biogeographic diversification within Eurasia. Mol Phylogenet Evol. 2010;57:1226–37. doi: 10.1016/j.ympev.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 9.King K, Torres RA, Zentgraf U, Hemleben V. Molecular evolution of the intergenic spacer in the nuclear ribosomal RNA genes of cucurbitaceae. J Mol Evol. 1993;36:144–152. doi: 10.1007/BF00166250. [DOI] [PubMed] [Google Scholar]
- 10.Volkov RA, Medina FJ, Zentgraf U, Hemleben V. Molecular cell biology: organization and molecular evolution of rDNA, nucleolar dominance, and nucleolus structure. Prog Bot. 2004;65:106–146. doi: 10.1007/978-3-642-18819-0_5. [DOI] [Google Scholar]
- 11.Komarova NY, Grimm GW, Hemleben V, Volkov RA. Molecular evolution of 35S rDNA and taxonomic status of Lycopersicon within Solanum sect. Petota. Plant Syst Evol. 2008;276:59–71. doi: 10.1007/s00606-008-0091-2. [DOI] [Google Scholar]
- 12.Saini A, Jawali N. Molecular evolution of 5S rDNA region in Vigna subgenus Ceratotropis and its phylogenetic implications. Plant Syst Evol. 2009;280:187–206. doi: 10.1007/s00606-009-0178-4. [DOI] [Google Scholar]
- 13.Tucker S, Vitins A, Pikaard CS. Nucleolar dominance and ribosomal RNA gene silencing. Cur Opin Cell Biol. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Layat E, Sáez-Vásquez J, Tourmente S. Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 2012;53(2):267–276. doi: 10.1093/pcp/pcr177. [DOI] [PubMed] [Google Scholar]
- 15.Matyasek R, Dobesova E, Huska D, Ježková I, Soltis DE, Soltis PS, et al. Interpopulation hybridization generates meiotically stable rDNA epigenetic variants in allotetraploid Tragopogon mirus. Plant J. 2016;85:362–377. doi: 10.1111/tpj.13110. [DOI] [PubMed] [Google Scholar]
- 16.Maluszynska J, Hasterok R, Weiss H. rRNA genes – their distribution and activity in plants. In: Małuszynska J, editor. Plant cytogenetics. Katowice: Silesian University Press; 1998. pp. 75–95. [Google Scholar]
- 17.Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, et al. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot. 2006;97:205–216. doi: 10.1093/aob/mcj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia S, Garnatje T, Kovarik A. Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma. 2012;121:389–394. doi: 10.1007/s00412-012-0368-7. [DOI] [PubMed] [Google Scholar]
- 19.Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- 20.Volkov R, Zanke C, Panchuk I, Hemleben V. Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theor Appl Genet. 2001;103:1273–1282. doi: 10.1007/s001220100670. [DOI] [Google Scholar]
- 21.Volkov RA, Komarova NY, Panchuk II, Hemleben V. Molecular evolution of rDNA external transcribed spacer and phylogeny of sect. Petota (genus Solanum) Mol Phylogenet Evol. 2003;29:187–202. doi: 10.1016/S1055-7903(03)00092-7. [DOI] [PubMed] [Google Scholar]
- 22.Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on non-homologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coen ES, Thoday JM, Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982;295:564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Shi L, Li D, Sun Y, Niu Y, Chen Z, et al. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS ONE. 2012;7(8):e43971. doi: 10.1371/journal.pone.0043971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahelka V, Kopecký D, Baum BR. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae) Mol Biol Evol. 2013;30:2065–2086. doi: 10.1093/molbev/mst106. [DOI] [PubMed] [Google Scholar]
- 26.Volkov RA, Komarova NY, Hemleben V. Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst Biodiv. 2007;5:261–276. doi: 10.1017/S1477200007002447. [DOI] [Google Scholar]
- 27.Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA. Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol. 2004;56:439–463. doi: 10.1007/s11103-004-4678-x. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/S1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 29.Dadejová M, Lim KY, Soucková‐Skalická K, Matyášek R, Grandbastien MA, Leitch A, et al. Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytol. 2007;174:658–668. doi: 10.1111/j.1469-8137.2007.02034.x. [DOI] [PubMed] [Google Scholar]
