Abstract
This protocol describes targetable reactive electrophiles and oxidants (T-REX)—a live-cell-based tool designed to (i) interrogate the consequences of specific and time-resolved redox events, and (ii) screen for bona fide redox-sensor targets. A small-molecule toolset comprising photocaged precursors to specific reactive redox signals is constructed such that these inert precursors specifically and irreversibly tag any HaloTag-fused protein of interest (POI) in mammalian and Escherichia coli cells. Syntheses of the alkyne-functionalized endogenous reactive signal 4-hydroxynonenal (HNE (alkyne)) and the HaloTag-targetable photocaged precursor to HNE (alkyne) (also known as Ht-PreHNE or HtPHA) are described. Low-energy light prompts photo-uncaging (t1/2 <1–2 min) and target-specific modification. The targeted modification of the POI enables precisely timed and spatially controlled redox events with no off-target modification. Two independent pathways are described, along with a simple setup to functionally validate known targets or discover novel sensors. T-REX sidesteps mixed responses caused by uncontrolled whole-cell swamping with reactive signals. Modification and downstream response can be analyzed by in-gel fluorescence, proteomics, qRT-PCR, immunofluorescence, fluorescence resonance energy transfer (FRET)-based and dual-luciferase reporters, or flow cytometry assays. T-REX targeting takes 4 h from initial probe treatment. Analysis of targeted redox responses takes an additional 4–24 h, depending on the nature of the pathway and the type of readouts used.
INTRODUCTION
Chemical redox signals have emerged as major small-molecule modulators in nearly all of life’s essential processes1. Contrary to their typical pathological role2, it is now believed that orchestrated transient fluxes of reactive electrophiles3 and/or oxidants4–6 at low concentrations promote physiological fitness. Unlike traditional second messengers such as phosphate signal carriers, redox signals find their target nonenzymatically, relying upon their inherent chemical reactivity, as well as that of their protein target(s). Such a mechanism necessitates a delicate balance and precise spatiotemporal relationship between the reactive small-molecule signal and the target protein(s) that undergo(es) selective chemical modification against an otherwise unperturbed proteome. These intricacies render specific redox responses difficult to model in living systems. Identification of genuine redox-sensor targets that respond to a specific reactive signal in an otherwise unperturbed cellular setting—comprising many endogenous signaling molecules at basal concentrations, such as oxidants, electrophiles and metabolites—also presents a challenging task.
The emerging significance of lipid-derived signaling electrophiles (LDEs)—such as 4-hydroxynonenal (HNE)—as endogenous redox-linked signal carriers7,8, and small-molecule redox-activator drugs with functional properties similar to these endogenous LDEs9–13, warrants a clear understanding of the role of signaling electrophiles in redox regulation. However, many important biological questions underpinning specificity and timing remain largely unresolved by currently used methods.
Redox responses have been studied by many elegant methods, such as mechanical stretching or shear stress (at the cell level)14–18, and specific growth-factor or metabolite-targeted stimulation under physiologic or pathologic settings19–21—e.g., H2O2 signaling through Nox enzymes22, and glucose-stimulated mitochondrial ROS production in hyperglycemia23–25.
In terms of a general pharmacological (i.e., small molecule–based) strategy, the physiologic impacts of nonenzymatic redox-linked protein modifications in living systems are conventionally studied using bolus dosing approaches: an excess of a reactive signal is administered and a specific phenotype of interest is read out7,8, or the gamut of proteins modified are identified by affinity purification followed by mass spectrometry7,26–34. These powerful approaches have shed light on the functional importance of LDE signaling, and they have identified novel LDE-sensitive targets and pathways. Importantly, these data collectively suggest that distinct LDEs are capable of driving vastly different pathways spanning inflammation35–38, immune response3,39–41, epigenetic signaling26,42–44, apoptotic signaling45–47 and so on.
Many conventional signaling pathways function through a gain of function or a dominant loss of function—mechanisms that are phenotypic even with low-stoichiometry modifications at a single signaling node. By analogy, it is likely that low-stoichiometry modification of intrinsically reactive proteins is important in redox signaling. Under bolus dosing conditions, these key modifications can often be lost in noise stemming from off-target modification of highly abundant proteins1,48,49. For example, quantitative proteomics studies have also shown that reactive signals such as HNE covalently label ~1,000 cysteine-active targets under typical global treatment regimens27,30,31. Accordingly, bolus dosing strategies with reactive signals typically recapitulate complete loss of function28,50, and they are a simple and versatile approach to modeling oxidative damage-associated pathological phenotypes51. Whereas modifications of a plethora of targets typically mark the end point of oxidative damage50–53, endogenous physiologic redox signaling is known to occur under localized generation of transient fluxes of basal redox signals3,6,21,38,54. Thus, because bolus dosing uses whole-specimen treatment conditions, it has been challenging to shed light on the specific impacts of specific redox events that are on their own necessary and/or sufficient to prompt signaling response downstream. Thus, a simple transposable method would strongly complement this approach to help researchers (i) identify bona fide sensitive proteins from global dosing experiments and (ii) begin to understand how these events specifically affect downstream phenotypes (Fig. 1). The latter is critically important because the broad target spectra of redox signals under super-physiological concentrations probably mask or suppress the phenotypic responses that occur when endogenous amounts of LDEs are generated. LDEs with different chemical structures can have different membrane permeability, stability, toxicity and target affinities, resulting in discrete bioactivities and pathway specificity, as well as differing levels of promiscuity8,29,41,44,47. This information can be obtained only by using methods that probe the specificity, timing and extent to which modification of the specific target alone is adequate to elicit a response. The method described herein can be used to (i) investigate the precise consequences of specific redox events on specific signaling response and (ii) enable targeted screening and discovery of novel redox-sensor genes from precision delivery of specific LDEs.
Figure 1.
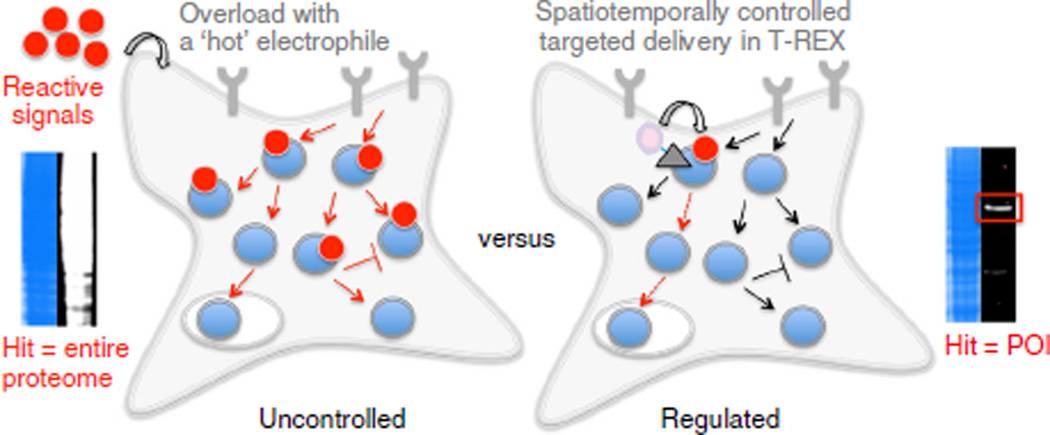
Strategies for studying cellular redox responses. The predominant small molecule–based strategy for studying cellular redox responses typically constitutes bolus dosing of a cell with reactive signals (left panel). T-REX offers a complementary and previously not recognized ‘on-demand redox targeting’ approach to the study of gain-of-function or dominant loss-of-function consequences of specific redox modifications with precise timing and target specificity (this protocol, right panel). Blue circles designate cellular proteins and red dots are reactive endogenous redox signals. All arrows illustrate representative redox pathways and trajectories (the regular and T-shaped arrows indicate direct/indirect activation and inhibition, respectively). Red arrows signify those that are being perturbed under the respective conditions. The gray triangle with the purple sphere designates a photocaged precursor (see Fig. 2 for chemical structure), and the gray Y-shaped objects represent cell-surface receptors. The accompanying gel-based data demonstrate the specificity in terms of targeted modification achieved in T-REX, which also offers temporal control through light-driven signal delivery. See Figures 2 and 3 for approaches to probe downstream response.
Development of the method: T-REX on-demand redox targeting Temporal control and target specificity
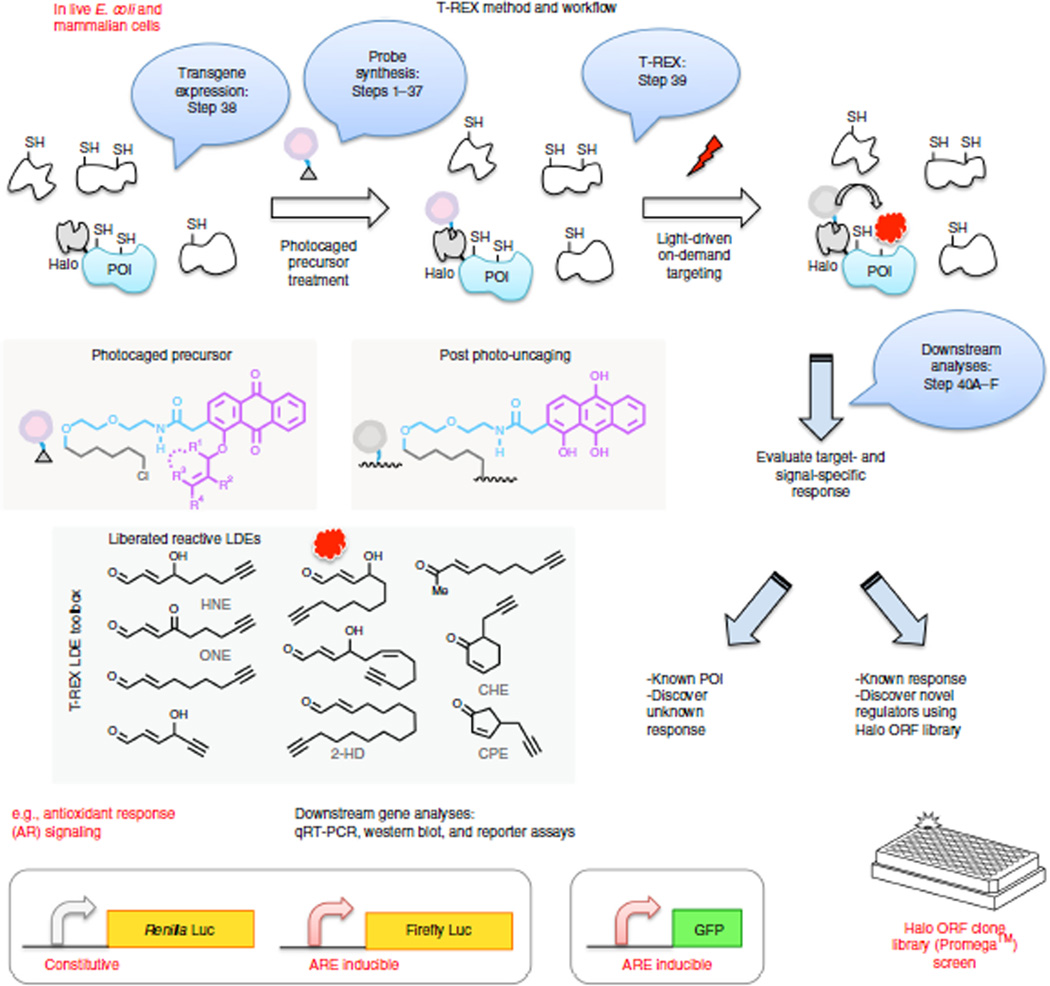
T-REX technology55–57 selectively modifies a specific redox-sensor POI, and it enables the decoding of the functional consequences of specific redox events, against the backdrop of an otherwise unperturbed proteome (Fig. 2). A potential redox-sensor protein is chosen based on previous lists of postulated LDE-modified proteins from global proteomics experiments, or putative redox-sensor POIs. The POI is genetically fused to a HaloTag domain. The T-REX assay uses a small organic molecule that is made up of a photocaged LDE and a chloroalkane recognition unit. The HaloTag enzyme58–61 rapidly and specifically conjugates to the chloroalkane recognition unit, resulting in an irreversible 1:1 Halo–small-molecule complex. The 15-atom linker between the chloroalkane function and the photocaged LDE renders the inert caged motif solvent exposed such that low-energy light illumination (0.3 mW/cm2, 365 nm; Supplementary Fig. 1 and Supplementary Videos 1 and 2) unleashes (t1/2 < 1–2 min) a maximum of one LDE molecule per caged precursor in vitro56,57 and in cells (t1/2 ~ 0.5 ± 0.3 min) (Supplementary Fig. 2a). Proximity enhancement62 enables targeted LDE modification of the redox-sensitive POI, whereas HaloTag itself does not react with the liberated LDE (Supplementary Fig. 2b).
Figure 2.
On-target, on-demand redox signaling enabled by T-REX. Bubbles indicate experimental steps described in the protocol. Either E. coli or mammalian cells expressing HaloTag-fused proteins of interest (POIs) are treated with designated photocaged precursors (5–25 µM, 2 h) to achieve a 1:1 covalent binding between the HaloTag and the photocaged probe. After rinsing cycles, exposure of the cells (for 3–20 min) to low-energy light (0.3 mW/cm2, 365 nm) at room temperature elicits rapid liberation of a reactive signal (lipid-derived electrophiles (LDEs), inset) from the photocaged probe bound to HaloTag. Proximity enhancement62 facilitates on-target, on-demand covalent modification of amino acid residue(s) on the POIs, typically cysteines. HNE is also known to be capable of modifying lysine and histidine (see text). Regardless of residue specificity, T-REX is able to ping one potential responsive protein with a precision dose of reactive lipid. Irrespective of residue identity, in-gel fluorescence analysis reports on the presence of HNE modification on the POIs. Residue specificity in POI modification is determined by LC–MS/MS analysis post cell lysis and resin-assisted enrichment (Fig. 5c). Once a specific sensor protein has been earmarked by T-REX, target- and residue(s)-specific post-translational modification can be directly linked to the signaling function of interest in an otherwise unperturbed cellular background. T-REX can (i) interrogate specific redox-linked signaling responses and (ii) discover novel regulators that upon selective lipidation are sufficient to elicit a biologically relevant response. Generality and scope in terms of both target and signal specificity are exemplified with distinct vertebrate sensor proteins (e.g., Keap1, RRM1, HSPB7) and structurally distinct LDEs (inset). Pathway activation is analyzed using dual-luciferase reporter assays or GFP reporter assays by flow cytometry. Endogenous downstream gene activation can be analyzed by qRT-PCR and western blotting.
We performed validation experiments that included the following:
Blocking experiments to check for specificity
Pretreatment of Halo–POI-expressing cells with a HaloTag-targetable photocaged LDE (‘photocaged precursor’ hereafter)—before the addition of tetramethylrhodamine (TMR) dye–conjugated chloroalkane and subsequent live imaging—confirmed that the photocaged precursors saturate the Halo protein binding site within 2 h (ref. 55), consistent with HaloTag’s rapid second-order reaction60 (Box 1). Functionality of HaloTagged POIs was also assessed (Box 2). Both TMR-dye-conjugated chloroalkane and the photocaged precursor (Fig. 2, inset) labeled HaloTag exclusively. Hence, there is no reaction of caged precursors with other cellular targets or the POI, and the chloroalkane appendage is stable55–57. Such a result is common because eukaryotic cells and most bacteria, including E. coli58,59,61, do not express haloalkane dehydrogenases, conferring excellent bio-orthogonality.
Box 1. Live imaging: assessment of complete HaloTag conjugation (i.e., blocking experiment with HaloTag TMR ligand) ● TIMING 2 d.
Transient transfection using TransIT-2020
-
Split HEK-293 cells in two 35-mm glass-bottom dishes. For each dish, seeding ~4 × 105 cells in 2.0 ml of total cell culture medium should result in cells that are ~40–50% confluent after 24 h, which is optimal.
▲ CRITICAL STEP A cell density that is too low or too high may result in excessive cell death or poor transfection efficiency, respectively. Higher cell density is also not optimal for imaging.
Transfect cells with the HaloTag-conjugated POI using TransIT-2020 according to the manufacturer’s protocol.
The subsequent steps should be performed 24–36 h post transfection.
Blocking with HaloTag TMR (Halo-TMR) ligand and imaging
▲ CRITICAL STEP The following steps should be performed under dim light.
Treat the cells in one dish with HtPHA in 2 ml of serum-free medium. The protocol for treatment with the T-REX photocaged precursor is identical to that in Step 39A(i–ii).
Treat the second dish with serum-free medium containing DMSO instead.
Rinse both sets of cells three times (each time with 1.5 ml of serum-free medium) every 30 min over 1.5 h.
-
After the third rinse cycle, remove the rinse medium from both dishes and replace it with 2 ml of serum-free medium containing 3 µM Halo-TMR ligand (both dishes).
▲ CRITICAL STEP Make sure that the Halo-TMR ligand is thoroughly mixed by pipetting up and down at least 8–10 times.
Incubate the cells for 1–2 h at 37 °C.
Rinse 3× with 1.5 ml of serum-free medium.
Image cells using a confocal microscope according to the instruments protocol. (In our case, a Zeiss 710 confocal microscope was used for image acquisition. Images were analyzed using ImageJ software.)
Box 2. Functional assay for Halo-Keap1 binding to Nrf2 ● TIMING 2 d.
Transient transfection using TransIT-2020
-
Split HEK-293 cells in two 35-mm glass-bottom dishes. For each dish, seeding ~4 × 105 cells in 2.0-ml total cell culture medium should result in cells that are ~40–50% confluent after 24 h, which represents optimal cell density.
▲ CRITICAL STEP A cell density that is too low or too high may result in excessive cell death or poor transfection efficiency, respectively. Higher cell density is also not optimal for imaging.
Transfect one plate with 1,500 ng of eGFP-Nrf2 plasmids and transfect the second plate with 750 ng of eGFP-Nrf2 and 750 ng of pMIR-DsRed-IRES-Halo-Keap1. Transfect with TransIT-2020 according to the manufacturer’s protocol.
Incubate the cells at 37 °C for 24–36 h in a humidified incubator in the presence of a 5% CO2 atmosphere, and then proceed with imaging and data analysis.
Imaging and data analysis
Image cells using a confocal microscope according to the instrument’s protocol. (In our case, a Zeiss 710 confocal microscope was used for image acquisition.)
-
Images were analyzed using ImageJ software. Briefly, the average green fluorescence intensity in the nucleus (Fnucleus) was quantified by tracing a free-hand circle around the nucleus. Next, the average green florescence intensity of the cytosol was measured by tracing a free-hand circle around the cytosol, excluding the nucleus. The ratio of nuclear to cytosolic green fluorescence was subsequently calculated.
▲ CRITICAL STEP To get reliable results, it is important to collect images from at least 100 individual cells per condition.
Cytotoxicity and off-target stress responses
The caged precursors were nontoxic, as judged by AlamarBlue and trypan blue viability assays55,56. UV light exposure under T-REX conditions also does not elicit upregulation of γ-H2AX63 and does not perturb other stress-sensitive pathways such as the NF-κB pathway (ref. 64), markers for DNA damage and inflammatory signaling, respectively (Supplementary Fig. 3). In T-REX, the maximum LDE signal delivered is equal to the concentration of the HaloTag fusion protein (Fig. 2). Thus, side reactions of the T-REX-liberated LDE with proteins other than the target POI occur at a much lower rate than whole-cell flooding. By contrast, global treatment with reactive LDEs induces time- and dose-dependent cytotoxicity. The effector concentration for half-maximum response (EC50) of viability, for instance, even for the robust cell line HEK-293T, is ~31 µM over 18 h of treatment8,56. Because the typical concentrations used in the literature for redox signaling studies with LDEs, for instance HNE, are above 20 µM and can reach as high as 1 mM over prolonged treatment, users are encouraged to carefully evaluate the extent of loss of cell viability under these conditions and consider associated off-target responses.
Quantification of the extent of modification
Alkyne functionalization enables fluorescence-based quantification of the amount of LDE signal delivered to the POI, and that remains unliberated on the HaloTag (Fig. 2)56. The low background signal to the overall proteome, along with the fact that targeting is not achieved when the HaloTag is expressed separately with the POI (the ‘nonfused’ system, Box 3)56,57, collectively led us to the conclusion that the majority of liberated lipid electrophile that does not hit its intended target is likely intercepted by small-molecule thiols such as glutathione65–68. The percentage of delivery (i.e., the amount of signal that is delivered to the POI with respect to the total initially present in the photocage) is assayed post cell lysis by a series of steps involving TEV (Tobacco etch virus) protease–mediated separation of HaloTag from the POI, click coupling69 reaction with Cy5-azide and in-gel fluorescence analysis (Fig. 3). Western blotting of a housekeeping protein (e.g., actin) and the target POI, respectively, normalizes for loading and transfection efficiency across all samples against no-light-exposed and/or no-TEV-treated controls. Subtraction of the amount of signal associated with unreacted photocage on HaloTag accounts for the true percentage of POI molecules modified in the cells (see equation in Step 40A(ix)). The value obtained from this method is broadly similar to that estimated by ion peak integration post liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis56.