- 30.Guo X, Han F. Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell. 2014;26:4311–4327. doi: 10.1105/tpc.114.129841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borisjuk NV, Davidjuk YM, Kostishin SS, Miroshnichenco GP, Velasco R, Hemleben V, et al. Structural analysis of rDNA in the genus Nicotiana. Plant Mol Biol. 1997;35(5):655–660. doi: 10.1023/A:1005856618898. [DOI] [PubMed] [Google Scholar]
- 32.Fulnecek J, Lim KY, Leitch AR, Kovarik A, Matyasek R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88:19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- 33.Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long‐term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytol. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- 34.Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 1976;58:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- 35.Hizume M, Sato S, Tanaka A. A highly reproducible method of nucleolus organizing regions staining in plants. Stain Technol. 1980;55:87–90. [PubMed] [Google Scholar]
- 36.Gerlach W, Dyer T. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucl Acids Res. 1980;8:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unfried I, Gruendler P. Nucleotide sequence of the 5.8 S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucl Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borisjuk N, Momot V, Gleba Y. Novel class of rDNA repeat units in somatic hybrids between Nicotiana and Atropa. Theor Appl Genet. 1988;76:108–112. doi: 10.1007/BF00288839. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. Comp Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 41.Gruendler P, Unfried I, Pascher K, Schweizer D. rDNA intergenic region from Arabidopsis thaliana structural analysis, intraspecific variation and functional implications. J Mol Biol. 1991;221:1209–1222. doi: 10.1016/0022-2836(91)90929-Z. [DOI] [PubMed] [Google Scholar]
- 42.Perry KL, Palukaitis P. Transcription of tomato ribosomal DNA and the organization of the intergenic spacer. Mol Gen Genet. 1990;221:102–112. doi: 10.1007/BF00280374. [DOI] [PubMed] [Google Scholar]
- 43.Grabiele M, Debat HJ, Moscone EA, Ducasse DA. 25S–18S rDNA IGS of Capsicum: molecular structure and comparison. Plant Syst Evol. 2012;298:313–321. doi: 10.1007/s00606-011-0546-8. [DOI] [Google Scholar]
- 44.Bauer N, Horvat T, Biruš I, Vičić V, Zoldoš V. Nucleotide sequence, structural organization and length heterogeneity of ribosomal DNA intergenic spacer in Quercus petraea (Matt.) Liebl. and Q. robur L. Mol Genet Genom. 2009;281:207–221. doi: 10.1007/s00438-008-0404-8. [DOI] [PubMed] [Google Scholar]
- 45.Davidjuk YM, Hemleben V, Volkov RA. Structural organization of 5S rDNA of eggplant, Solanum melongena L. Biol Syst. 2010;2:3–6. [Google Scholar]
- 46.Kitamura S, Inoue M, Shikazono N, Tanaka A. Relationships among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theor Appl Genet. 2001;103:678–686. doi: 10.1007/s001220100643. [DOI] [Google Scholar]
- 47.Tynkevich Y, Volkov R. Structural organization of 5S ribosomal DNA in Rosa rugosa. Cytol Genet. 2014;48:1–6. doi: 10.3103/S0095452714010095. [DOI] [PubMed] [Google Scholar]
- 48.Douet J, Tourmente S. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity. 2007;99:5–13. doi: 10.1038/sj.hdy.6800964. [DOI] [PubMed] [Google Scholar]
- 49.Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM. A molecular phylogeny of the solanaceae. Taxon. 2008;57:1159–1181. [Google Scholar]
- 50.Sheidai M, Mosallanejad M, Khatamsaz M. Karyological studies in Hyoscyamus species of Iran. Nord J Bot. 1999;19:369–374. doi: 10.1111/j.1756-1051.1999.tb01130.x. [DOI] [Google Scholar]
- 51.Lavania UC, Kushwaha JS, Lavania S, Basu S. Chromosomal localization of rDNA and DAPI bands in solanaceous medicinal plant Hyoscyamus niger L. J Genet. 2011;89:493. doi: 10.1007/s12041-010-0071-5. [DOI] [PubMed] [Google Scholar]
- 52.Tu TY, Sun H, Gu ZJ, Yue JP. Cytological studies on the Sino‐Himalayan endemic Anisodus and four related genera from the tribe hyoscyameae (solanaceae) and their systematic and evolutionary implications. Bot J Linn Soc. 2005;147:457–468. doi: 10.1111/j.1095-8339.2005.00384.x. [DOI] [Google Scholar]
- 53.Chiarini FE, Barboza GE. Karyological studies in Jaborosa (solanaceae) Bot J Linn Soc. 2008;156:467–478. doi: 10.1111/j.1095-8339.2007.00734.x. [DOI] [Google Scholar]
- 54.Moscone EA, Loidl J, Ehrendorfer F, Hunziker AT. Analysis of active nucleolus organizing regions in Capsicum (Solanaceae) by silver staining. Am J Bot. 1995;82:276–87. doi: 10.2307/2445534. [DOI] [Google Scholar]