Box 3. Blockage of pathway activation by HaloTag nonfused control: analysis using western blotting ● TIMING 4 d.
Transient transfection using polyethylenimine (PEI; PolyScience) ● TIMING 1 d
-
Plate HEK-293 cells in a 35-mm (8 cm2 surface area) dish. For a 35-mm dish, seeding ~7 × 105 cells in 1.5 ml of total cell culture medium should result in cells that are ~70% confluent after 24 h, which is optimal for transfection with PEI.
▲ CRITICAL STEP A cell density that is too low or too high may result in excessive cell death or poor transfection efficiency, respectively.
Transfect the cells with 500 ng of each of the following plasmids: pMIR-Dsred-IRES-Halo-HA, pMIR-DsRed-IRES-His-Keap1 and pcDNA3.1 myc-Nrf2. For transient transfection using PEI, see Section 2, Step 38A above.
Treatment of cells with T-REX photocaged precursor, photo-uncaging and harvest of cells
Perform Section 2, Step 39A, except harvest the cells 4 h post illumination.
SDS–PAGE and western blotting
-
Lyse cells by adding 30 µl of freshly prepared and prechilled lysis buffer and subjecting them to 3 flash freeze–thaw cycles.
▲ CRITICAL STEP Make sure that the cells are resuspended well in lysis buffer.
Remove debris by centrifugation (18,000g, 8 min) at 4 °C. Determine protein concentration using the Bradford assay using BSA as standard. Analyze ~30 µg of lysate protein by SDS–PAGE and western blotting using a standard protocol.
Figure 3.
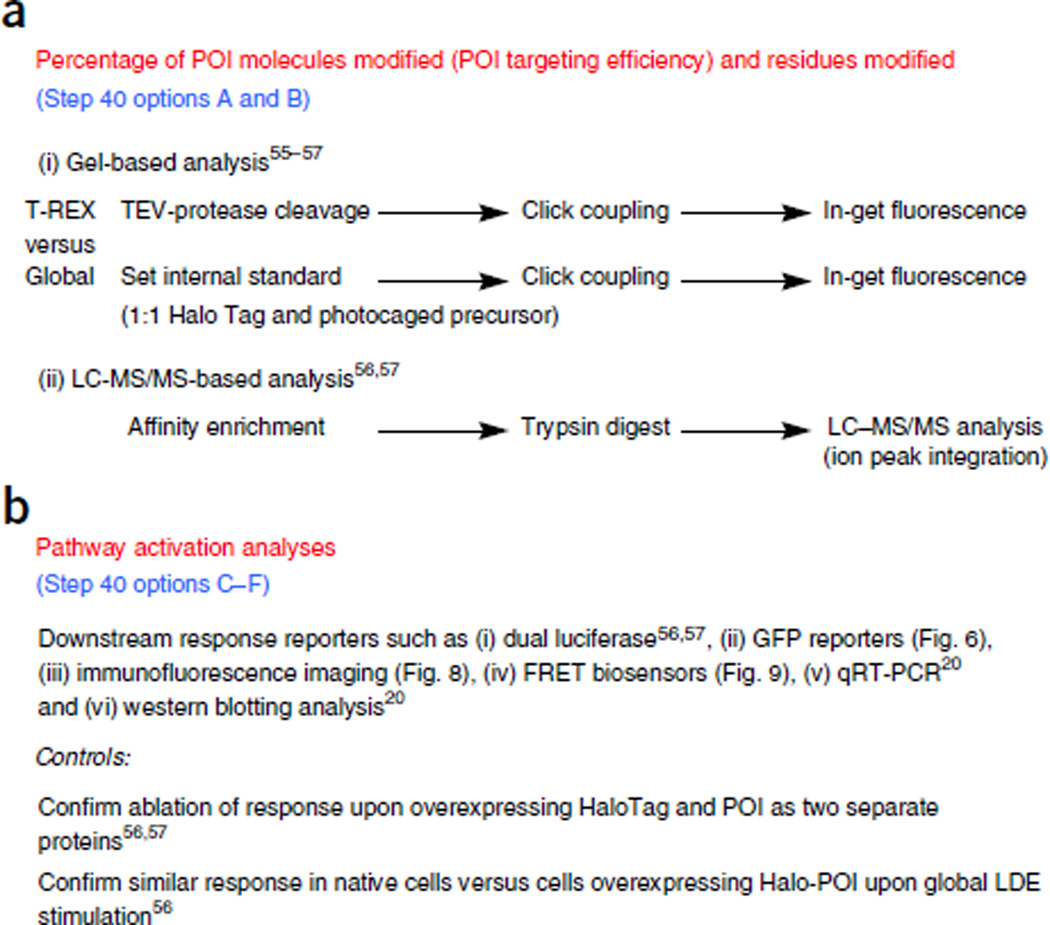
T-REX approach allows flexibility while enabling quantification of modification and response at numerous points. (a) Validation of protein as redox-sensitive. Biochemical information expected is as follows: (i) identification of the percentage of LDE modification and (ii) residue specificity. (b) Evaluation of pathway activation alongside recommended controls. Functional information expected: (i) global transcriptional response; (ii) cell-to-cell transcriptional response; (iii) changes in endogenous biological species; (iv) perturbation of signaling activities; and (v) alterations in mRNA abundance and (vi) protein levels of downstream genes.
Generality in the scope of targetable LDEs
Tolerance of HaloTag to a range of sterically demanding groups appended to chloroalkane ligands permits versatile functionalization of the caged precursor56, which makes it feasible to deliver LDEs of varying chemical architectures (Fig. 2, inset). For all the LDEs studied, expressing Halo and POI as two separate proteins, in place of the Halo–POI fusion protein, resulted in no labeling of the POI in the cells, confirming that proximity-based targeting was in operation56,57. In vitro kinetic analyses56 suggest a two-step targeting mechanism: formation of an initial target–signal encounter complex followed by covalent Michael adduction with Cys residue(s) on the target. Labeling efficiency for a given target is governed by partitioning between the rate of covalent adduct formation and diffusion of the LDE signal out of the coordination shell of the target POI55,56.
A platform for targeted screening and discovery of bona fide sensor genes
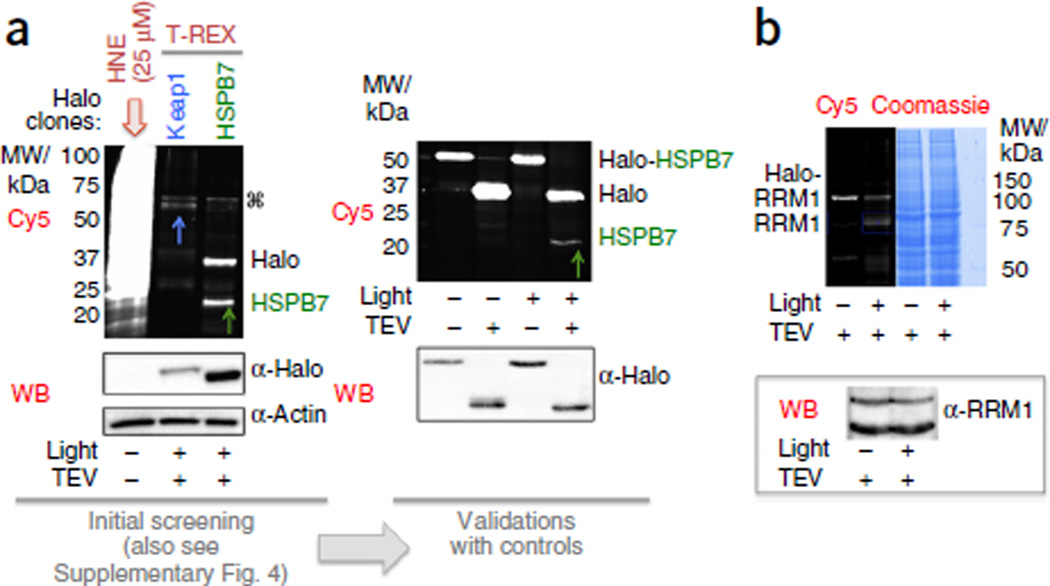
One of the major benefits of T-REX is the commercially available HaloTag human and mouse full-length ORF (open reading frame) clone libraries (Kazusa Collection, Promega). This gives an added dimension because it makes screening of potential electrophile-sensitive gene products very simple. As proof of concept, an in-house screen of ten HaloTag proteins allowed us to identify two proteins that are ‘first responders’ to basal amounts of HNE (Fig. 4 and Supplementary Fig. 4). The majority of the candidates we chose were previously identified as potentially LDE-sensitive by global proteomic profiling26–28,30,31,34 and include the following: (i) human ribonucleotide reductase (RNR) subunits RRM1 and RRM2 (and its isoform p53R2)—each subunit pair, RRM1/RRM2 or RRM1/p53R2, constitutes an active RNR complex that is essential for nuclear or mtDNA replication, respectively70; (ii) PI3K and PRKCD—two of several kinases that regulate the Nrf2-transcription-factor-driven antioxidant response (AR) pathway in mammals71; (iii) Cul3—a ligase that mediates proteasomal degradation of mammalian Nrf2 (ref. 72); and (iv) DCAF11—a mammalian analog of a stress-responsive protein in Caenorhabditis elegans. We also screened zebrafish HSPB7—a member of the small heat-shock protein family that is highly and selectively expressed in the heart73,74. hspb7 is not upregulated by heat shock75, and thus it probably has other regulation mechanisms that are as yet unidentified. Keap1—a redox-sensitive negative regulator of the Nrf2–AR pathway—served as a positive control in the screen56,57. Expression of these proteins was assessed by blotting for Halo protein (assumed to be present in a 1:1 ratio with the fused POI). By this metric, most proteins were successfully expressed, although expression varied. However, in addition to the positive control Keap1, only two proteins from this screen—RRM1 and HSPB7—were modified by HNE (Fig. 4a and Supplementary Fig. 4) under the conditions in which HNE signals were delivered in controlled amounts.
Figure 4.
Commercial HaloTag library allows discovery and validation of ‘first responders’ to a specific LDE using T-REX. The screen first identified first responders to basal amounts of HNE (Supplementary Fig. 4a). This was coupled with T-REX secondary validations using appropriate controls for effects of light alone with or without separation of Halo and POI domains during processing. As an example, T-REX-targeted HNEylation using a panel of ten distinct Halo ORF clones identified (a) zebrafish HSPB7 (theoretical MW ~18 kDa) and (b) human RRM1 (theoretical MW ~90 kDa) as novel HNE-sensitive targets. (a) The established Keap1 (theoretical MW ~70 kDa) targeting was used as a diagnostic positive control (also see Supplementary Fig. 4a). Global HNE shows comparison with established protocols (left panel in a). ⌘ marks a nonspecific band in the data set from a representative rapid initial screen of multiple HaloTag clones (Supplementary Fig. 4a). Secondary validation of HSPB7 HNEylation was performed with a full set of controls (right panel in a). HSPB7 protein identity was confirmed by pulldown (Supplementary Fig. 4b). Actin was used as loading control. Halo antibody was used to evaluate the expression level of Halo fusion proteins. (b) Secondary validation of RRM1 HNEylation (also see Supplementary Fig. 4a for an initial screen). RRM1 protein identity was confirmed by western blotting (top band, Halo-RRM1; bottom, RRM1 post TEV protease–assisted separation of Halo and RRM1). (Note: the expression plasmid vector for Halo-Keap1, Halo-HSPB7 and Halo-RRM1 encodes bicistronic expression of an internal fluorescent protein control, DsRed, alongside the Halo-tagged POI, explaining the fluorescent band at 27 kDa in these gels.) WB, western blotting.
As p53R2 and RRM1 expression was similar and RRM2 (a protein known to have a short half-life70) was also detectable, these data show that RRM1 is probably the HNE-sensitive subunit of active RNR complexes—RRM1/RRM2 and RRM1/p53R2 heterodimers. Other proteins were not appreciably HNEylated. Remarkably, RRM1, p53R2 and PRKCD—previously identified HNE-sensitive hits from global treatment approaches26–28,30,34—had expression similar to that of Keap1; yet T-REX-assisted HNE delivery was markedly different. By contrast, whole-cell HNE treatment led to nonspecific targeting under otherwise identical conditions (Fig. 4a and Supplementary Fig. 4a). Although the reasons behind these differences are likely to be multifactorial and system- and/or context-dependent, when an entire cell is swamped with reactive LDE in excess, the time-dependent nature of the underlying covalent chemistry in LDE modification typically controls the extent of off-target labeling, and thus unresponsive proteins in the T-REX screen may react too slowly with HNE to serve as ‘first responders’. Less reactive subunits or targets could start to react when HNE is in excess. In a multisubunit protein complex such as RNR, HNE transfer to other subunits could also occur under these circumstances. On the other hand, T-REX releases a maximum of one LDE molecule per HaloTag–POI unit55–57, and the POI is substoichiometrically modified by the liberated LDE55–57. We discuss aspects of existing methods and considerations for potential artifacts below.
Determination of residue specificity
Once the positive result of LDE sensitivity has been established by gel-based analysis, the identity of specific amino-acid residues modified can be determined by standard affinity enrichment followed by LC–MS/MS characterizations (Fig. 3). For Keap1, the position of the HaloTag (N or C terminus) exerted no influence on cysteine residue labeling by T-REX (Fig. 5 and Supplementary Table 1)56. Furthermore, as similar levels of signaling responses are achieved by T-REX and whole-cell LDE stimulation (see Applications section), the ability of T-REX to elicit a response indicates that functionally relevant residues are targeted56,57.
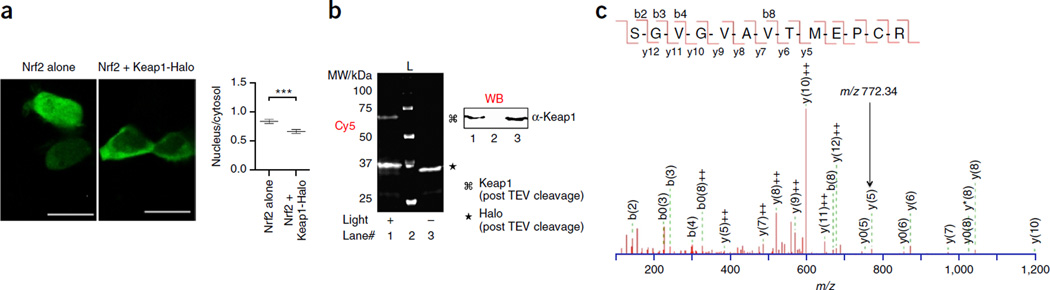
Figure 5.
Assessment of N- versus C-terminal HaloTagging on T-REX functionality, exemplified by Keap1 LDE targeting. Results with Halo-Keap1 were previously reported56,57. (a) Live imaging analysis shows that Keap1-Halo promotes Nrf2 nuclear exclusion as with Halo-Keap1 (ref. 57). Inset shows quantification performed using ImageJ (NIH). ***, P < 0.001. Scale bars, 20 µm. (b) In-gel fluorescence analysis shows that targeted HNEylation of Keap1 in HEK-293 cells using the Keap1-Halo construct is equally as efficient as using Halo-Keap1 (refs. 56,57). L, MW ladder. (c) Ionization spectrum of Keap1 peptide modified by a representative cyclohexenone-derived LDE (CHE, Fig. 2 inset) as a result of T-REX on HEK-293 cells expressing C-terminal HaloTagged Keap1, subsequent enrichment of modified Keap1-Halo from T-REX-treated cells and LC-MS/MS analysis. The same Cys residue (C613) was modified in the corresponding experiment in which N-terminally HaloTagged-Keap1 was used56. Also see Supplementary Tables 1 and 2. Arrow points to the diagnostic m/z peak for C613 modification.
Selection of specific Cys residues is likely to be dominated by individual Cys nucleophilicity in its native microenvironment. For example, LC–MS/MS analysis showed C613 modification of Keap1 by a cyclohexenone-derived LDE (namely, CHE, Fig. 2 inset), regardless of N- or C-terminal HaloTag fusion (Fig. 5c and Supplementary Table 1). Mutagenesis studies previously suggested that other Cys residues within Keap1 can compensate for the lack of C613, underscoring functional redundancy across multiple Cys residues on Keap1 (refs. 56,57). Interestingly, global treatment of cells with CHE also resulted in the modification of the same Cys residue on the Halo-Keap1 protein (Supplementary Table 2). Our previous LC–MS analysis of Keap1 modifications by HNE, an LDE of much higher reactivity than CHE, under T-REX versus global conditions resulted in nonoverlapping residues, as well as a wider scope of residues modified.
Versatility in both mammalian cells and E. coli
We also showed that the method can afford similar precision targeting of reactive LDEs in bacteria, using E. coli as proof of concept. In this example, recombinantly expressed human Keap1 genetically encoded with HaloTag at the N terminus was selectively reacted with HNE(alkyne) using T-REX (Fig. 6). As in the case of mammalian cells, photocaged precursors did not show adverse effects on the growth rate of E. coli, and they were able to permeate the E. coli cells within 2 h during the logarithmic growth phase when Halo-Keap1 expression was induced at 19 °C. The procedures used for photo-uncaging and downstream labeling analysis for mammalian cell samples were also transferable to E. coli.
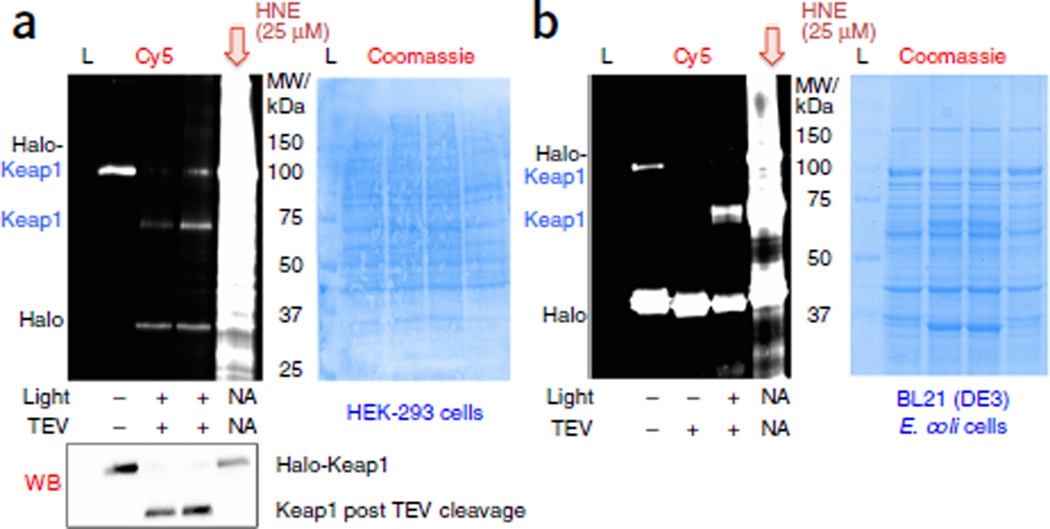
Figure 6.
T-REX targeting is equally efficient in human (HEK-293) cells and E. coli. Halo-Keap1 human protein55–57 is used as a model protein. Coomassie-stained membrane is used to evaluate uniform loading of total lysate proteins in each lane. Independent duplicates of Keap1 targeting results are presented in a. Partial cleavage of the fusion protein (into separate Halo and Keap1 proteins) was often observed during standard E. coli growth conditions, accounting for the observed Cy5 signal on the Halo band (see no-light, no-TEV sample lane in b). Theoretical MWs: Halo, ~33 kDa; Keap1, ~70 kDa. L, MW ladder; TEV, TEV protease.
Applications of the method
Redox targeting: establishing target-specific biological sufficiency in specific redox events
A major advantage of T-REX is that it has the potential to decode the gain-of-function (or dominant loss-of-function) consequences of specific redox events in living systems in a time-resolved manner. This benefit is not offered by any existing tool, despite the growing interest from both the academic and pharmaceutical communities. One critical pathway regulated by redox signaling is the Nrf2–AR axis. The conserved Nrf2–AR pathway is a gatekeeper for the expression of hundreds of detoxification and antioxidant genes that are essential for cytoprotective defense in all cell types in metazoa. This pathway also has an essential role in aspects of physiology, such as organogenesis and life-span regulation, and, conversely, in various disease states, such as tumor metastasis and drug resistance71,76,77. There are many electrophilic pharmacophores (e.g., tecfidera, bardoxolone, sulforaphane, curcumin)9–13,78 with chemical reactivity similar to that of the endogenous AR stimulator HNE—which is a reactive signaling compound known to have >800 cysteine targets under bolus dosing conditions27,30,31,34. These electrophiles are believed to confer therapeutic benefits by upregulating, among others, the Nrf2–AR pathway9–13,78. The use of T-REX in cultured human cells enables targeted Nrf2–AR pathway activation with precise timing and without perturbing other redox-sensor protein networks55–57. T-REX has shown that targeted HNEylation of one of the many redox-sensitive AR regulators, Keap1, with low stoichiometry is sufficient to stimulate maximal AR within the complex multisensor protein networks regulating Nrf2. (‘HNEylation’ is defined as a post-translational modification of a protein by HNE through covalent chemical conjugation to any residue. The target residue is most often cysteine, but HNE can also react with lysine and histidine residues30,79–81. Our gel-based analysis of T-REX targeting assessment shows no prejudice regarding residue specificity, nor the specific chemical identity of adducts formed.) In this way, we were also able to rule out the proposed HNE-sensing ability of Nrf2 itself, as co-overexpression of Nrf2 (which directly binds Halo-Keap1) does not result in HNEylation of Nrf2 in cells57. In addition, because reactive LDEs such as HNE will react with any isolated protein bearing Cys (and also His and Lys residues, depending on incubation time and concentration3), T-REX is an ideal method for determining functionally relevant modification events that are sufficient to trigger signaling.
Gel-based fluorescence quantification shows that the amount of LDE, exemplified by the cyclohexenone analog CHE (Fig. 2 inset), reacted with Keap1 under whole-cell LDE treatment conditions (25 µM, 20 min, EC50 (viability) ~90 µM) is ~6-fold higher than that achieved under T-REX56. However, global treatment provides no additional bonus in terms of the magnitude of AR upregulation. The percentage of Keap1 molecules modified under T-REX conditions can be determined easily based on two independent methods of quantification: in-gel fluorescence (vide supra) and ion peak integration56. For a representative targeted modification of Keap1 with cyclohexenone-derived LDE (Fig. 2, inset), the two methods yielded percentage targeting efficiencies of 19% and 15%, respectively56. Controls showed that the Nrf2–AR upregulation phenotype is not due to T-REX affecting the proteasomal pathway that regulates steady-state Nrf2 protein levels57. The observed Nrf2–AR upregulation was also not due to untargeted delivery because AR activation did not occur when HaloTag and the target POI, Keap1, were overexpressed as two separate proteins56,57. Keap1 is unusually cysteine-rich, and it reacts rapidly with electrophiles, so this experiment gives a high degree of confidence that T-REX does not perturb other sensor proteins that regulate Nrf2–AR in the cell. In fact, the result allowed us to postulate that T-REX proceeds via a target–signal encounter complex formed specifically because the electrophile is juxtaposed to the target upon photo-uncaging56.
Redox targeting: interrogating signal-specific and on-target signaling strength
We have also developed an integrated electrophile toolbox that enables targeted delivery of various linear enal, enone and cyclic-enone-based LDEs to specific sensor proteins in cells (Fig. 2, inset)56. Targeting efficiency is not largely influenced by intrinsic electrophilicity of the reactive signals. This observation is consistent with in vitro kinetic data, which show largely similar initial on-rates of LDE adduction of Keap1 (ref. 56). Thus, T-REX probably creates a microenvironment that behaves as if the target POI has been transiently treated with saturating LDEs. The ability of the T-REX LDE toolbox to provide a range of signaling LDEs opens a new avenue to quantitatively dissect how a specific sensor protein or pathway deals with specific reactive LDEs. Using Keap1 as a model sensor protein in cells, T-REX provides a means to elucidate how reactive LDE modifications directly translate to the strength of Nrf2–AR activation. In addition, the case with a cyclopentenone-based LDE (namely, CPE, Fig. 2 inset)56 helps exemplify the potential utility of T-REX in the identification of novel small molecules that elicit pathway activation only through targeted delivery, but fail to activate AR from whole-cell flooding before toxicity56.
Quantification of signaling response in subpopulations
We have described various methods that report on average increases in AR signal across the whole population. We stress that to identify new signaling effects of POI-targeted redox modifications on any other transcriptional pathways of interest users can simply replace the Nrf2–AR activation reporter plasmids used herein with any of the tens of signal transduction reporter plasmids that are commercially available in both luciferase- and GFP-reporter formats (e.g., one commercial source that we have used is the Cignal 45-Pathway Reporter Array from Qiagen). So far, we have shown pathway modulation using the dual-luciferase assay, which reports transcriptional activation of Nrf2-driven AR (refs. 56,57), as well as qRT-PCR (ref. 57) and western blotting analyses, which evaluate AR-driven downstream genes at the mRNA and protein levels, respectively56,57. These data that look at AR upregulation in ensembles of cells on the whole show little or no difference in the manner in which T-REX and whole-cell HNE exposure stimulate AR. Herein, we report an orthogonal flow cytometry assay that shows the extent of AR-driven GFP expression (‘the GFP reporter assay’ hereafter) (Fig. 7). This assay allows us to compare the effects of whole-cell HNE flooding and the T-REX approach on AR on a cell-by-cell basis.
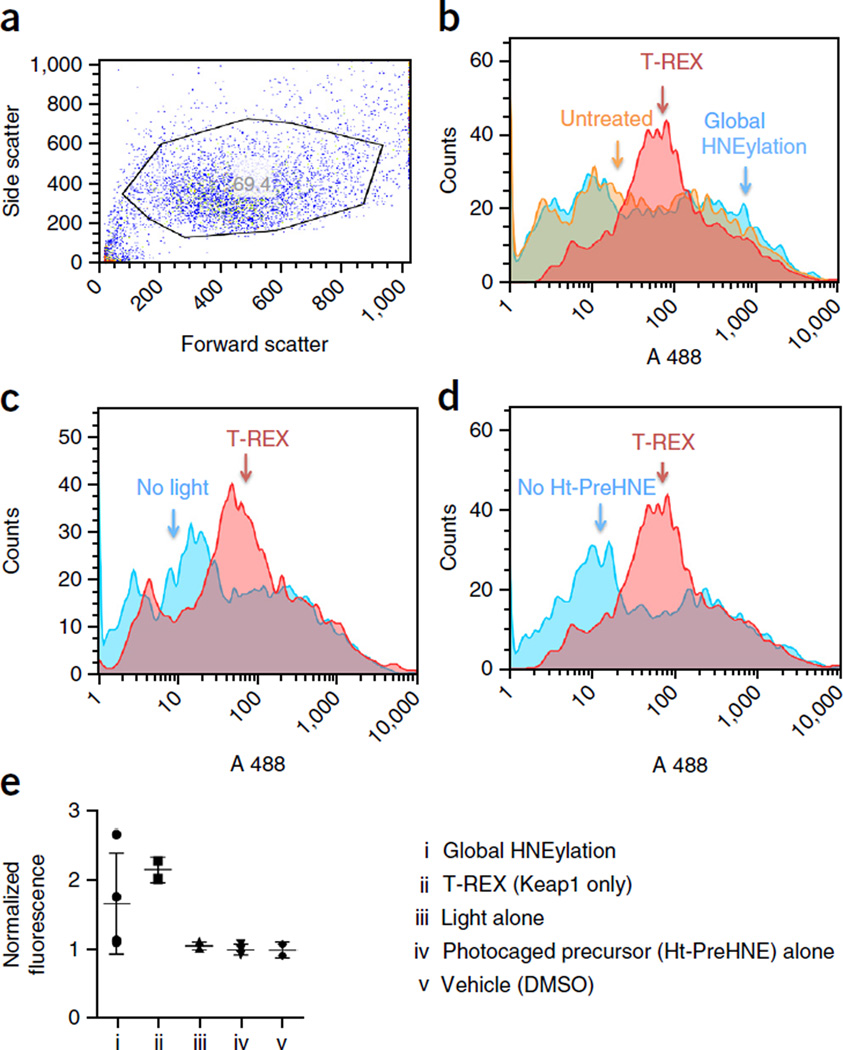
Figure 7.
Flow-cytometry-based ARE-GFP reporter assay quantifying T-REX-mediated activation of antioxidant response in a subpopulation of live HEK-293 cells. (a) Representative GFP expression level in cells transiently transfected with plasmids encoding Halo-Keap1, Nrf2 and ARE-GFP. (b–d) Representative single-parameter (GFP signal) histograms obtained from cells transiently transfected with plasmids encoding Halo-Keap1, Nrf2 and ARE-GFP that have been treated as indicated. (b) Comparison of the results between whole-cell HNE stimulation (blue), T-REX-assisted Keap1-specific activation (red) and no treatment (no photocaged precursor and no light) (orange). (c) Comparison between T-REX (red) and ‘no light exposure’ control (blue). (d) Comparison between T-REX (red) and ‘no photocaged precursor’ control (blue). (e) Representative data from total fluorescence analysis of GFP signal. Error bars designate s.d. (n = 3).
In this flow cytometry assay, live cells are first gated by forward (size) and side scatter to give a general population of healthy cells (Fig. 7a). This specific gating is applied to each data set (e.g., photocaged precursor Ht-PreHNE alone, light alone and so on). This population should be a single group, and it should be the largest single population for each data set. We gated the scatter group in several ways, and similar downstream results were obtained, but it is important that each data set (including all appropriate controls) be gated the same way. Analyzing this scatter group for green fluorescence (AR reporter) told a story that was slightly different from that painted by our previous ensemble experiments56,57. Intriguingly, whole-cell HNE treatment strongly increases AR in a subset of cells, principally those showing a medium level of AR in the ground state. Notably, there is little change in cells with low basal AR (Fig. 7b).
On the other hand, with T-REX, the increase in GFP signal stemmed from an increase in AR in all but the cells with the highest basal AR (Fig. 7b–d). We attribute the lack of effect on cells with a high basal AR to the fact that these cells express a lower level of Keap1 protein with respect to Nrf2, rendering them less susceptible to specific AR upregulation by T-REX. Nonetheless, T-REX enables the initiation of AR in a larger fraction of the cell pool than does global HNEylation, and it does not hyperstimulate AR. Given that T-REX shows that most cells can respond to low-occupancy HNE stimulation through Keap1 modification, it is likely that the small number of responders seen with HNE is a result of the compensatory suppression of AR due to alkylation of multiple proteins. Such an observation would be consistent with T-REX being able to mimic endogenous LDE signaling, and it is thus further consistent with the low off-target spectrum associated with T-REX and the fact that T-REX faithfully reports on AR signaling selectively through Keap1 (refs. 56,57).
Generality across other redox-sensor targets and pathways
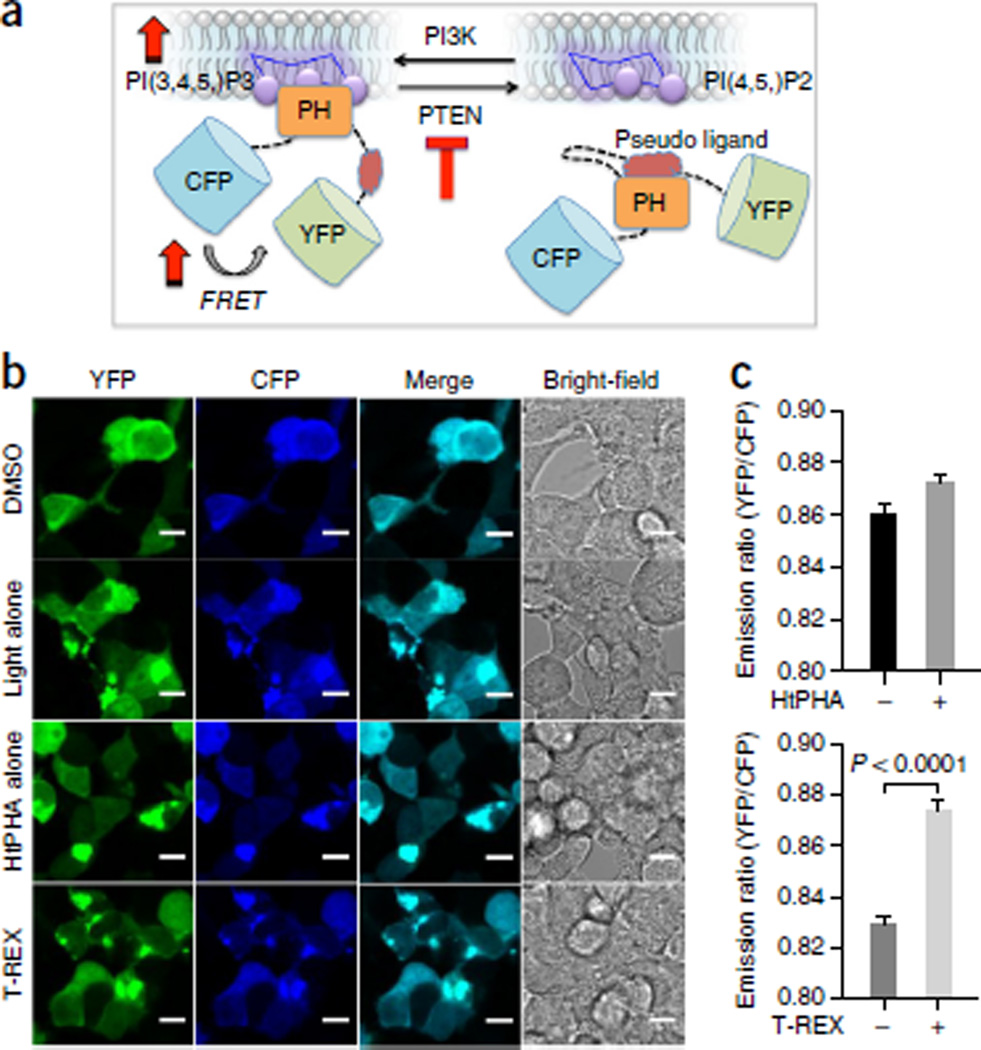
We further validated the application of T-REX beyond the targeted perturbation of Nrf2–AR signaling axis by selective downregulation of a key redox-sensitive tumor suppressor protein, PTEN82. Oxidation or alkylation of PTEN by LDEs is known to inactivate PTEN phosphatase activity83–85. PTEN modifications elicit dominant loss of function of PTEN86–88, and thus minor modifications can result in measurable accumulation of its cellular substrate, phosphatidylinositol 3,4,5-triphosphate (PIP3). Building on our previous work that establishes T-REX-assisted HNEylation of PTEN55, we here showed that T-REX also offers a means to temporally modulate the PTEN signaling. Two different orthogonal and established readouts—immunofluorescence (IF) analysis of endogenous PIP3 levels in fixed cells (Fig. 8) and FRET-based ‘lnPAkt’ (indicator of phosphoinositide based on Akt) reporter assay89,90 in live cells (Fig. 9)—were used. The representative images of fixed cells in Figure 8 and live cells in Figure 9 both underscore that T-REX coupled with either IF or FRET assays is nonintrusive to cellular integrity—an aspect that we encourage users to be aware of before proceeding to data quantification. Although the fold changes in the measured FRET signals (Fig. 9) are small, they are within the range previously established for growth factor–induced or pharmacological perturbation of the same pathway using the identical lnPAkt FRET reporter plasmid89,90 (Figs. 8 and 9). These outcomes also suggest that as in Nrf2–AR signaling56,57, single-target redox modulation events can be important physiological events that can fully recapitulate a variety of cellular redox processes.
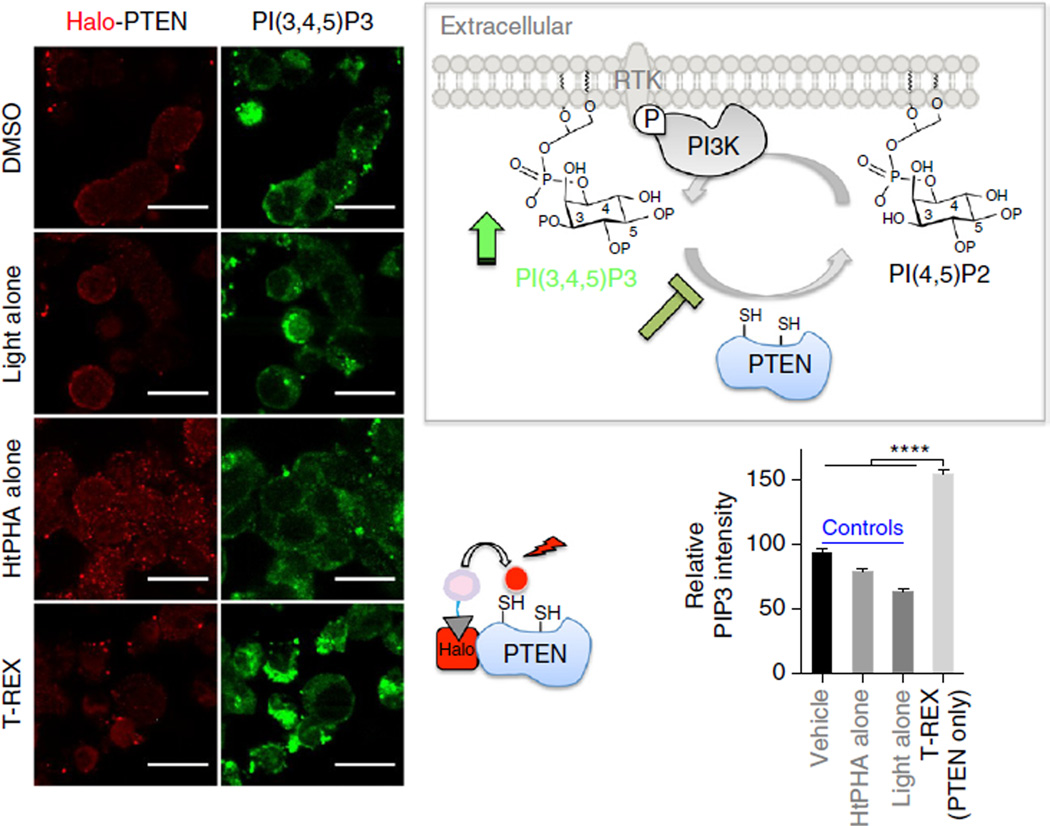
Figure 8.
Immunofluorescence analysis of endogenous PIP3 phosphoinositide in fixed cells subsequent to PTEN-targeted redox modification enabled by T-REX in live cells. Live HEK-293 cells expressing Halo-PTEN were subjected to T-REX-targeted HNEylation of PTEN55. Dominant loss-of-function inactivation of PTEN results in upregulation of PIP3. The cells were fixed and immunostained by anti-PIP3 (green) and anti-Halo (red). Error bars represent the standard error of the mean (N = 86). Scale bars, 20 µm. Inset: schematic of the PI3K/PTEN signaling. Partial inactivation of PTEN raises the levels of PIP3. ****, P < 0.0001.
Figure 9.
FRET-based biosensor assay in live cells, reporting the levels of endogenous PIP3 subsequent to PTEN-targeted redox modification enabled by T-REX. (a) Live HEK-293 cells expressing ‘lnPAkt’ FRET biosensor88 and HaloPTEN were subjected to T-REX conditions that enabled substoichiometric HNEylation of PTEN55. Dominant loss-of-function inactivation of PTEN upregulates the membrane-bound PIP3 phosphoinositide. Increase in cellular PIP3 competitively binds the pleckstrin homology (PH) domain of Akt, displacing the ‘pseudo ligand’88. Conformational change associated with the membrane recruitment results in an increase in FRET signal88. (b) Representative live-cell images and (c) quantification of the YFP:CFP emission ratio. Scale bars, 20 µm. Top: Control: HtPHA treatment alone did not perturb the emission ratio appreciably. Bottom: T-REX redox targeting of PTEN selectively enhances FRET signal (right bar) as compared with that of samples exposed to light alone (left bar). Error bars designate the standard error of the mean (N = 170 cells). a adapted with permission from Ananthanarayanan B., Ni, Q. & Zhang, J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc. Natl. Acad. Sci. USA 102, 15081–15086 (2005). Copyright 2005 National Academy of Sciences, USA.
Identification of novel sensor genes: discovering novel redox regulators sufficient for a specific response
This can be accomplished using the procedure for the representative HaloTag ORFclone library screen (Fig. 4 and Supplementary Fig. 4) and subsequent probing of transcriptional response (Fig. 2). The initial screen, target validation and downstream response studies are all built on T-REX.
Identification of novel sensor genes: precise assessment of redox sensitivity in combination with proteomics and transcriptomics-based target ID approaches
T-REX, in unison with these existing technologies, provides unparalleled opportunities for accurate characterizations of their precision response to specific LDE signals delivered to a specific sensor protein at a specific time (Fig. 4a and Supplementary Fig. 4a).