- 55.Stiefkens L, Peñas MLL, Bernardello G, Levin RA, Miller JS. Karyotypes and fluorescent chromosome banding patterns in southern African Lycium (Solanaceae) Caryologia. 2010;63:50–61. doi: 10.1080/00087114.2010.10589708. [DOI] [Google Scholar]
- 56.Schubert I, Wobus U. In situ hybridization confirms jumping nucleolus organizing region in Allium. Chromosoma. 1985;92:143–148. doi: 10.1007/BF00328466. [DOI] [Google Scholar]
- 57.Shishido R, Sano Y, Fukui K. Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol Gen Genet. 2000;263:586–591. doi: 10.1007/s004380051205. [DOI] [PubMed] [Google Scholar]
- 58.Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D. Quantitative karyotyping and dual-color FISH mapping of 5S and 18S–25S rDNA probes in the cultivated Phaseolus species (Leguminosae) Genome. 1999;42:1224–33. doi: 10.1139/g99-070. [DOI] [PubMed] [Google Scholar]
- 59.Gleba YY, Momot V, Okolot A, Cherep N, Skarzhynskaya M, Kotov V. Genetic processes in intergeneric cell hybrids Atropa + Nicotiana. Theor Appl Genet. 1983;65:269–276. doi: 10.1007/BF00276562. [DOI] [PubMed] [Google Scholar]
- 60.Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol Gen Genet. 1993;240:159–169. doi: 10.1007/BF00277053. [DOI] [PubMed] [Google Scholar]
- 61.Lim KY, Kovarik A, Matýăsek R, Bezděek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- 62.Kovarik A, Matyasek R, Lim K, Skalická K, Koukalova B, Knapp S, et al. Concerted evolution of 18–5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol J Linn Soc. 2004;82:615–625. doi: 10.1111/j.1095-8312.2004.00345.x. [DOI] [Google Scholar]
- 63.Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol J Linn Soc. 2004;82:485–501. doi: 10.1111/j.1095-8312.2004.00335.x. [DOI] [Google Scholar]
- 64.Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci U S A. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Navashin M. Chromosome alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia. 1934;5:169–203. doi: 10.1508/cytologia.5.169. [DOI] [Google Scholar]
- 66.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Cur Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 67.Pikaard CS, Reeder RH. Sequence elements essential for function of the Xenopus laevis ribosomal DNA enhancers. Mol Cell Biol. 1988;8:4282–4288. doi: 10.1128/MCB.8.10.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doelling JH, Gaudino RJ, Pikaard CS. Functional analysis of Arabidopsis thaliana rRNA gene and spacer promoters in vivo and by transient expression. Proc Natl Acad Sci U S A. 1993;90:7528–7532. doi: 10.1073/pnas.90.16.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hemleben V, Zentgraf U. Structural organization and regulation of transcription by RNA polymerase I of plant nuclear ribosomal RNA genes. Results Probl Cell Differ. 1994;20:3–24. doi: 10.1007/978-3-540-48037-2_1. [DOI] [PubMed] [Google Scholar]
- 70.Georgiev S, Papazova N, Gecheff K. Transcriptional activity of an inversion split NOR in barley (Hordeum vulgare L.) Chrom Res. 2001;9:507–514. doi: 10.1023/A:1011640830891. [DOI] [PubMed] [Google Scholar]
- 71.Chandrasekhara C, Mohannath G, Blevins T, Pontvianne F, Pikaard CS. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Gen Dev. 2016;30:177–190. doi: 10.1101/gad.273755.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mishima M, Ohmido N, Fukui K, Yahara T. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae) Chromosoma. 2002;110:550–8. doi: 10.1007/s00412-001-0175-z. [DOI] [PubMed] [Google Scholar]
- 73.García-Olmedo F, Carbonero P, Aragoncillo C, Salcedo G. Loss of redundant gene expression after polyploidization in plants. Experientia. 1978;34:332–333. doi: 10.1007/BF01923018. [DOI] [Google Scholar]
- 74.Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:21.1–21.19. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 75.Buggs RJA, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE. The legacy of diploid progenitors in allopolyploid gene expression patterns. Phil Trans R Soc. 2014;B369:20130354. doi: 10.1098/rstb.2013.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. doi: 10.1023/A:1006392424384. [DOI] [PubMed] [Google Scholar]
- 78.Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos: KF492694, KF496929-KF496945, KX196202-KX196207 and KY126357-KY126368.