Comparison with other methods
Genetic and chemoproteomics approaches
Redox signaling is a chemical signaling paradigm that is radically different from enzyme-assisted post-translational modifications such as phosphorylation. Accordingly, classic genetic approaches are not optimal for the study of the temporal and spatial dynamics underpinning redox signaling. Targeted knockdown and/or knock-in approaches91,92 assume the presence of one protein will be necessary for the desired response to occur. Even though a specific protein may be necessary for a particular signaling event under endogenous signaling conditions, such a scenario is unlikely to be the case under typical bolus dosing because of functional redundancy among sensor proteins and the pathways that they regulate. Knockdown also often disrupts protein–protein interactions that are essential for functional intercommunication within multicomponent signaling networks. Specifically, protein expression levels fluctuate so drastically during dynamic physiological processes such as development93 that redox perturbation under steady-state conditions often elicits pleotropic effects that are challenging to interpret. The modifications are also largely nonsequence- and nonsite-specific1, and many redox-sensor proteins have multiple functionally redundant Cys residues1,49. Mutagenesis strategies are thus not always effective. Innovative quantitative proteomics platforms, on the other hand, have opened exciting doors to profiling relative Cys reactivity within the human proteome26,28–30,32,33,94–96. Chemical biology methods for site-specific analysis and global mapping of cysteine modifications onto the redoxome are also established26,28,30,32,33,94–96. Despite the powerful capability to rank reactivities and define the sites of modifications, the chemoproteomics strategies with global LDE exposure provide no ability to perturb specific targets on demand. Downstream validation is difficult because it typically involves replicating the swamping experiments in knockdown cells and measuring changes to the pleiotropic response, conditions in which temporal and target resolution are both low. T-REX is an exciting starting point in addressing these outstanding biological questions; it generates answers to a subset of important questions that strongly complement those addressed by existing whole-cell probing and profiling methods.
Limitations and considerations
HaloTagging versus endogenous protein targeting
Although the monomeric nature of HaloTag prevents unintended self-association58– 61, this 33-kDa protein adds steric bulk to the target POI. Despite the availability of functionally validated HaloTag clones from Promega, HaloTagging of a protein may affect complex protein function in an unpredictable way. This factor should be evaluated for individual target POIs under study. For example, we have confirmed that HaloTag does not perturb functional integrity of proteins thus far investigated in our laboratory. For instance, Halo-Keap1 is, as expected, a dimer57, and with both N- (ref. 57) and C-terminal (Fig. 5a) fusion of the tag the engineered protein binds Nrf2 and maintains cytosolic localization similar to that of native Keap1 (Box 2). Likewise, Halo-RRM1 has reductase activity largely similar to that of its non-HaloTagged counterpart.
To date, T-REX targeting has been demonstrated only with HaloTagging55–57. However, one would expect T-REX to function equally well on other similar fusion proteins (e.g., SNAP or CLIP tags97), provided that the appropriate ligands were available. By extension, in cases in which proteins of interest have high-specificity and/or high-affinity ligands that tolerate chemical modification, a specific T-REX photocaged precursor targeting an endogenous POI could be tailor-made. In other words, the chloroalkane recognition unit within the photocaged precursor (Fig. 10) could be replaced with a known ligand of the endogenous target POI under study. However, one must first ensure that the modified ligand can interact with the POI and determine photo-uncaging efficiency.
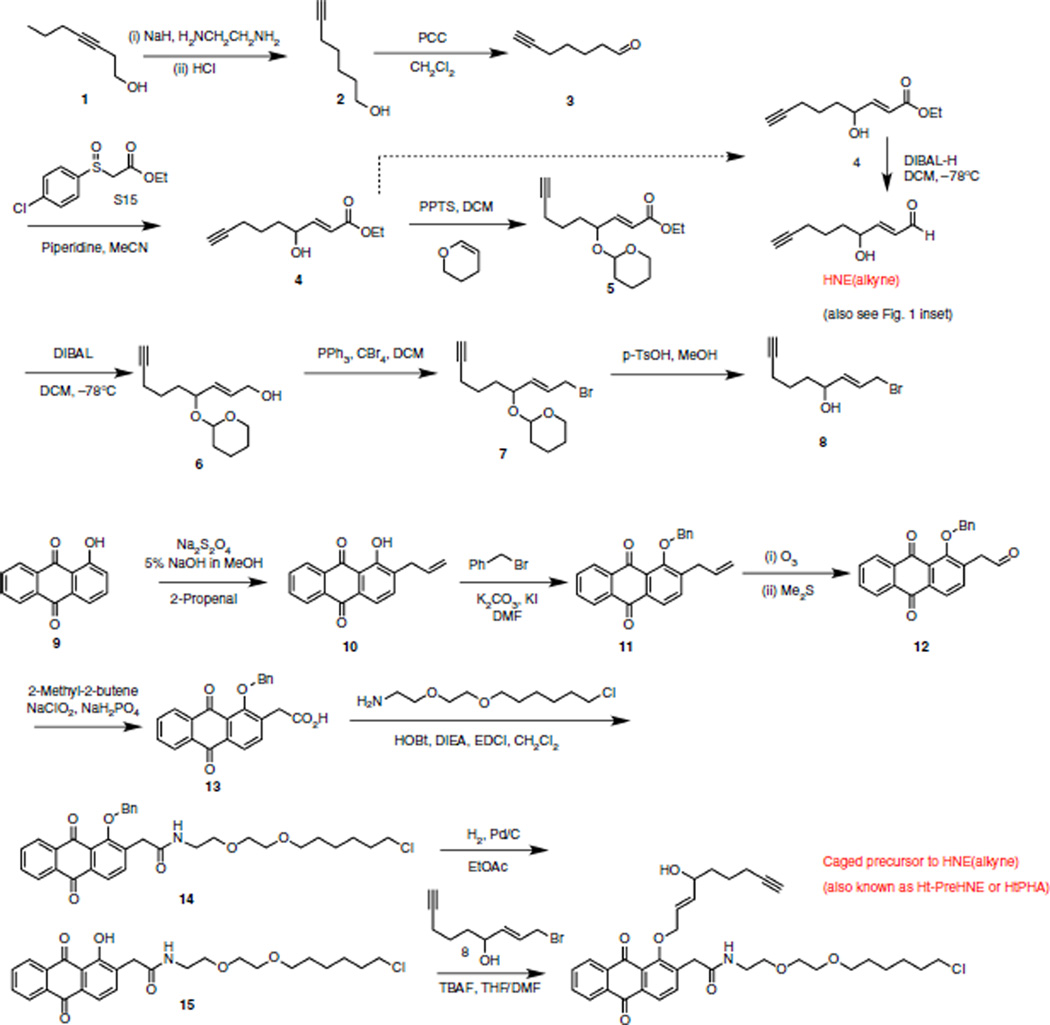
Figure 10.
Chemical syntheses of HNE(alkyne)55 (also see Fig. 1 inset), and HaloTag-targetable caged precursor to HNE(alkyne)55,56, also known as Ht-PreHNE56 or HtPHA55.
Overexpression and nonspecific response
Although the technique currently uses overexpressed proteins, overexpression does not appear to bias the outcomes in favor of delivery55–57—an outcome that was part of the initial design concept and must hold for a pseudo-intramolecular delivery mechanism. Nonetheless, we recommend independent case-by-case assessments using controls similar to the following. In the study of the Nrf2–AR pathway, we ruled out the contribution of untargeted delivery to T-REX-mediated pseudo-intramolecular delivery, using three independent lines of evidence56. The best and most general control is to simultaneously overexpress HaloTag and the target POI as two separate proteins (termed the non-fused system) and to replicate the experiments and look for loss of downstream response56,57 (Box 3). To confirm that the photocaged precursor molecule interaction with HaloTag is required for delivery, the D106A HaloTag point mutant60, which is unable to form a covalent bond with the chloroalkane unit, is recommended. We have also found empirically that overexpression levels of many proteins can be dialed down by selecting for cell lines that have integrated the plasmid post transfection. This approach also limits potential variability due to transient transfection of the POI56. One can also use less powerful or inducible promoters in mammalian cells or E. coli, which lack a specific importer (lacZY), allowing IPTG concentrations to be more accurately titrated (Tuner, Novagen). Alternatively, overexpression of Halo proteins may be executed in a null background using cells in which the endogenous variant is knocked out91.
Comparison with existing methods
Most small-molecule-based methods for identifying HNE-sensitive proteins rely to some extent on bolus dosing27,30,31. The principal alternative to T-REX is activity profiling. This has been carried out mostly in lysates26,28, but also more recently in cells29,30. Importantly, several differences between lysate-based and cell-based data have been delineated30, which underscores the need for better methods to probe reactivity in biologically relevant contexts. Powerful approaches for profiling both serine and cysteine residues—the latter being the most likely HNE-modified residue26,28,30—exist in the literature. Histidine and lysine—residues that may react with HNE79–81—are currently not able to be profiled. Thus, activity profiling can identify many potential LDE-reactive cysteines (~1,000) in a high-throughput manner26,28,30: addition of an excess of LDE that competitively binds to the profiled cysteines can be detected because those cysteines that bind the LDE are lost from the profiling pool. Profiling thus has benefits over T-REX in that it can identify many specific cysteine targets on specific proteins rapidly (T-REX can be used to identify specific cysteines, but it is more time-consuming). Furthermore, because the proteomics profiling method profiles activity, it can potentially report on cysteines or enzymes that are not HNEylated but that are functionally coupled to an off-target HNEylation event, e.g., through changes in complexation or cysteines that are on enzymes whose stability is compromised under reaction conditions. Depending on the goal of the experiment, such an outcome may or may not be desired. However, ultimately, a hit on profiling does not necessarily mean that a bona fide HNEylation event has occurred, and thus multiple downstream validations are required after the initial hits have been identified. Furthermore, profiling is limited by the number of cysteines that can be identified by the activity probe used, which, although high, is not exhaustive. Many well-known reactive enzymes, such as Keap1, are rarely observed in reactivity profiling.
By contrast, the whole ORFeome of mouse and human is available as Halo-tagged clones (Kazusa collection, Promega), which makes high-throughput screening with T-REX possible. Although initial screening is more laborious than profiling, T-REX is streamlined to allow downstream pathway interrogation using reporter assays such as dual-luciferase- and GFP-based transcriptional reporters (shown in Steps 40C and 40D of the PROCEDURE). We believe that a combination of profiling (to generate potential hits) and T-REX (for validation and downstream signal interrogation) is the most powerful approach.
Alternatives to profiling include direct identification of HNEylated proteins by MS35,37,43,44 pulldown assays using radiolabeled HNE98,99 and in vitro HNEylation43,80. These methods are all relatively low throughput and do not lend themselves to downstream signaling pathway interrogation. However, similar to T-REX, they do identify a specific modification of a specific protein, but under uncontrolled swamping conditions.
Further modifications of the LDE signal
LDE signals themselves can be modified by reduction3,98, oxidation3, alkylation27,98 and other secondary processes. This is an intrinsic property of LDEs, and for this reason in all methods using LDEs one cannot assume that the active species is the specific LDE added. Although little work has been carried out to compare how faithfully each method reports on modification by the intended electrophile as opposed to a metabolite thereof, because of the low dose of LDE generated and the ‘faster than diffusion’ kinetics required for T-REX to occur, it seems likely that T-REX will be relatively less susceptible to chemical modification of the LDE than approaches based on bolus dosing (where the electrophile is in excess).
Relevance to ‘real-life’ situations
T-REX is a tool used to identify and interrogate a single (or potentially a small number of) specific protein modification(s) at a time through a native reactive chemical signal. It is best used to model redox signaling, in which modest perturbations to a pre-existing cellular reactive lipid electrophile pool elicit a beneficial3,38,54, typically cytoprotective response, such as AR3,38,54,71,77. ‘Signaling’ can occur because changes in an LDE upregulation can be compartmentalized, or ‘directed’ to a specific target and because second-order rates of association with an LDE vary widely for different enzymes26,27,34. Furthermore, because LDE levels (and hence targeted labeling by T-REX) are low, signaling events probably occur through gain of function or dominant loss of function. (If LDE modification causes inhibition of enzymatic activity, unless this is a dominant phenotype (as is the case for PTEN82,86–88), the modification may not be phenotypic during redox signaling because of the low concentrations of HNE, leading to low target protein occupancy.) Because T-REX can label only a small percentage of the total target protein present, the requirements for observing a response are similar to those for lipid signaling. Furthermore, T-REX ‘directs’ HNE to a target enzyme in a manner similar to endogenous signaling, thereby mimicking ‘redox signaling’ reasonably well. Individual pathological effects of overproduction of LDEs can also be interrogated, in principle, using T-REX, for example, to interrogate the extent to which HNEylation of a specific protein may elicit apoptosis. However, as pathological effects stem from hyperproduction of LDEs, in which complete loss of function could occur, ancillary factors or a high percentage of modifications of the target may be required to recapitulate these scenarios, which would render T-REX less useful.
LDE chain length
HaloTag is unreactive to the reactive electrophiles, and it thus generally serves as a good point source of reactive signals56. However, we have found that the hydrophobic surface of Halo can interfere with efficient release of long-chain (> ~15 carbons) fatty-acid-derived LDEs—for instance, 2-HD56 (Fig. 2, inset). This was presumed to occur because 2-HD binds nonspecifically to Halo, allowing noncovalent association to occur after photo-uncaging. Consistent with this assertion, in vitro 2-HD release assays in the presence of 1% (wt/vol) SDS led to efficient liberation, whereas no liberation was observed without SDS56. In principle, the problem may be solved by the use of alternative tags, such as CLIP and SNAP tags, in place of HaloTag, along with modification of the chloroalkane unit of the photocaged precursors to the benzyl-cytosine or benzyl-guanine motif—the covalent recognition units for CLIP- and SNAP tags, respectively97.
Number of reactive motifs on LDEs
If the LDE signal houses more than one reactive group, as in the case with 4-oxononenal (ONE, Fig. 2 inset), the specificity will be lost because a reactive enone moiety is exposed within the photocaged precursor itself before photo-uncaging56. Dual photocaging of the ketone, as well as of the aldehyde of ONE, is a viable solution. For instance, protecting the aldehyde function with the anthraquinone and the ketone with an o-nitrobenzyl-derived acetal would enable simultaneous uncaging of both ketone and aldehyde. Alternatively, one could use a semistable protecting group for the ketone motif—such as an acetal—with a half-life longer than the 2-h incubation time. In this way, after 2-h incubation of cells with the photocaged precursor, light exposure would liberate the aldehyde, and unmasking of the ketone motif would happen on a similar time scale, only after photo-uncaging of the aldehyde.
Cell type and viability
Although the light source is of low energy (Fig. 2, Supplementary Fig. 1, and Supplementary Videos 1 and 2) and does not affect cellular viability within the time scale and types of cells thus far used (assessed by AlamarBlue and Trypan blue assays56 in HEK-293, COS-1 and E. coli), it is important to independently validate potential effects on cell viability.
MATERIALS
REAGENTS
Reagents for chemical synthesis
Ethylenediamine (Sigma-Aldrich, cat. no. E26266)
Sodium hydride (Fisher Chemical, cat. no. S318 10)
3-Heptyn-1-ol (Sigma-Aldrich, cat. no. 630845)
1 N HCl (Sigma-Aldrich, cat. no. 38283 Fluka)
Magnesium sulfate, anhydrous (JT Baker, cat. no. J41620)
Hexanes (Sigma-Aldrich, cat. no. 227064)
Ethyl acetate (EtOAc; Sigma-Aldrich, cat. no. 270989)
Diethyl ether (Et2O; Sigma-Aldrich, cat. no. 673811)
Dichloromethane (DCM, CH2Cl2; Sigma-Aldrich, cat. no. 270997)
Pyridinium chlorochromate (PCC; Sigma-Aldrich, cat. no. 190144)
1-Hydroxyanthraquinone (TCI-America, cat. no. H0354)
2-Propenal (Sigma-Aldrich, cat. no. 01680)
2-(2-(6-chlorohexyloxy)ethoxy)ethanamine (Promega, cat. no. P6711)
Sodium thiosulfate (Na2S2O4; Sigma-Aldrich, cat no. 72049)
Calcium chloride (CaCl2; Sigma-Aldrich, cat no. C1016)
Sulfuric acid (H2SO4; Sigma-Aldrich, cat. no. 339741)
Celite (Sigma-Aldrich, cat. no. 22140)
Piperidine (Sigma-Aldrich, cat. no. 411027)
Methyl-2-sulfinyl-acetate (Sigma-Aldrich, cat. no. 237582)
Acetonitrile (ACN; Sigma-Aldrich, cat. no. 271004)
Dihydropyran (DHP; Sigma-Aldrich, cat. no. D106208)
Pyridinium p-toluenesulfonate (PPTS; Alfa-Aesar, cat. no. A15708)
Sodium bicarbonate (NaHCO3; Sigma-Aldrich, cat. no. S6014)
Sodium sulfate (Sigma-Aldrich, cat. no. S9627)
Toluene (Sigma-Aldrich, cat. no. 244511)
Diisobutylaluminum hydride (1.0 M solution in hexanes; DIBAL-H; Sigma-Aldrich, cat. no. 190306)
Tetrabromomethane (CBr4; Sigma-Aldrich, cat. no. C11081)
Triphenyl phosphine (PPh3; Sigma-Aldrich, cat. no. 93092)
p-Toluenesulfonic acid (p-TsOH) monohydrate (Sigma-Aldrich, cat. no. 402885)
Methanol (MeOH; Sigma-Aldrich, cat. no. 322415)
Benzyl bromide (Sigma-Aldrich, cat. no. B17905)
Potassium carbonate (K2CO3; Sigma-Aldrich, cat. no. P5833)
Potassium iodide (KI; Sigma-Aldrich, cat. no. P2963)
Acetone (Sigma-Aldrich, cat. no. 34850)
Dimethyl sulfide (Me2S; Sigma-Aldrich, cat. no. 274380)
2-Methyl-2-butene (Sigma-Aldrich, cat. no. 86262)
tert-Butanol (t-BuOH; Sigma-Aldrich, cat. no. 471712)
Sodium phosphate monohydrate (NaH2PO4 · H2O; Sigma-Aldrich, cat. no. S9638)
Sodium chlorite (NaClO2; Sigma-Aldrich, cat. no. 71388)
Hydroxybenzotriazole (HOBt; Sigma-Aldrich, cat. no. 157260)
N,N-Diisopropylethyleneamine (DIEA; Sigma-Aldrich, cat. no. D125806)
1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDCI; Sigma-Aldrich, cat. no. E6383)
Palladium on carbon (Pd/C; Sigma-Aldrich, cat. no. 205699)
Tetra-n-butylammonium fluoride (TBAF; Sigma-Aldrich, cat. no. 241512)
Tetrahydrofuran (THF; Sigma-Aldrich, cat. no. 401757)
Dimethylformamide (DMF; Sigma-Aldrich, cat. no. 227056)
Potassium permanganate (KMnO4; Sigma-Aldrich, cat. no. 223468)
Silica gel (Silicycle; cat. no. SiliaFlash P60)
Dry ice
Trifluoroacetic acid (TFA; JT Baker, cat. no. 9470-01)
4× Laemmli sample buffer (BioRad, cat. no. 1610747)
Reagents for T-REX experiments in E. coli
pet28a-Halo-Keap1 plasmid (available from the author upon request)
E. coli BL21 Codon plus (DE3) RIL competent cells (Agilent, cat. no. 230245)
Tryptone (IBI Scientific, cat. no. IB49182)
Yeast extract (Fisher, cat. no. BP1422-2)
Agar (Fisher, cat. no. EC232-658-1)
Chloramphenicol (Goldbio, cat. no. C-105-5)
Kanamycin (Goldbio, cat. no. K-120-5)
LB-KAN agar plates (kanamycin 50 µg/ml)
IPTG (Gold Biotechnology, cat. no. I2481C)
TCEP-HCl (Goldbio, cat. no. TCEP1)
Sulfo-Cy5 azide (Lumiprobe, cat. no. B3330)
Copper Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (Cu-TBTA; Lumiprobe, cat. no. 21050)
HaloTag-targetable precursor to HNE(alkyne) (also known as Ht-PreHNE or HtPHA) (Fig. 10) (Steps 1–37 and Reagent Setup)
HNE(alkyne) (Fig. 10) (Steps 1–11A and Reagent Setup)
HEPES (Fisher, cat. no. BP310-1)
OmniPur lysozyme from egg white (EMD Millipore, cat. no. 5950)
Dnase-I from bovine pancrease (EMD Millipore, cat. no. 260913)
DMSO (Fisher, cat. no. D128-500)
t-Butanol (Fisher, cat. no. A401-1)
SDS (Teknova, cat. no. S9974)
Copper sulfate pentahydrate (Sigma, cat. no. 209198-100G)
β-Mercaptoethanol (BME; Sigma-Aldrich, cat. no. M6250)
TEV protease (see Box 4)
Standard reagents for protein gel electrophoresis
Box 4. His6-TEV-S219V expression and purification ● TIMING 4–5 d.
Expression of His6-TEV-S219V in E. coli
Transform chemically competent BL21 (DE3) RIL cells with His6-TEV-S219V (Addgene, Plasmid no. 8827), and plate it on an LB–ampicillin–agar (100 µg/ml) plate. Incubate the plate overnight at 37 °C.
Pick a single colony from the plate and inoculate it with 5 ml of LB–ampicillin–chloramphenicol medium. Shake the flask at 200 r.p.m. at 37 °C overnight.
Dilute the 5-ml overnight culture in 1 liter of LB–kanamycin medium in a 2-liter flask. Shake the culture flask at 200 r.p.m. at 37 °C until the OD value reaches 0.6–0.8.
-
Induce the culture with IPTG to a 1 mM final concentration. Shake the culture at 37 °C for 6 h at 200 r.p.m.
▲ CRITICAL STEP For maximal protein yield, it is important to induce expression when the OD is 0.6–0.8
-
Harvest the cells by centrifugation at 7,000g for 10 min at 4 °C. Discard the supernatant by decanting. Keep the cell pellet on ice.
■ PAUSE POINT The cell pellets can be flash-frozen and stored at −80 °C for at least 1 month.
Preparation of buffers
All buffers can be prepared beforehand, filtered using a 0.22-µm filter and stored without the addition of reducing agents at 4 °C for up to 2 weeks.
-
Preparation of His6-TEV-S219V lysis buffer: prepare His6-TEV-S219V lysis buffer by mixing 50 mM Na2HPO4, pH 8.0, 100 mM NaCl, 10 mM imidazole, 5% glycerol and 5 mM BME.
▲ CRITICAL STEP Freshly add BME just before use.
-
Preparation of His6-TEV-S219V wash buffer: prepare His6-TEV-S219V wash buffer by mixing 50 mM Na2HPO4, pH 8.0, 200 mM NaCl, 25 mM imidazole, 5% glycerol and 5 mM BME.
▲ CRITICAL STEP Freshly add BME right before use.
-
Preparation of His6-TEV-S219V elution buffer: prepare His6-TEV-S219V elution buffer by mixing 50 mM Na2HPO4, pH 8.0, 200 mM NaCl, 125 mM imidazole, 5% glycerol and 5 mM BME.
▲ CRITICAL STEP Freshly add BME right before use.
-
Preparation of His6-TEV-S219V storage buffer: prepare His6-TEV-S219V storage buffer by mixing 50 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 10% glycerol and 3 mM TCEP.
▲ CRITICAL STEP Freshly add TCEP before use. Readjust the pH to 7.5 after adding TCEP.
Purification of His6-TEV-S219V using a TALON cobalt-affinity column
▲ CRITICAL STEP Maintain the cell pellet, cell lysate and all buffers on ice at all times to minimize loss of protein activity.
-
Pipette out 14 ml of TALON resin (7-ml bed volume (BV)) in an Econo column. Let the buffer flow through. Wash the resin with 10 BV of water followed by 5 BV of lysis buffer.
▲ CRITICAL STEP Do not allow the resin to dry.
Pre-equilibrate a GE Healthcare Hiload 26/60 Superdex 200 prep grade column (ID no. 0823027) with 320 ml of storage buffer.
-
If the pellet is frozen, thaw it on ice. Resuspend the cells in 5 ml of His6-TEV-S219V lysis buffer per gram of cell pellet.
▲ CRITICAL STEP Cell pellets need to be resuspended vigorously. Pipette up and down until no clumps are visible. Clumps of cells can clog the cell disruptor.
-
Lyse cells by passing the cell suspension twice through the cell disruptor at 13,000 p.s.i.
▲ CRITICAL STEP Maintain the lysate on ice at all times.
Centrifuge the lysate at 30,000g for 30 min at 4 °C. Collect the supernatant in clean glassware.
-
To remove DNA, add streptomycin sulfate solution to a final concentration of 2%.
▲ CRITICAL STEP Prepare streptomycin sulfate solution by dissolving the solid in 8–10 ml of chilled ddH2O. Add streptomycin sulfate solution drop by drop while stirring the supernatant gently at 4 °C. Vigorous stirring can lead to loss of protein activity. You should observe a viscous yellow DNA precipitate upon adding streptomycin sulfate. Discontinue adding streptomycin sulfate white protein precipitate is observed.
Centrifuge the supernatant at 30,000g for 30 min at 4 °C. Collect the supernatant in clean glassware.
-
Resuspend the pre-equilibrated TALON resin with the supernatant and transfer it to the clean glassware. Incubate the supernatant with the resin for 50 min–1 h at 4 °C while gently stirring.
▲ CRITICAL STEP A long incubation time can lead to decay of protein activity. A very short incubation can result in inefficient binding and therefore reduced protein yield.
Add the cell lysate–resin mixture back to the Econo column and let the lysate flow through. Avoid letting the column run dry.
Wash the resin three times, each with 2 BV of His6-TEV-S219V wash buffer.
-
Elute His6-TEV-S219V from the column using His6-TEV-S219V elution buffer. Collect 1-ml fractions and check for the presence of protein by measuring A280 values. Pool the protein-containing fractions. Concentrate the protein in a 10-kDa-cutoff concentrator to bring the total volume to 10–12 ml.
▲ CRITICAL STEP A concentration that is too high can result in protein precipitation. If the starting volume is too large, protein can be loaded in two batches on the Superdex 200 column.
Load the protein on a pre-equilibrated column for buffer exchange at a flow rate of 1 ml/min.
Collect fractions corresponding to the protein peaks. Pool the fractions. Concentrate to a final concentration of 4–5 mg/ml. Measure protein concentration using standard protein concentration measurement assays.
Confirm the purity of His6-TEV-S219V using protein gel electrophoretic analysis. The molecular weight of His6-TEV-S219V is 26 kDa.
-
Divide His6-TEV-S219V into aliquots, flash-freeze them and store them at −80 °C.
■ PAUSE POINT His6-TEV-S219V aliquots are stable at −80 °C for at least a year.
Additional reagents for recombinant protein expression in E. coli (optional)
Streptomycin B sulfate (Goldbio, cat. no. S-150-100)
TALON metal affinity resin (Clontech, cat. no. 635502)
Reagents for T-REX experiments in cultured mammalian cells
pMIR-DsRed-IRES-His6-Halo-Keap1 plasmid, available from Addgene (ID no. 58240)
Halo-ORF clone library in pFN21a vector, available from Promega
pcDNA3.1 myc3 Nrf2 plasmid, available from Addgene (ID no. 21555)
pcDNA3 eGFP-Nrf2, available from Addgene (ID no. 21549)
pMIR-Halo-PTEN plasmid, available from Addgene (ID no. 58241)
pcDNA3 InPAkt plasmid (materials transfer from J. Zhang, UCSD)
HEK-293 cells (ATCC, cat. no. CRL-1573) ! CAUTION The cell lines used in your research should be regularly checked to ensure that they are authentic and that they are not infected with mycoplasma. ▲CRITICAL We expect that the procedure will work with other cell types as well.
TransIT-2020 (Mirus, cat. no MIR5400)
Polyethyleneimine (PEI; Polyscience, cat. no. 23966-2; see Reagent Setup)
Standard medium for cell culture
HaloTag TMR ligand (Promega, cat. no. G8251)
Luciferase plasmid (Promega, cat. no. E3641)
pCMV-Renilla luciferase plasmid (Promega, cat. no. E6931)
ARE-GFP plasmid (Promega, CCS-0020G)
HaloTag targetable photocaged precursor to HNE(alkyne) (also known as Ht-PreHNE or HtPHA; Fig. 10; Steps 1–37 and Reagent Setup)
HNE(alkyne) (Fig. 10; Steps 1–11A and Reagent Setup)
Quickstart Bradford 1× dye (Bio-Rad, cat. no. 5000205)
BSA as standard 2 mg/ml (Pierce, cat. no. 23209)
TCEP-HCl (Goldbio, cat. no. TCEP1)
Sulfo-Cy5 azide (Lumiprobe, cat. no. B3330)
Copper Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (Cu-TBTA; Lumiprobe, cat. no. 21050)
DMSO (Fisher, cat. no. D128-500)
t-Butanol (Fisher, cat. no. A401-1)
SDS (Teknova, cat. no. S9974)
Copper sulfate pentahydrate (Sigma, cat. no. 209198-100G)
TEV protease (see Box 4)
Ammonium bicarbonate (Sigma-Aldrich, cat. no. 09830)
Formic acid (Sigma-Aldrich, cat. no. F0507)
Iodoacetamide (Sigma-Aldrich, cat. no. I1149)
Sequencing-grade modified trypsin (Promega, cat. no. V5113)
Standard reagents for protein gel electrophoresis
Standard reagents for western blotting
Milk for PVDF membrane blocking (Walmart, Great Value Nonfat Instant Dry Milk)
Mouse monoclonal anti-Keap1 primary antibody, 1:5,000 (Abcam, cat. no. Ab119403)
Rabbit polyclonal anti-HaloTag primary antibody, 1:2,000 (Promega, cat. no. 9281)
Rabbit polyclonal anti-RRM1 primary antibody, 1:2,000 (Abcam, cat. no. Ab81085)
Mouse monoclonal anti-phosphatidylinositol 3,4,5-triphosphate (PIP3, 1:500; Echelon Biosciences, cat. no. Z-P345)
Goat anti-rabbit IgG Alexa Fluor 647 preadsorbed, 1:1,000 (Abcam, cat. no. Ab150083)
Goat anti-mouse Ig, Human ads-FITC, 1:1,000 (Southern Biotech, cat. no. 1010-02)
4,6-Diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich, cat. no. D9542)
Rabbit polyclonal anti-GFP primary antibody, 1:1,000 (Santa Cruz, cat. no. sc-8334)
Mouse monoclonal anti-actin, 1:30,00 (Sigma-Aldrich, cat. no. A47000)
Secondary antibody to mouse, 1:5,00 (Abcam, cat. no. Ab67890)
Secondary antibody to rabbit, 1:8,000 (Abcam, cat. no. Ab97051)
Reagents for making Firefly and Renilla luciferase substrates (optional)
HEPES (Fisher, cat no. BP310-1)
Magnesium sulfate (JT Baker, cat. no. J41620)
DTT (Goldbio, DTT100)
EDTA (Fisher Chemical, cat. no. BP120-1)
ATP disodium salt hydrate (Fisher Chemical, cat. no. AC102800500)
Coenzyme A (Avanti, cat. no. 870700P)
d-Luciferin Firefly (Goldbio, cat. no. L-123-250)
Tris base (Fisher Chemical, cat. no. BP152 10)
Trans-1,2-Diaminocyclohexane-N,N,N′,N′-tetraacetic acid monohydrate (CDTA; Alfa Aesar, cat. no. B22928-14)
BSA (Fisher Chemical, cat. no. BP9703-100)
Goat Serum (Sigma-Aldrich, cat. no. G9023)
Triton X-100 (Fisher, cat. no. BP-151-100)
Glycerol (Fisher Chemical, cat. no. BP229 4)
Sodium acetate anhydrous (USB, cat. no. 21608)
Sodium sulfate (Sigma-Aldrich, cat. no. S9627)
Sodium pyrophosphate (Fisher Chemical, S390-500)
2-(4-aminophenyl)-6-methylbenzothiazole (APMBT; Enamine, cat. no. EN300-17374)
Methanol anhydrous (Sigma-Aldrich, cat. no. 322415)
Coelenterazine (Goldbio, cat. no. CZ2.5)
EQUIPMENT
Equipment for chemical synthesis
Fume hood
Weighing balance
Weighing paper
Spatulas
Round-bottom flasks
Separatory funnel
Graduated cylinders
Pasteur pipettes
Tweezers
Magnetic stir plate
Syringes and needles
Magnetic stir bars
Glass-backed thin-layer silica chromatography plates
365-nm UV lamp (Spectroline E-Series)
Column for flash chromatography
Vacuum pump
Rotary evaporator
Ozonator
Nitrogen and Argon gas
Schlenk line
Equipment for T-REX experiments in E. coli
Standard equipment for E. coli cell culture
Standard equipment for protein gel electrophoresis
Cell density meter for OD measurement
Centrifuge capable of spinning culture tubes
Microcentrifuge tubes
Temperature-adjustable shaker incubator
Handheld UV lamp with 365-nm light (Spectroline ENF 240C)
ChemiDoc-MP imaging system (Bio-Rad)
Econo column (Bio-Rad, cat. no. 7374251)
Amicon Ultra-15, MWCO 10 kDa (Millipore, cat. no. UFC901024)
GE Healthcare Hiload 26/60 Superdex 200 prep grade column (ID no. 0823027)
Äkta FPLC system (GE Healthcare)
Equipment for T-REX in cultured mammalian cells
Standard equipment for mammalian cell culture
Hand-held UV lamp with 365-nm light (Spectroline ENF 240C)
Sterile 48-well cell culture plates
White, opaque, flat-bottom 96-well plate for luminescence measurement (Corning)
Glass-bottom dishes for imaging (In Vitro Scientific, 1.5N, D35-20-1).
Confocal microscope with appropriate filters
Plate reader for measuring luminescence (Biotek Cytation3)
Flow cytometer with appropriate lasers and filters (BD LSRII)
Biosafety level 2 hood
ChemiDoc-MP imaging system (Bio-Rad)
Capillary liquid chromatography (CapLC) system (Waters Co.)
QSTAR XL (ABSciex)
Everest C18 (5 µm, 500 µm i.d. × 15 mm (Grace)
Jupiter C18 (3 µm, 100 µm i.d. × 150 mm (Phenomenex)
Analyst QS 1.1 software (ABSciex)
REAGENT SETUP
Luria broth medium
To make Luria broth (LB) medium, add 10 g of NaCl, 10 g of tryptone and 5 g of yeast extract per liter. Autoclave to sterilize. Store at room temperature (15–22 °C) up to 1 week.
LB agar
To make LB agar, add 10 g of NaCl, 10 g of tryptone, 5 g of yeast extract and 15 g of agar per liter. Autoclave to sterilize. Add desired antibiotic and pour into a Petri dish once the LB agar is cool to touch. Store the Petri dish at 4 °C for up to 1 week.
Lysis buffer A
Lysis buffer A is 50 mM HEPES, pH 7.6, 5 mM imidazole and 5 mM BME. Store all buffers at 4 °C for up to 1 week. Add reducing agent right before use.
Wash buffer A
Wash buffer A is 50 mM HEPES, pH 7.6, 100 mM NaCl, 10 mM imidazole and 5 mM BME.
Wash buffer B
Wash buffer B is 50 mM HEPES, 100 mM NaCl, 20 mM imidazole and 5 mM BME, pH 7.6.
Elution buffer
The elution buffer is 50 mM HEPES, pH 7.6, 100 mM NaCl, 200 mM imidazole and 5 mM BME.
LB–ampicillin–chloramphenicol medium
LB–ampicillin–chloramphenicol medium is 100 µg/ml of ampicillin and 30 µg/ml chloramphenicol in the desired volume of LB medium.
LB–kanamycin medium
LB–kanamycin medium contains 50 µg/ml of kanamycin in the desired volume of LB medium.
HaloTag-targetable precursor to HNE(alkyne) (also known as Ht-PreHNE or HtPHA)
Make a stock of 150–200 mM HtPHA in DMSO. Determine the concentration using a UV-visible spectrophotometer (at 25 °C, ε366 = 3,950 M−1 cm−1). One-shot aliquots can be stored protected from light at −80 °C for >6 months.
HNE(alkyne)
Make a stock of 150–200 mM HNE(alkyne) in DMSO. Determine the concentration using a UV-visible spectrophotometer (at 25 °C, ε225 = 16,00 M−1 cm−). One-shot aliquots can be stored at −80 °C for >6 months.
CuSO4 solution
Make a 100 mM CuSO4 · 5H2O solution in ddH2O. The solution can be stored for >1 year at 4 °C.
TCEP solution
Make a 100 mM TCEP-HCl solution in 50 mM HEPES (pH 7.6). Aliquots can be stored at −20 °C for up to 6 months. Avoid multiple freeze–thaw cycles.
Cy5 azide
Make a 0.5 mM Cy5 azide solution in DMSO. Divide the solution into aliquots and store them at −20 °C for >6 months protected from light as one-shot aliquots.
20% (wt/vol) SDS
Make an aqueous stock containing 10 g of SDS in 50 ml of ddH2O. Vortex the mixture to dissolve the contents.
Kanamycin
Make an aqueous stock containing kanamycin at a concentration of 50 mg/ml in autoclaved ddH2O (1,000×) and filter-sterilize (0.22-µm filter) the solution. Aliquots can be stored at −20 °C for >6 months.
Chloramphenicol
Dissolve chloramphenicol at a concentration of 30 mg/ml in 200-proof ethanol (1,000×). Aliquots can be stored at −20 °C for >6 months.
IPTG
Make a 1 M solution of IPTG (1,000×) in autoclaved ddH2O, and filter-sterilize (0.22-µm filter) the solution before use.
Polyethyleneimine (25,000 MW linear chain)
Make an aqueous stock containing polyethyleneimine (PEI) to a concentration of 1 mg/ml in autoclaved ddH2O by heating it at 80 °C. Let the solution cool to room temperature. Adjust the pH of the solution to 7.0. Filter-sterilize (0.22-µm filter) the solution. Aliquots can be stored at −20 °C for >6 months. Avoid multiple freeze–thaw cycles.
Blocking buffer
Add 20 µl of Triton X-100 to 9.5 ml of 50 mM HEPES, pH 7.6. Vortex the mixture to mix the contents. Add 0.5 ml of goat serum and mix it well.
Incubation buffer
Add 20 µl of Triton X-100 to 100 ml of 50 mM HEPES, pH 7.6. Vortex the mixture to mix the contents. Add 1 ml of goat serum and mix the contents well.
DAPI
Make a 5 mg/ml stock DAPI solution in DMSO.
5× Passive lysis buffer
Make an aqueous stock containing 125 mM Tris, pH 7.8, 10 mM 1,2-CDTA, 10 mM DTT, 5 mg/ml BSA, 5% (vol/vol) Triton X-100 and 50% (vol/vol) glycerol in ddH2O. 5× passive lysis buffer (PLB) can be stored at −20 °C for at least 2 months100.
1× Firefly luciferase substrate
Make an aqueous stock containing 75 mM HEPES, pH 8.0, 4 mM MgSO4, 20 mM DTT, 0.1 mM EDTA, 0.53 mM ATP, 0.27 mM coenzyme A and 0.47 mM d-luciferin firefly in ddH2O. Divide the substrate into aliquots in amber tubes. The substrate can be stored at −80 °C for at least 2 months100.
1× Renilla luciferase buffer
Make an aqueous stock containing 7.5 mM sodium acetate, pH 5.0, 400 mM sodium sulfate, 10 mM CDTA, 15 mM sodium pyrophosphate and 0.025 mM APMBT in ddH2O. Divide the buffer into aliquots in microcentrifuge tubes. The buffer can be stored at −20 °C for at least 3 months101.
100× Renilla luciferase substrate
Dilute coelenterazine to ~ 0.5 mg/ml in anhydrous methanol immediately upon receipt. Determine the concentration of the substrate using a UV-visible spectrophotometer. Blank the spectrophotometer with dry methanol. Measure A345. At 25 °C, ε345 = 9,800 M−1 cm−1 in methanol. Calculate the concentration. Further dilute the stock in dry methanol to make a final concentration of 0.55 mM coelenterazine. Aliquots of stock Renilla luciferase substrate can be stored at −80 °C for at least 3 months101. Note: Premade substrates for luciferase assay are also available commercially (Dual-Luciferase Reporter Assay System; Promega, cat. no. E1910).
▲ CRITICAL Moisture can lead to decay of coelenterazine; avoid exposure to wet methanol, for example.
EQUIPMENT SETUP
ChemiDoc-MP Imaging system setup for Cy5 florescent gel imaging
Set the Cy5 excitation source as red epi illumination and emission filter as 695/55 filter.
Automated plate reader protocol for luciferase reporter assays for AR pathway activation
Set up the plate reader with the following commands: inject 50 µl of firefly substrate; shake the plate for 2 s; pause for 2 s; and read luminescence for 10 s. Subsequently, run the protocol. Repeat the protocol, except inject 50 µl of 1× Renilla substrate into 50 µl of Renilla luciferase buffer. The gain for the detector will need to be optimized based on the signal intensity.
Flow cytometer settings for GFP reporter assays for AR pathway activation
Run calibration and cleaning based on the manufacturer’s instructions. On the BD LSR-II, GFP is detected in the 488-1 channel with a 488-nm excitation laser, 525/50 filter and 505LP mirror for all of the experiments.
LC–MS/MS setup for the identification of LDE-modified sites on the POI
For the LC step, separate the peptides on a CapLC system coupled to a QSTAR XL. Desalt onto an Everest C18 (5 µm, 500 µm i.d. × 15 mm) with solvent A (97:3 H2O:ACN with 0.1% (vol/vol) formic acid and 0.01% (vol/vol) TFA) at 40 µl/min. After a 6-min wash, separate the peptides on a Jupiter C18 (3 µm, 100 µm i.d. × 150 mm) using a 40-min linear gradient of 10% to 40% solvent B (85% ACN/10% isopropanol + 0.1% (vol/vol) formic acid + 0.0075% (vol/vol) TFA) at 250 nl/min. In our laboratory, MS/MS data acquisition was performed using Analyst QS 1.1 software in positive ion mode for information-dependent acquisition analysis. To do this, set the nanospray voltage to 2.1 kV for all experiments in a positive ion mode. Use nitrogen as the curtain (value of 20) with heated interface at 130 °C. Set the declustering potential at 80 eV and Gas1 as 5 (arbitrary units). In information-dependent acquisition analysis, full-scan MS data are acquired after each survey scan from m/z 350 to 1,300. The three highest-intensity ions above the predefined threshold of 28 counts per s with multiple charge states (+2 and +3) are selected for tandem MS (MS/MS), with rolling collision energy applied for detected ions based on different charge states and m/z values. Each MS/MS acquisition is completed and switched back to survey scan when the precursor intensity falls below a predefined threshold or after a maximum of 65 s acquisition.
PROCEDURE
▲ CRITICAL All chemical reactions are conducted in oven-dried glassware under an atmosphere of nitrogen unless otherwise stated. Concentration involves removal of solvents by means of a rotary evaporator (equipped with a 37–40 °C water bath) attached to a diaphragm pump (15–60 Torr), followed by removal of residual solvents at <1 Torr with a vacuum pump. Flash chromatography is performed on silica gel 60 (230–400 mesh): a typical purification procedure for 5 g of crude product uses 50 g of silica gel in a 4 × 30 cm (diameter × length) column, and 10-ml fractions are collected.
Synthesis of HNE (alkyne) ● TIMING 4 d
-
1|
Add 2.76 g of NaH (69 mmol) to 26.8 ml of ethylenediamine (40 mmol) at 0 °C under an atmosphere of nitrogen. Stir the mixture for 1 h at room temperature and then for another hour at 60 °C (Fig. 2 inset and Fig. 10).
! CAUTION NaH releases a flammable gas (hydrogen) on contact with water and other protic solvents. Keep it away from naked flames and use it in a fume hood.
-
2|
Cool the mixture to 45 °C and add 2 ml of 1 (16.4 mmol). Heat the reaction back to 60 °C and stir it for 1 h.
-
3|
Slowly add 20 ml of 1 N HCl at 0 °C, extract it with ether (50 ml ×3) and dry it with magnesium sulfate (200 mg or more until the newly added powder no longer clumps upon swirling).
-
4|
Purify the residue after concentration in vacuo by using flash chromatography with hexanes:EtOAc (2:1 vol/vol) as the eluent to obtain alcohol 2. (Rf = 0.5 hexanes:EtOAC 2:1).
? TROUBLESHOOTING
■ PAUSE POINT Alcohol 2 can be stored in a sealed glass vial in a −20 °C freezer for a minimum of 6 months.
-
5|
Dissolve 1.44 g of the product 2 (28 mmol) in 40 ml of CH2Cl2. Add 5.53 g of PCC (56 mmol) and stir the mixture at room temperature for 1 h.
-
6|
Filter the reaction mix through Celite (~10 g).
-
7|
Concentrate the filtrate and isolate the aldehyde 3 after separation via flash chromatography using hexanes:EtOAc (2:1 vol/vol) as the eluent (Rf = 0.45 hexanes:EtOAc 5:1).
? TROUBLESHOOTING
■ PAUSE POINT Aldehyde 3 is best used immediately. If a pause is needed, 3 can be stored strictly under air-free conditions with minimal exposure to moisture, at −80 °C for a few days.
-
8|
Add 1.16 g of aldehyde 3 (10.5 mmol) and 1.73 ml of piperidine (17.6 mmol) to a solution of 1.74 g of methyl 2-phenylsulfinylacetate (8.78 mmol) in 40 ml of CH3CN.
-
9|
Stir the mixture overnight at room temperature. Add aqueous ammonium chloride (10 ml) and extract the aqueous phase with CH2Cl2 (70 ml ×3).
-
10|
Collect the organic layer, dry it, concentrate it and isolate ester 4 using flash chromatography with hexanes:EtOAc (10:1 vol/vol) as the eluent (Rf = 0.5 hexanes:EtOAc 10:1; stain with KMnO4; impurity at 0.95 shows as red under UV).
■ PAUSE POINT Ester 4 can be stored in a sealed glass vial in a −20 °C freezer for a minimum of 6 months.
-
11|Compound 4 can be used to prepare either HNE(alkyne) or compound 8 by following the steps in options A and B, respectively.
- Preparation of HNE(alkyne)
- HNE(alkyne) can be prepared from 4 by reduction of ester with DIBAL-H. To do this, dissolve 0.4 g of ester 4 (2.2 mmol) in 20 ml of CH2Cl2. Cool the mixture to −80 °C.
- Dissolve 4.45 ml of DIBAL-H (1 M in hexane, 4.4 mmol) in 20 ml of CH2Cl2 and add it dropwise to the mixture. Stir it for 1 h.
- Add 24.7 ml of 1 N HCl, extract the aqueous phase with diethyl ether, dry it and concentrate it in vacuo.
-
Isolate the pure product via flash chromatography using hexanes:Et2O (4:1 vol/vol) as the eluent (Rf = 0.5 in hexanes: Et2O 4:1).▲ CRITICAL STEP HNE(alkyne) is highly unstable in regard to air oxidation and polymerization. The purified material should be characterized promptly, and during this time it may be stored under argon at room temperature. For long-term storage (at least 1 year), HNE(alkyne) should be stored as one-shot aliquots in DMSO at −80 °C and must be used immediately once thawed.? TROUBLESHOOTING
- Preparation of Compound 8 ● TIMING 4 h
- To 0.4 g of the ester 4 (2 mmol), add 1.1 ml of 3,4-dihydropyran (12 mmol) and 0.08 g of pyridinium p-toluenesulfonate (0.32 mmol). Stir the mixture for 24 h at room temperature under argon.
-
Add saturated NaHCO3 aqueous solution (5 ml), extract the aqueous phase with DCM (10 ml ×3). Wash twice with water (10 ml), followed by once with brine (10 ml) and dry with Na2SO4 (40 mg or more, until the newly added powder no longer clumps upon swirling) to obtain 5.■ PAUSE POINT 5 can be stored in a sealed glass vial in a −20 °C freezer for at least 6 months.
- Dissolve 0.5 g of the protected ester 5 (1.8 mmol) in 20 ml of toluene. Cool the mixture to −80 °C.
- Add two equivalents of DIBAL-H (1 M in hexane) dropwise to the mixture. Stir it for 1 h and then add 0.5 ml of 3 M NaOH.
-
Purify the desired product using flash chromatography with hexanes:EtOAc (1:1 v/v) as the eluent to obtain 6.■ PAUSE POINT 6 can be stored in a sealed vial in a −20 °C freezer for 1 month.
- Brominate 0.35 g of the resulting alcohol 6 (1.4 mmol) in 20 ml of distilled DCM at 4 °C by adding 0.5 g of CBr4 (1.54 mmol) and 0.44 g of PPh3 (1.68 mmol). Stir the mixture for 15 min.
- Add saturated NaHCO3 (50 ml), extract the aqueous phase with DCM (40 ml ×3), dry it with Na2SO4 (200 mg or more, until the newly added powder no longer clumps upon swirling) and concentrate it.
-
Isolate the desired product 7 after flash chromatography using hexanes:EtOAc (6:1 vol/vol) as the eluent.? TROUBLESHOOTING■ PAUSE POINT 7 should be promptly used in the subsequent step. Temporary storage (<1 week) is possible in moisture-free conditions and at low temperature (≤ −20 °C).
- To deprotect the THP group, weigh out 0.26 g of 7 (0.87 mmol), add 0.66 g of pTsOH (0.35 mmol) and 25ml of MeOH. Stir the mixture overnight.
- Add NaHCO3 (50 ml), and extract the aqueous phase with EtOAc (40 ml ×3). Wash it with water (40 ml) and concentrate it in vacuo.
-
Isolate the desired product 8 after flash chromatography using hexanes:EtOAc (4:1 vol/vol) as the eluent.■ PAUSE POINT 8 is immediately used in the subsequent step. Temporary storage (<2 d) is possible under air-and moisture-free conditions at low temperature (≤ −20 °C).
Synthesis of the anthrahydroquinone cage ● TIMING 3 d
-
12|
Weigh out 1 g of 1-hydroxyanthraquinone (9) (4.46 mmol). Dissolve it in 100 ml of 5% NaOH in 1:1 MeOH:ddH2O under argon (Fig. 10).
-
13|
Add 4.3 g of Na2S2O4. Heat the mixture to 70–75 °C for 10 min, and then add 3.0 ml of 2-propenal (44.6 mmol). Heat the reaction mix and reflux it overnight.
-
14|
Let the mix cool and then add it to a cold 200-ml solution of 2.5% H2SO4. Extract the aqueous phase with 3 × 50 ml of CH2Cl2, combine the organic phases and dry the combined organic phases over CaCl2 (10 g, 48 h).
-
15|
Concentrate the sample in vacuo and purify the residue via flash chromatography using 2:1 hexanes:CH2Cl2 as the eluent to obtain 10 (ref. 102).
-
16|
Weigh out 0.2 g of 10 (0.76 mmol) and dissolve it in 10 ml of DMF.
-
17|
Add 0.27 ml of benzyl bromide, 0.628 g of potassium carbonate and 0.038 g of potassium iodide. Stir the mixture for 1 h at 65 °C and subsequently cool it to room temperature.
A color change from dark purple to orange should be observed as the reaction progresses.
-
18|
Dilute the mixture with 50 ml of water and extract the organic layer using 50 ml of EtOAc.
-
19|
Wash the organic layer with 50 ml of water, 50 ml of brine and 50 ml of 1N HCl. Dry and concentrate the mixture in vacuo to yield 11 as a yellow solid (Rf = 0.1 hexanes:EtOAc 30:1; benzyl bromide 0.8).
■ PAUSE POINT 11 can be stored in a sealed glass vial in a −20 °C freezer for at least 6 months.
-
20|
Dissolve 0.23 g of compound 11 (0.65 mmol) in 50 ml of CH2Cl2. Cool the solution to −78 °C in an ice bath made using acetone and dry ice.
-
21|
Bubble O3 through the mixture using an ozonator for 15 min, followed by the addition of 4.5 ml of Me2S. Allow the mixture to warm up to room temperature and let it stir for 10 h.
! CAUTION Me2S has a pungent stench and must be used in a fume hood.
-
22|
Concentrate the reaction mix in vacuo. Dilute the reaction mix with 50 ml of EtOAc and wash it with 50 ml of water.
-
23|
Collect the organic layer, dry it with Na2SO4 (100 mg or more until the newly added powder no longer clumps upon swirling) and concentrate it to yield 12 (Rf = 0.6 hexanes:EtOAc 3:1).
■ PAUSE POINT 12 should ideally be promptly used in the subsequent step. If a pause is required, the purified material can be stored for up to 3 d at −80 °C in a moisture-free argon atmosphere.
-
24|
Dissolve 0.23 g of 12 (0.645 mmol) and 9 ml of 2-methyl-2-butene in 37.5 ml of t-BuOH and cool it to 0 °C. Add a solution of 0.633 g of NaH2PO4·H2O (4.59 mmol) and 0.524 g of NaClO2 (5.79 mmol) in water dropwise to the reaction mix.
-
25|
Warm up the resulting mix at room temperature and let it stir overnight.
-
26|
Add 150 ml of 0.1 N aqueous HCl and extract the aqueous phase with EtOAc (200 ml ×3). Wash the organic extract with water (200 ml) and brine (200 ml), dry it with Na2SO4 (200 mg or more, until the newly added powder no longer clumps upon swirling) and concentrate it in vacuo to yield 13 as a yellow solid (Rf = 0.05 hexanes:EtOAc 3:1).
■ PAUSE POINT 13 can be stored in a sealed glass vial in a −20 °C freezer for a minimum of 1 year.
Coupling of Halo linker and the anthrahydroquinone cage ● TIMING 1 d
-
27|
Dissolve 0.24 g of 13 (0.65 mmol) and 0.14 g of 2-(2-(6-chlorohexyloxy)ethoxy)ethanamine (0.65 mmol) in 15 ml of CH2Cl2, and cool the mixture to 0 °C (Fig. 10).
-
28|
Sequentially add 0.13 g of HOBt (0.78 mmol), 0.22 ml of DIEA (1.95 mmol) and 0.14 g of EDCI (0.91 mmol). Let the mixture warm to room temperature, and stir it overnight.
-
29|
Add water (20 ml) and then CH2Cl2 (20 ml). Extract the organic layer with CH2Cl2 (20 ml ×3) and concentrate it.
-
30|
Purify the residue via flash chromatography using hexanes:EtOAc (1:2 vol/vol) as the eluent to yield amide 14 as a yellow oil (Rf = 0.3 hexanes:EtOAc 1:2).
? TROUBLESHOOTING
■ PAUSE POINT 14 can be stored in a sealed glass vial in a −20 °C freezer for at least 2 years.
Synthesis of HaloTag-targetable precursor to HNE (alkyne) (also known as Ht-PreHNE or HtPHA) ● TIMING 1 d
-
31|
To remove the benzyl-protecting group, dissolve 0.16 g of 14 (0.277 mmol) in 21 ml of EtOAc in a round-bottom flask and add 10% Pd/C (0.028 g, 0.028 mmol; Fig. 10).
? TROUBLESHOOTING
-
32|
Degas the mixture by sequential vacuum exposure, and then by purging with nitrogen (three times); refill the flask with hydrogen gas (1 atm) at room temperature and stir it for 1 h.
▲ CRITICAL STEP Rigorously degassed solvent must be used to expel dissolved oxygen.
-
33|
Filter the reaction mixture through a 4-cm-deep pad of Celite (5 g), and concentrate it to yield 15 as a yellow solid (Rf = 0.5 hexanes:EtOAc 1:6).
! CAUTION Activated Pd/C is pyrophoric; do not let the powder completely dry out when filtering (chase with excess EtOAc), and store waste in a dedicated container that is wetted with water.
-
34|
Dissolve 0.1 g (0.2 mmol) of phenol 15 and 0.112 g of TBAF (0.4 mmol) in 2 ml of THF and 2 ml of DMF.
-
35|
Add 0.177 g of bromide 8 (0.6 mmol) to the mixture, and stir it at room temperature overnight.
-
36|
Add 10 ml of water, and extract the organic layer with ethyl acetate (20 ml ×3).
? TROUBLESHOOTING
-
37|
After concentration in vacuo, purify the residue via flash chromatography using hexanes:EtOAc (1:5 vol/vol) as the eluent to yield HtPHA as a yellow solid (Rf = 0.6 hexanes:EtOAc 1:6).
■ PAUSE POINT HtPHA should be stored as one-shot aliquots in DMSO at −80 °C for up to 6 months protected from light; it should be used immediately once thawed, and should be handled in dim light.
T-REX in live mammalian cells or in E. coli
▲ CRITICAL The two steps of this section are transfection (Step 38) and the T-REX experiment (Step 39). For both steps, option A will give instructions for mammalian cells and option B will give instructions for E. coli
-
38|Express HaloTagged POIs in the cells following the steps in option A for transfecting mammalian cells and option B for transfecting E. coli cells. Halo-Keap1 with a TEV cleavage site between Halo and Keap1 is used as an example of Halo fusion protein in this protocol. A hexa-histidine tag is placed before HaloTag57. Option A is carried out using HEK-293 cells cultured in an 8-cm2 adherent culture dish in a humidified atmosphere of 5% CO2 at 37 °C. The amounts of reagents used in this procedure can be scaled up or down according to the size of the adherent culture dish that is used in experiments.
- Transient transfection of mammalian cells with PEI ● TIMING 1 h
- Allow cells to reach 60–70% confluence at the point of transfection.
-
Mix 6 µl of 1 mg/ml PEI stock solution with 1.5 µg of plasmid DNA (pMIR-CMV-Dsred-IRES-Halo-Keap1 in 150 µl of antibiotic-free, serum-free medium in a microcentrifuge tube. Incubate this solution for 15 min at room temperature.▲ CRITICAL STEP This step will need to be modified depending on the requirements of the experiment. For LC–MS/MS analysis experiments (see Step 40B), the experiment should be done in a 4 × 21 cm2 adherent culture dish. For each 21-cm2 dish, mix 12 µl of 1 mg/ml PEI stock solution with 6.0 µg of plasmid DNA (pMIR-CMV-Dsred-IRES-His6-Halo-Keap1) in 200 µl of antibiotic-free, serum-free medium.
- Meanwhile, aspirate the old medium from the dishes and replace it with 1.5 ml of fresh medium. ▲ CRITICAL STEP If you are using 21-cm2 dishes, replace the medium with 3 ml of fresh medium.
- Subsequent to a 15-min incubation period, add the plasmid DNA solution dropwise to the cells.
- Subsequent experiments are performed 24 h post transfection. To do this, proceed to Step 39A.
- Transformation and cell growth of E. coli cells for T-REX ● TIMING 2 h
- Transform chemically competent BL21 codon plus (DE3) RIL cells with pet28a-Halo-Keap1, and plate them on an LB–kanamycin plate. Incubate the plate overnight at 37 °C.
- Pick one colony from the plate and inoculate 5 ml of LB–kanamycin–chloroamphenicol medium (50 µg/ml kanamycin and 30 µg/ml chloramphenicol) with the colony. Shake the flask at 200 r.p.m. at 37 °C overnight.
- Dilute the 5-ml overnight culture in another 5 ml of LB–kanamycin medium to a final OD of 0.1–0.2. Shake the culture flask at 200 r.p.m. at 37 °C until the OD value reaches 0.6–0.8.
-
Induce the culture with IPTG at 19 °C to a 0.5 mM final concentration. Shake the culture at 19 °C overnight at 200 r.p.m.▲ CRITICAL STEP It is important to induce expression when the OD is 0.6–0.8.
- Proceed to Step 39B.
-
39|Perform the T-REX experiment by performing the steps in option A for mammalian cells and those in option B for E. coli.
-
Treatment of mammalian cells with T-REX photocaged precursor, photo-uncaging and harvest of cells ● TIMING 4 h▲ CRITICAL STEP The subsequent steps should be done under dim light.
-
Treat the cells with HtPHA for 2.5 h in 1.5 ml of serum-free medium at a final concentration of 25 µM.▲ CRITICAL STEP Remove the old medium. Medium that contains serum affects the uptake of T-REX photocaged precursors by cells and thus should be avoided. The solubility of the T-REX photocaged precursor is low; therefore, vigorous mixing during dilution of the compound (from the DMSO stock aliquot) in 37 °C serum-free medium is required to make sure that the solution is homogeneous.
-
Rinse three times (each time with 1.5 ml of serum-free medium) every 30 min over 1.5 h.▲ CRITICAL STEP Medium should be added slowly along the side wall of the dish to prevent cells from detaching. Adding and removing the medium should be done as gently as possible to avoid cells detaching from the culture dish, and it should be done as efficiently as possible to avoid cells being dried out during this step. Marking the point of the plate where you will add and aspirate the medium and performing all operations at that point minimizes loss of cells.
-
For the samples designated for light exposure, remove lids from the plates and irradiate monolayered adherent cultures with 365-nm UV light for 20 min at room temperature (Supplementary Fig. 1 and Supplementary Video 1), and subsequently re-incubate at 37 °C for a further 5 min.! CAUTION Wear UV-protective safety eyeglasses while shining light.
- Trypsinize the cells with 500 µl of trypsin, transfer them to microcentrifuge tubes and harvest them by centrifugation at 500g for 8 min at room temperature.
-
Wash the cell pellets twice with 1× PBS. Flash-freeze the cell pellets in liquid N2.■ PAUSE POINT The cell pellets can be stored at −80 °C for up to 3 d.
-
-
Treatment of bacterial cells with T-REX photocaged precursor, photo-uncaging and harvest of cells ● TIMING 4 h▲ CRITICAL STEP The subsequent steps should be done in dim light.
- After overnight growth, dilute the cells to an OD of 0.3–0.4 in LB–kanamycin medium.
- Treat the cells with 25 µM HtPHA for 2 h while shaking them at 200 r.p.m. at 19 °C.
- Remove 1 ml of cell suspension and transfer it to a microcentrifuge tube. Centrifuge the mixture at 5,000g for 5 min at 4 °C. Discard the supernatant. Wash the cell pellet by resuspending it in 1 ml of PBS and centrifuging to collect the pellet.
- Repeat the wash step four additional times.
-
After the fifth rinse, resuspend the cells in 500 µl of PBS and irradiate them by placing the samples under a 365-nm UV light source for 30 min at room temperature, while constantly shaking the samples at 80–100 r.p.m. (Supplementary Video 2).! CAUTION Wear UV-protective safety eyeglasses while shining light.▲ CRITICAL STEP The microcentrifuge tubes must be uncapped during irradiation.
- Incubate the mixture for an additional 10 min post light shining at room temperature.
-
Collect the cells by centrifugation at 5,000g, for 5 min at 4 °C.■ PAUSE POINT Cell pellets can be stored at −80 °C for up to 3 d.
-
Downstream analyses
-
40|
The nature of the downstream analyses used will depend on the objectives of the experiment. Using Nrf2–AR signaling as a model response pathway, here we describe six different methods for analyzing the effects of protein-specific modification with LDEs. Option A describes how to quantify targeting efficiency using click coupling; the first step in this process is to lyse the cells (mammalian or bacterial) obtained at the end of Step 39.
Option B describes steps for liquid-chromatography-coupled tandem mass spectrometry (LC–MS/MS) analysis to determine which residue(s) on the POI are modified by HNE(alkyne). His6-Halo-Keap1, with a TEV cleavage site between Halo and Keap1, is used as an example of a Halo fusion protein that contains cysteines to be modified in this protocol. The following protocol is carried out with HEK-293 cells in a 4 × 21 cm2 adherent culture dish. The amounts of reagents used in this protocol can be scaled up or down according to the size of the adherent culture dish used.
Options C and D describe how to evaluate the extent of downstream transcriptional activation as a consequence of targeted redox modification on a specific sensor POI upstream, either by dual-luciferase reporter assays with the use of a plate reader detecting bioluminescence in cell lysates (option C) or by GFP reporter assays with the use of a flow cytometer detecting GFP fluorescence in live cells (option D). In both cases, the Nrf2-AR transcriptional activation is used as an example readout as a result of Keap1-targeted HNEylation.
Options E and F describe how to probe the functional downstream impact on endogenous biological entities triggered by targeted redox modification of an upstream sensor POI, either by antibody staining of the endogenous species in fixed cells (option E) or by FRET-based biosensor readout reporting cellular levels directly in live cells (option F). The measurement of changes in endogenous PIP3 phosphoinositides is used as an example of the result of targeted HNEylation of PTEN lipid phosphatase that is coupled to the accumulation of cellular PIP3 levels.- Quantification of targeting efficiency by click coupling ● TIMING 6 h
-
For mammalian cells, add 15 µl of lysis buffer, consisting of 50 mM HEPES, pH 7.6, 0.3 mM TCEP and 1% Nonidet p-40, to mammalian cell pellets from Step 2A above and subject them to three cycles of freeze–thaw.For E. coli cells, resuspend the cell pellet in 100 µl of lysis buffer consisting of 50 mM HEPES, pH 7.6, 2 mM TCEP, 1% Nonidet p-40, 150 µg/ml Lysozyme and 5 µg/ml DNAse-I. Incubate the mixture for 20 min at room temperature with agitation.▲ CRITICAL STEP (for mammalian cells) Make sure that the cells are resuspended well in lysis buffer.▲ CRITICAL STEP (for E. coli cells) Do not vortex the cells after adding lysozyme or DNAse-I.
- Remove debris by centrifugation (18,000g, 8 min) at 4 °C.
- Measure the lysate concentration by Bradford assay relative to BSA as standard.
-
Dilute a portion of the clarified lysate to 1 mg/ml in a final volume of ~25 µl containing 50 mM HEPES, pH 7.6, 0.3 mM TCEP and 0.2 mg/ml His6-TEV-S219V (prepared as described in Box 4). Incubate the mixture at 37 °C for 45 min.▲ CRITICAL STEP The optimal concentration of lysate protein is ~1.0 mg/ml. A high concentration of lysate protein causes failure of click coupling. Precipitates may appear upon adding His6-TEV-S219V. Gentle resuspension is necessary to ensure the success of click coupling. For samples without light exposure, His6-TEV-S219V should be omitted.
-
For click coupling in a final volume of ~30 µl, add the following reagents to a final concentration of 1.7 mM TCEP, 5% (vol/vol) t-BuOH, 1% (wt/vol) SDS, 1 mM CuSO4, 0.1 mM TBTA, 10 µM Cy5 azide and the TEV protease–treated lysate above. Incubate the resulting mixture at 37 °C for 30 min.▲ CRITICAL STEP All the concentrations above are critical to the success of click coupling. Mix well to make sure that the solution is homogeneous. Generally, SDS is needed to obtain good results.
-
Quench with 5 µl of 4× Laemmli buffer that contains 6% (vol/vol) BME, and further incubate for 5 min at 37 °C. Load 20 µl into each well of a 10% (wt/vol) polyacrylamide gel, and resolve by electrophoresis.▲ CRITICAL STEP 4× Laemmli buffer should be warmed in advance to ensure homogeneity. Fresh SDS–PAGE buffer should be used to reduce the background signal. It is recommended that the wells of polyacrylamide gel (remove buffer in the wells using a P-200 Pipetman with a loading tip, repeat for 4–5 times) be rinsed before loading the samples with fresh SDS–PAGE buffer to enhance the signal-to-noise ratio.
-
Upon completion of the gel electrophoresis, rinse the gel with 20 ml of ddH2O (×2, 5 min) and analyze for a Cy5 signal using a ChemiDoc-MP imaging system (Bio-Rad; see Equipment Setup). Any alternative fluorescence gel imager platforms can be used in this step.▲ CRITICAL STEP Rinsing reduces the background signal. It is recommended that the gel be rinsed several times and that it be analyzed after each rinse to obtain the optimal result (highest signal-to-noise ratio).
- Transfer the gel to a PVDF membrane for western blot analysis. After transfer is complete, block the membrane with 10% (wt/vol) milk in Tris-buffered saline containing 0.2% (vol/vol) Tween 20 (i.e., TBST), and then probe it with anti-Keap1 and anti-actin antibodies (see materials list for dilutions).
- Use the following equation to calculate the targeting efficiency:
where Cy5x is the amount of Cy5 signal from the target protein in the sample exposed to light; WBx is the amount of western blot signal on the target protein in the sample exposed to light; Cy5y is the amount of Cy5 signal on the Halo-fusion protein in the sample not exposed to light; Cy5Halo is the amount of Cy5 signal on the Halo protein in the sample exposed to light; WBy is the amount of western blot signal on the Halo-fusion protein in the sample not exposed to light. -
In our laboratory, Bio-Rad Image Lab software is used to quantify the intensities of the Cy5 and western blot signals. If you are also using this software, open your Cy5 or western blot image using the software. From the ‘Analysis Tool Box’, pick ‘Volume Tools’ followed by ‘Rectangle’. Draw rectangles of the same sizes around each of the desired bands. Draw another rectangle of the same size in an area without any bands, and use this for background subtraction. (Designate this rectangle as the background by double-clicking and choosing ‘background’.) Under the ‘Subtraction Method’ menu, choose ‘Global’. Click on ‘Analysis table’ to generate the quantitated signal intensities corresponding to your selected bands. Export the analysis table to Excel. Use the ‘Adjusted volume’ values as the signal intensities of your desired bands.? TROUBLESHOOTING
-
-
LC–MS/MS analysis of modified cysteines ● TIMING 2–3 d▲ CRITICAL STEP The starting material for option B is prepared as described in Steps 38A and 39A.
-
Enrichment of (His-tagged) protein from mammalian cells. Harvest confluent monolayer cultures of cells from 4 × 21 cm2 cultured plates.■ PAUSE POINT While immediate lysing is recommended, as HNE modifications are unstable, if a pause is needed the cell pellets can be flash-frozen in liquid N2 and stored at −80 °C for up to 3 d.
-
Pool the pellets and lyse them in 100 µl of lysis buffer A with 3 cycles of rapid freeze–thaw.▲ CRITICAL STEP Make sure that the cells are resuspended well in lysis buffer A.
- Remove debris by centrifugation at 18,000g for 8 min at 4 °C.
- Equilibrate a 20-µl bed volume of TALON resin by washing it three times with 500 µl of lysis buffer A.
- Determine the lysate concentration using a Bradford assay with BSA as standard.
-
Dilute the lysate to 1.0 mg/ml with lysis buffer A, and add it to the TALON resin in a 1.7-ml centrifuge tube.Incubate the suspension in the dark at 4 °C for 1.5 h with end-over-end rotation.▲ CRITICAL STEP A high concentration of lysate protein enhances nonspecific binding, and therefore incorporates impurities into the final pulled-down Keap1. A portion of clarified lysate (typically 5–10 µl) should be saved before treatment with TALON resin to confirm the modification of cysteines by electrophiles using click coupling. See Steps 3 and 40A above.
-
Centrifuge the sample at 500g for 3 min at 4 °C. Remove the supernatant and add 360 µl of wash buffer A to the resin. Incubate the suspension in the dark at 4 °C for 3 min with end-over-end rotation.▲ CRITICAL STEP Do not remove any resin in this step or subsequent steps.
- Remove the supernatant after centrifugation at 500g for 3 min at 4 °C. Add 240 µl of wash buffer B, followed by incubation in the dark at 4 °C for 3 min with end-over-end rotation.
- Repeat Step 40B(viii) above.
-
Remove the wash buffer after centrifugation at 500g for 3 min at 4 °C. Add 25 µl of elution buffer and incubate the resin in the dark at 4 °C for 5 min with end-over-end rotation.▲ CRITICAL STEP The supernatant from the wash step should be removed thoroughly to maximize the yield of His6-Halo-Keap1 in the elution.
- Collect the eluent after centrifugation at 18,000g for 3 min at 4 °C. Mix the eluent with 4× Laemmli buffer with 6% (vol/vol) BME, incubate it for 5 min at 37 °C and resolve it on a 10% SDS–PAGE gel. Stain the gel with Coomassie R-250 stain (or freshly prepared Colloidal Coomassie G-250 stain for enhanced sensitivity) for 24–48 h until the desired sensitivity is achieved.
-
Rinse the gel with ddH2O for 30 min, and excise the band corresponding to His6-Halo-Keap1.■ PAUSE POINT The gel slices can be stored at −80 °C with 50 µl of ddH2O for up to 2 weeks.
- LC-MS/MS analysis. Wash the gel pieces with 100 µl of ddH2O. Remove and discard water.
- Add 100 µl (50:50) of 100 mM NH4HCO3(pH 7.8):acetonitrile to the gel pieces and let them sit for 10 min. Remove and discard the liquid.
- Add 50 µl of acetonitrile and let the sample sit for 5 min. Remove and discard acetonitrile. Dry the gel pieces in a fume hood for 10 min.
-
Reduce the proteins with 5 mM TCEP in 50 mM NH4HCO3 solution, pH 7.8, for 45 min at 37 °C and alkylate with 20 mM iodoacetamide in 50 mM NH4HCO3 in the dark for 45 min.▲ CRITICAL STEP Use of TCEP instead of DTT is very important, as DTT can reduce labeling of proteins by LDEs.
- Repeat Step 40B(xiii–xv).
- Rehydrate the gel pieces by adding 60 µl (10 µg/ml solution) of trypsin in 50 mM NH4HCO3, pH 7.8; keep on ice for 30 min and then at 37 °C overnight.
- To stop the enzymatic reaction, add formic acid to a final concentration of 1%. Remove and save the supernatant.
- Add 120 µl of 50% acetonitrile containing 5% (vol/vol) formic acid to the trypsinized gel pieces. Let the sample sit for 45 min. Sonicate the sample for 5 min. Remove the supernatant and combine it with the supernatant from Step 40B(xix).
- Repeat Step 40B(xx).
- Add 90% acetonitrile containing 5% (vol/vol) formic acid. Let the sample sit for 5 min. Remove and combine the supernatants. Dry the supernatant under vacuum.
- Resuspend peptides in 60 µl of 0.1% (vol/vol) formic acid and inject the solution into the LC system described in the Equipment Setup section of the Protocol.
-
Collect LC–MS/MS data using the setup described in the Equipment Setup section of the Protocol.▲ CRITICAL STEP To prevent loss of modification, the ionization temperature should not be too high (in this case it should be 130 °C).
- After data acquisition, combine the individual MS/MS spectra acquired for each of the precursors within a single LC run, smoothen, de-isotope using an Analyst ‘script’, mascot.dll, to create a peak list and save the peak list to a file.
- Subsequently, use the peak list file to query the NCBI human subdatabase and contaminations using Mascot 2.4 from Matrix Science (London, UK) with the following parameters: peptide mass tolerance, 0.3 Da; MS/MS ion mass tolerance, 0.3 Da. Allow up to two missed cleavages. In our case, several variable modifications were applied, including methionine oxidation and cysteine carbamidomethylation, along with electrophile Michael adduct, reduced electrophile Michael adduct, dehydrated electrophile Michael adduct, dehydrated electrophile (1,2)-addition adduct or electrophile Michael adduct in Schiff-based form on cysteine residues56. Only significant scores for the peptides, defined by Mascot probability analysis (http://www.matrixscience.com/help/scoring_help.html#PBM) as greater than ‘identity’ with 95% confidence, should be considered for the peptide identification and modification site determinations.
- Manually inspect and validate all MS/MS spectra for the identified peptides with HNE-type modifications.
-
To approximate the extent of modifications on a given site, extract the modified peptide signal (XIC) from the chromatogram and compare it with the XIC of the unmodified peptide.? TROUBLESHOOTING
-
-
Analysis using Luciferase reporter assay for evaluating AR activation ● TIMING 3 d▲ CRITICAL STEP Option C starts with a new transfection.
-
Transient transfection using TransIT-2020. Plate HEK-293 cells in a 48-well plate (0.9-cm2 surface area). For a single well, seeding ~ 5 × 104 cells in 300 µl of complete cell culture medium should result in cells that are 50–60% confluent after 24 h, which is optimal for transfection with TransIT-2020. It is important to have at least triplicate samples for each condition. If you are seeding in multiple wells, make a stock of 1.5 × 105 cells/ml and add 300 µl of the stock to each well after careful suspension.▲ CRITICAL STEP A cell density that is too low or too high may result in excessive cell death or poor transfection efficiency, respectively.
- Premix ARE-Firefly Luciferase plasmid and pCMV-Renilla luciferase plasmid in a 40:1 ratio by mass. Transfect the cells with the following plasmids: premixed 40:1 ARE-Firefly:pCMV Renilla luciferase plasmids, pCMV Halo-Keap1 and pcDNA3.1 myc-Nrf2.
-
For transient transfection of cells in a single well of a 48-well plate, in 26 µl of antibiotic-free, serum-free medium, add 120 ng of each plasmid (Halo-Keap1 and myc-Nrf2) and 120 ng of the ARE-Firefly:pCMV Renilla luciferase plasmid mix (360 ng total), followed by the addition of 0.78 µl of TransIT-2020. The solution should be mixed gently and incubated at room temperature for 20 min.▲ CRITICAL STEP The amounts of DNA and TransIT-2020 reagent may need to be optimized for different cell lines.
- Subsequent to the 20-min incubation, The resultant lipoplex should be added dropwise to the cells in complete medium.
- Subsequent steps should be performed 24 h post transfection.
-
Treatment of cells with T-REX photocaged precursor and photo-uncaging. Treat the cells as described in Step 39A, except:Cells should be treated with the T-REX photocaged precursor at a concentration of 25 µM in 300 µl of serum-free medium; rinsing should be performed with 300 µl of serum-free medium; and post light shining, the cells should be incubated for 18 h before measuring luciferase activity.
-
Measurement of luciferase activity. To analyze the level of AR activation using a luciferase reporter assay, wash the cells with 150 µl of 1× PBS.▲ CRITICAL STEP PBS should be added dropwise along the side walls of the wells to prevent cells from detaching.
-
Lyse cells by adding 65 µl of 1× PLB (ref. 101) and incubating them at room temperature on a shaker for 15–20 min.▲ CRITICAL STEP Lysate should not be left at room temperature for longer than 1 h.
- Meanwhile, thaw the Firefly substrate and prepare 1× Renilla luciferase substrate by adding the 100× coelenterazine stock in methanol in 1× Renilla luciferase buffer (ref. 102).
-
Pipette out 20 µl of the well-mixed lysate into a 96-well white opaque plate (Corning) for measuring chemiluminescence. Read the Firefly luciferase signal on a plate reader (in our case, a BioTek Cytation 3-cell imaging multimode microplate reader was used) after adding 50 µl of Firefly luciferase substrate. Subsequently, read the Renilla luciferase signal after adding 50 µl of the 1× Renilla substrate.▲ CRITICAL STEP If handling multiple wells simultaneously, an automated dispenser can be used to add Firefly and Renilla luciferase substrates in order to minimize decay of signal intensity.
-
Analyze the data by calculating the ratio of Firefly luciferase signal intensity to Renilla luciferase signal.? TROUBLESHOOTING
-
-
Analysis using a GFP reporter assay for evaluating AR activation ● TIMING 3 d▲ CRITICAL STEP Option D starts with a new transfection.
-
Transient transfection using TransIT-2020. Perform the transfection as described in Step 40C(i).A brief description is as follows: transfect the cells with ARE:GFP, pFN21a-Halo-Keap1 and pcDNA3.1 myc-Nrf2 in a 3:1:1 ratio by mass. For transient transfection of cells in a single well of a 48-well plate, to 26 µl of antibiotic-free, serum-free medium, add 156 ng of ARE::GFP, 52 ng of pFN21a-Halo-TEV-Keap1 and 52 ng of pcDNA3.1 myc-Nrf2 (260 ng total), followed by the addition of 0.78 µl of TransIT-2020. The lipoplex should be mixed gently and incubated at room temperature for 20 min.▲ CRITICAL STEP It is important to use plasmids that do not express another fluorescent protein.
- Subsequent to 20 min of incubation, the lipoplex should be added dropwise onto the cells.
- The following steps are performed 24 h post transfection.
- Treatment of cells with T-REX photocaged precursor and photo-uncaging. Treat the cells with photocaged precursor, as described in Step 40C(vi).
- Measurement of the GFP fluorescence using flow cytometry. Set up the flow cytometer equipment.
- Harvest the cells by adding 100 µl of trypsin TrypLE and incubating them at room temperature for 1 min. Transfer the cell solution to an FACS tube and rinse the well with another 900 µl of FACS buffer. Resuspend the culture several times to ensure a single-cell suspension before loading onto the flow cytometer.
- Perform the flow cytometry experiment to determine the GFP signal. Measure at least 10,000 events per well.
-
Data analysis. Any cytometry software can be used to process the data. Here, we used FlowJo (v 10).? TROUBLESHOOTING
-
- Immunofluorescence analysis ● TIMING 4 d
-
Transient transfection using TransIT-2020. Split HEK-293 cells into four 35-mm glass-bottom dishes. For each dish, seeding ~4 × 105 cells in 2.0 ml of total cell culture medium should result in cells that are ~40–50% confluent after 24 h, which represents optimal cell density.▲ CRITICAL STEP A cell density that is too low or too high may result in excessive cell death or poor transfection efficiency, respectively. Higher cell density is also not optimal for imaging.
- Transfect each plate with 2,000 ng of Halo-PTEN plasmid. Transfect with TransIT-2020 according to the manufacturer’s protocol.
- The subsequent steps should be performed 24–36 h post transfection.
- Treatment of cells with T-REX photocaged precursor, photo-uncaging and cell fixation. Treat the cells with photocaged precursor, as described in Step 39A. The amounts of reagents used should be scaled up for a 4 × 35 mm adherent culture dish.
- After shining light on the sample, incubate the plates at 37 °C for 10 h.
- Aspirate old medium and wash the sample once gently with 1.5 ml of 1× PBS. Fix the cells by adding 1.5 ml of 2% formaldehyde (prechilled at 4 °C). Incubate the plates for 20 min at 4 °C.
-
Aspirate formaldehyde. Add 1.5 ml of 1× PBS to the plates.■ PAUSE POINT Fixed cells can be stored at 4 °C for up to 2 weeks.
-
Permeabilization of cell membrane and antibody binding. Aspirate PBS from the plates.Add 1.5 ml of blocking–permeabilization buffer to each plate. Incubate the plates at 37 °C for 1 h.
- Meanwhile, prepare antibodies to Halo (1:1,000) and PIP3 (1:500) in 600 µl of incubation buffer.
- Remove blocking–permeabilization buffer, wash the cells once with 50 mM HEPES, pH 7.6, and add 150 µl of primary antibody solution. Add primary antibody solution only in the recessed region in order to minimize the amount of antibody usage. Incubate the mixture for 2 h at room temperature.
- Gently remove the primary antibody solution using a pipette. The solution can be stored at 4 °C and reused.
- Wash the cells by adding 1.5 ml of 1× PBS to the plates. Incubate them at 37 °C for 5 min. Aspirate the PBS and repeat the rinse step two additional times.
-
Prepare 1:1,000 dilutions of fluorophore-conjugated secondary antibodies in 600 µl of PBS with 0.02% Triton X-100.▲ CRITICAL STEP Avoid exposure of fluorophore-conjugated antibodies to stray light.
- Add 150 µl of secondary antibody solution to each plate. Incubate the cells for 1 h at room temperature, protected from light.
- Remove the secondary antibody solution and add 1.5 ml of 1× PBS to the plates. Incubate the cells at 37 °C for 5 min. Aspirate the PBS and repeat the rinse step two additional times.
-
Prepare a 1 µg/ml dilution of DAPI in 1× PBS. Add 1.5 ml of DAPI solution to the plates. Incubate the cells for 1 min at room temperature. Aspirate and rinse the plates once with 1× PBS.■ PAUSE POINT Cells can be imaged immediately or they can be stored protected from light at 4 °C in 1× PBS for 1–2 d.
- Imaging and data analysis. Image cells using a confocal microscope according to the instruments protocol. (In our case, a Zeiss 710 confocal microscope was used for image acquisition.)
-
Images should be analyzed using ImageJ software (NIH). Briefly, the average green fluorescence intensity (intensity of signal due to PIP3) for each cell should be determined by drawing a free-hand circle around the image. This should be repeated for each condition such that 50–100 cells are analyzed each time from multiple different frames. A global average of the green fluorescence intensity should be calculated for each condition, and then these values can be plotted and analyzed using Prism.▲ CRITICAL STEP To get reliable results, it is important to collect images from at least 50 individual cells per condition.
-
-
FRET analysis ● TIMING 4 d▲ CRITICAL STEP Option F starts with a new transfection.
- Transient transfection using TransIT-2020. See Step 40E(i and ii).
- Transfect each plate with 1,000 ng of Halo-PTEN and 1,000 ng of InPAkt reporter plasmid. Transfect with TransIT-2020 according to the manufacturer’s protocol.
- The subsequent steps are performed 24–36 h post transfection.
- Treatment of cells with T-REX photocaged precursor, photo-uncaging and cell fixation. Treat the cells with photocaged precursor, as described in Step 39A. The amounts of reagents used should be scaled up for a 4 × 35 mm adherent culture dish.
-
Irradiate the cells with UV light for only 3 min. Incubate the plates at 37 °C for 10 h.▲ CRITICAL STEP A longer irradiation time will result in photobleaching of the FRET reporter proteins.
- Imaging and data analysis. Image cells using a confocal microscope according to the previously described protocol89. In our laboratory, a blue laser (408 nm) is used for excitation. Record the signals in the cyan channel (463–498 nm) and the yellow channel (525–620 nm).
-
Analyze the images using ImageJ software. To do this, determine the average cyan and yellow fluorescence intensity for each cell by drawing a free-hand circle around the image. Repeat this for each condition such that 150–200 cells are analyzed each time from multiple different frames. Calculate the ratio of mean yellow to mean cyan fluorescence intensity for each condition, and then plot these values and analyze the results using Prism.▲ CRITICAL STEP To get reliable results, it is important to collect images from at least 150 individual cells per condition.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| step | problem | possible reason | solution |
|---|---|---|---|
| 4 | Low yield of alcohol 2 |
Ethylenediamine is old Reaction interfered with by O2 and moisture Product destroyed during reaction quenching |
Use either re-distilled ethylenediamine or fresh out of the bottle Make sure that the reaction is protected under dry N2 or Ar The reaction quenching is exothermic; add the quencher (1 N HCl) slowly and make sure that the flask is well cooled |
| 7 | Low yield of aldehyde 3 |
Aldehyde 3 is volatile | Avoid excess heating (above 35 °C)of the rotary evaporator water bath |
| 11A(iv) | Low yield of HNE(alkyne) |
HNE(alkyne) is not stable | It is prone to air oxidation and polymerization. Carry on nonstop through steps involving reaction workup, rotary evaporator, chromatography, to final storage |
| 11B(viii) | Product 7 partially deprotected |
One of the isomers of product 7 is not stable |
Product 8 (deprotected 7) can be separated from 7 by chromatography on silica gel, or it can be carried through to the next step |
| 30 | Low yield of 14 | Reaction yield can be reduced by adventitious O2 and/or moisture |
Make sure that all the solvents are dry and that the reaction is protected under dry N2 or Ar. Thoroughly degas the solvents used |
| 31 | Incomplete deprotection of 14 |
Some of the catalyst (10% Pd/C) may be deactivated |
Add more catalyst (10% Pd/C) and extend the reaction time. Typically, high catalyst loading can be tolerated. Ensure that the reaction is tightly sealed and that an atmosphere of hydrogen is maintained throughout |
| 36 | Difficult to remove impurities from HtPHA |
Polarity of impurities is close to that of HtPHA |
Run flash chromatography with gradient eluent, from 1:3 to 1:5 (vol/vol, hexanes: EtOAc) |
| Final product decomposition |
Light-induced photo- uncaging of the final product |
During the synthesis (especially from 14 to 15, and 15 to HtPHA), protect the reaction flask from stray light |
|
| 40A(x) | No Cy5 signal on gel | Failure of click coupling | Check lysate protein concentration. The concentration should be around 1.0 mg/ml Check each reagent in the click coupling step. Make sure that all of them are freshly prepared and that the concentrations are correct. Mix the reaction well Use cells treated globally with HNE(alkyne) as a positive control |
| High Cy5 background | Old SDS-PAGE running buffer | Use fresh SDS-PAGE running buffer Rinse the gel several times and analyze the gel after each rinse to obtain the optimal result with highest signal-to-noise ratio Let the dye font run out completely before imaging the gel |
|
| Incomplete TEV cleavage |
Loss of TEV activity | Use a fresh aliquot. Avoid multiple freeze–thaw cycles Mix well after adding His6-TEV-S219V Increase TEV amount and incubation time for TEV cleavage |
|
| 40B(xxviii) | Low yield of pulldown protein |
Affinity protein purification condition is not optimal Protein is not eluting or eluting prematurely Protein is unstable after modification103 |
This protocol is optimized for His6-Halo-TEV-Keap1. Further optimization may be required for other proteins Monitor the protein in washes through SDS–PAGE. If premature elution is observed, decrease the concentration of imidazole in the wash buffers. Conversely, if no elution is observed, increase imidazole concentration to 200–400 mM or elute with Laemelli buffer to validate binding Add a proteasome inhibitor (bortezomib) in the lysis buffer and/or to cells |
| Modification not found |
Targeting is not efficient. Modification is reduced Inefficient MS conditions |
Check modification of protein by Cy5 labeling, as described in Step 3A Make sure that TCEP is used instead of DTT in the sample preparation Adjust the ionization temperature to obtain optimal results. A low ionization temperature may lead to poor ionization and decrease sensitivity. A high ionization temperature may cause loss of modification |
|
| 40C(xi); 40D(viii) |
Low Firefly/Renilla/ GFP signal intensity |
Significant loss of cells during rinsing Excessive cell death Low transfection efficiency Instrument setting is not optimal Response timing is not optimal |
Perform the rinsing with care. Add the medium slowly along the side wall of the culture dish. Adding and removing medium should be done as gently as possible but also as efficiently as possible: wash plates sequentially (2 or 3 plates at a time) to avoid drying out. Mark the position on the plate where cells are washed Use the suggested cell density for transfection. If using a different transfection reagent or different cells, optimize transfection conditions Optimize transfection conditions for the cell type and the reagent used Run a positive control. For flow cytometry, cells transfected with GFP can be used. Adjust gain (i.e., laser power) if necessary Set up a time course for measurements (recommended time points for pilot trials: 4 h, 12 h, 18 h post T-REX light exposure) |
| Batch-to-batch variability in targeting/ARE results |
Difference in experimental setup and execution |
Count cells and seed the numbers as specified in the protocol. Transfect and perform experiments at similar confluence Use cells at lower passage number (lower than 6–7 continuous passages) |
|
| Activation of ARE-luciferase with light alone or with pre-HNE alone |
Release of pre-HNE due to stray light Stressed cells |
Protect photocaged-precursor-treated samples from stray light Count cells before seeding. A cell density that is too low or too high can stress the cells, leading to higher background |
● TIMING
Steps 1–11A, chemical synthesis of HNE(alkyne): 4 d
Step 11B, preparation of compound 8: 4 h
Steps 12–26, synthesis of the anthrahydroquinone cage: 3 d
Steps 27–30, coupling of Halo linker and the anthrahydroquinone cage: 1 d
Steps 31–37, synthesis of HaloTag-targetable precursor to HNE(alkyne) (also known as Ht-PreHNE or HtPHA): 1 d
Step 38A, transient transfection of mammalian cells with polyethylenimine (PEI): 1 h
Step 38B, transformation and cell growth of E. coli cells for T-REX: 2 h
Step 39A, treatment of mammalian cells with T-REX photocaged precursor, photo-uncaging and harvest of cells: 4 h
Step 39B, treatment of bacterial cells with T-REX photocaged precursor, photo-uncaging and harvest of cells: 4 h
Step 40A, targeting efficiency quantification: 6 h
Step 40B, LC–MS/MS analysis of modified cysteines: 2–3 d h (including enrichment of (his-tagged) protein from mammalian cells, 4 h; and LC–MS/MS analysis, 2–3 d)
Step 40C, dual-luciferase reporter assays evaluating AR pathway activation: 3 d (including transient transfection using TransIT-2020, 1 h; treatment of cells with T-REX photocaged precursor and photo-uncaging, 4 h; and measurement of luciferase activity, 2 h)
Step 40D, analysis using a GFP reporter assay for evaluating AR pathway activation: 3 d (including transient transfection using TransIT-2020, 1 h; treatment of cells with T-REX photocaged precursor and photo-uncaging, 4 h; and measurement of GFP fluorescence using flow cytometry, 2 h)
Step 40E, immunofluorescence analysis, 4 d (including transient transfection using TransIT-2020, 1 h; treatment of cells with T-REX photocaged precursor, photo-uncaging and cell –fixation, 4 h; permeabilization of cell membrane and antibody binding, 6 h; and imaging and data analysis, 6 h)
Step 40F, FRET analysis: 4 d (including transient transfection using TransIT-2020, 1 h; treatment of cells with T-REX photocaged precursor, photo-uncaging and cell fixation, 4 h; and imaging and data analysis: 6 h)
ANTICIPATED RESULTS
Analytical data
HNE(alkyne) (see Fig. 2 inset and Fig. 10 for chemical structure)
1H-NMR (300 MHz) δ 1.50–1.76 (4H, m), 1.92 (1H, t, J = 2.7 Hz), 2.19 (2H, dt, J = 2.7, 6.3 Hz), 2.61 (1H, br), 4.39–4.44 (1H, m), 6.25 (H, ddd, J = 1.2, 7.8, 15.9 Hz), 6.79 (1H, dd, J = 4.5, 15.6 Hz), 9.50 (1H, d, J = 7.5 Hz). 13C-NMR (75 MHz) δ 18.1, 24.0, 35.1, 69.1, 70.4, 83.8, 130.7, 159.2, 193.9.
HtPHA (also known as Ht-PreHNE) (see Fig. 10 for chemical structure)
1H-NMR spectroscopy (0.02 g, 24% yield): 1H-NMR (300 MHz) δ 1.59–1.69 (4H, m), 1.96 (1H, t, J = 2.1 Hz), 2.23–2.26 (2H, m), 3.57 (2H, d, J = 6.6 Hz), 4.23–4.29 (1H, m), 4.55 (2H, d, J = 5.7 Hz), 5.10–5.18 (2H, m), 5.91–6.16 (3H, m), 7.64 (1H, d, J = 7.8 Hz), 7.75–7.79 (2H, m), 8.10–8.12 (1H, d, J = 8.1 Hz), 8.24–8.29 (2H, m). 13C-NMR (75 MHz) δ 18.3, 24.3, 34.4, 35.9, 68.6, 71.7, 74.3, 84.2, 117.2, 123.6, 124.7, 126.2, 126.7, 127.2, 131.2, 131.4, 133.5, 134.2, 134.8, 135.6, 135.8, 137.1, 142.7, 157.2, 182.7, 183.1. LRMS (LDI) calculated for C26H24O4 400.2 (M+), found 400.1.
TEV purification
The typical yield of TEV protease is 0.5 mg of pure protein per gram of cell pellet.
In-gel fluorescence to determine labeling
Typical labeling results with HNE are shown in Figures 4a, 5b and 6a and Supplementary Figure 4a. Targeting efficiency (calculated using the equation shown in Step 40A(ix)) ranges from 10 to 40%.
Supplementary Material
Acknowledgments
We thank the laboratories of T. Evans (Weill Cornell Medicine, New York) and J. Zhang (University of California, San Diego) for plasmids encoding zebrafish hspb7 and the lnPAkt (PIP3-reporter) construct, respectively. We acknowledge all of the Aye Laboratory members who have contributed to T-REX redox targeting protocols, particularly J. Li, X. Fang and Y. Fu, as well as Q. Lin of the State University of New York at Albany for assistance with the LC–MS/MS analysis of modifications on the protein pulled down from cells. Funding was provided by the NIH Director’s New Innovator award (1DP2GM114850), the National Science Foundation (NSF) CAREER award (CHE-1351400), the Beckman Young Investigator award and the Sloan Research Fellowship (to Y.A.) and the Burroughs Wellcome Funds CTRG (to principal investigator (PI) Y.A. and host T. Evans). S.P. is a Howard Hughes Medical Institute international predoctoral fellow (59108350). J.A.H. acknowledges the CBI training grant (T32GM008500, PI – H. Lin). V.N.P. thanks the Douglas family for an undergraduate research fellowship. D.K.L. thanks the Cornell University P3 scholars program. Imaging and flow cytometry data were acquired at the Cornell University Biotechnology Resource Center (NIH 1S10RR025502) and the Cornell University cytometry core (supported in part by the Empire State Stem Cell Fund), respectively. We acknowledge the NSF (NSF MRI: CHE-1531632 to Y.A. (PI)) for NMR instrumentation support at Cornell University.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS H.-Y.L. and Y.Z. were joint second authors of this work. S.P., M.J.C.L., H.-Y.L., Y.Z., J.A.H., V.N.P., D.K.L. and Y.A. developed protocols. S.P. and V.N.P. contributed to the data associated with T-REX in E. coli cells. M.J.C.L., Y.Z., J.A.H. and D.K.L. obtained the data associated with T-REX in cultured human cells. H.-Y.L. collected LC-MS/MS data. H.-Y.L. and Y.Z. contributed to chemical synthesis. S.P., M.J.C.L. and H.-Y.L. wrote the protocols. Y.A. wrote the manuscript with proofreading/editing contributions from S.P., M.J.C.L., Y.Z. and J.A.H.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Jacob C, Winyard PG. Redox Signaling and Regulation in Biology and Medicine. Wiley-VCH; 2009. [Google Scholar]
- 2.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 3.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 5.Murphy MP, et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011;13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmastro-Greenwood M, Freeman BA, Wendell SG. Redox-dependent anti-inflammatory signaling actions of unsaturated fatty acids. Annu. Rev. Physiol. 2014;76:79–105. doi: 10.1146/annurev-physiol-021113-170341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crunkhorn S. Deal watch: Abbott boosts investment in NRF2 activators for reducing oxidative stress. Nat. Rev. Drug Discov. 2012;11:96. doi: 10.1038/nrd3655. [DOI] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova AT, Kostov RV. Glucosinolates and isothiocyanates in health and disease. Trends Mol. Med. 2012;18:337–347. doi: 10.1016/j.molmed.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 2013;15:195–218. doi: 10.1208/s12248-012-9432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Fernandez B, Ortiz A, Gomez-Guerrero C, Egido J. Therapeutic approaches to diabetic nephropathy--beyond the RAS. Nat. Rev. Nephrol. 2014;10:325–346. doi: 10.1038/nrneph.2014.74. [DOI] [PubMed] [Google Scholar]
- 13.Bomprezzi R. Dimethyl fumarate in the treatment of relapsing–remitting multiple sclerosis: an overview. Ther. Adv. Neurol. Disord. 2015;8:20–30. doi: 10.1177/1756285614564152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann C, Zeiher AM, Dimmeler S. Shear stress inhibits H2O2-induced apoptosis of human endothelial cells by modulation of the glutathione redox cycle and nitric oxide synthase. Arterioscler Thromb. Vasc. Biol. 1997;17:3588–3592. doi: 10.1161/01.atv.17.12.3588. [DOI] [PubMed] [Google Scholar]
- 15.De Keulenaer GW, et al. Oscillatory and steady laminar shear stress differentially affect human endothelial redox state: role of a superoxide-producing NADH oxidase. Circ. Res. 1998;82:1094–1101. doi: 10.1161/01.res.82.10.1094. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham KS, Gotlieb AI. The role of shear stress in the pathogenesis of atherosclerosis. Lab. Invest. 2005;85:9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- 17.Kim SK, Woodcroft KJ, Oh SJ, Abdelmegeed MA, Novak RF. Role of mechanical and redox stress in activation of mitogen-activated protein kinases in primary cultured rat hepatocytes. Biochem. Pharmacol. 2005;70:1785–1795. doi: 10.1016/j.bcp.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Lehoux S. Redox signalling in vascular responses to shear and stretch. Cardiovasc Res. 2006;71:269–279. doi: 10.1016/j.cardiores.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 20.Bae YS, Oh H, Rhee SG, Yoo YD. Regulation of reactive oxygen species generation in cell signaling. Mol. Cells. 2011;32:491–509. doi: 10.1007/s10059-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brewer TF, Garcia FJ, Onak CS, Carroll KS, Chang CJ. Chemical approaches to discovery and study of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins. Annu. Rev. Biochem. 2015;84:765–790. doi: 10.1146/annurev-biochem-060614-034018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 24.Green K, Brand MD, Murphy MP. Prevention of mitochondrial oxidative damage as a therapeutic strategy in diabetes. Diabetes. 2004;53(Suppl. 1):S110–S118. doi: 10.2337/diabetes.53.2007.s110. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa T, Araki E. Impact of mitochondrial ROS production in the pathogenesis of diabetes mellitus and its complications. Antioxid. Redox Signal. 2007;9:343–353. doi: 10.1089/ars.2006.1458. [DOI] [PubMed] [Google Scholar]
- 26.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codreanu SG, et al. Alkylation damage by lipid electrophiles targets functional protein systems. Mol. Cell. Proteomics. 2014;13:849–859. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niphakis MJ, et al. A global map of lipid-binding proteins and their ligandability in cells. Cell. 2015;161:1668–1680. doi: 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Tallman KA, Porter NA, Liebler DC. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal. Chem. 2015;87:2535–2541. doi: 10.1021/ac504685y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HY, Tallman KA, Liebler DC, Porter NA. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell Proteomics. 2009;8:2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furdui CM, Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom. Rev. 2014;33:126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Carroll KS, Liebler DC. The expanding landscape of the thiol redox proteome. Mol. Cell Proteomics. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol. Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi A, et al. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 36.Straus DS, et al. 15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc. Natl. Acad. Sci. USA. 2000;97:4844–4849. doi: 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji C, Kozak KR, Marnett LJ. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J. Biol. Chem. 2001;276:18223–18228. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph TK, Freeman BA. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009;2:re7. doi: 10.1126/scisignal.290re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diab A, et al. Peroxisome proliferator-activated receptor-gamma agonist 15-deoxy-Delta(12,14)-prostaglandin J(2) ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2002;168:2508–2515. doi: 10.4049/jimmunol.168.5.2508. [DOI] [PubMed] [Google Scholar]
- 40.Uchida K. Redox-derived damage-associated molecular patterns: ligand function of lipid peroxidation adducts. Redox Biol. 2013;1:94–96. doi: 10.1016/j.redox.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cubillos-Ruiz JR, et al. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell. 2015;161:1527–1538. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doyle K, Fitzpatrick FA. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 2010;285:17417–17424. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritz KS, et al. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem. Res. Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galligan JJ, et al. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 2014;136:11864–11866. doi: 10.1021/ja503604t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo M, et al. 15-Deoxy-Delta(12,14)-prostaglandin J(2): the endogenous electrophile that induces neuronal apoptosis. Proc. Natl. Acad. Sci. USA. 2002;99:7367–7372. doi: 10.1073/pnas.112212599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J. Biol. Chem. 2009;284:9176–9183. doi: 10.1074/jbc.M808656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chipuk JE, et al. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 2012;148:988–1000. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 49.Foyer C, Faragher R, Thornalley P, editors. Redox Metabolism and Longevity Relationships in Animals and Plants. Taylor & Francis; 2009. [PubMed] [Google Scholar]
- 50.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem. Res. Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 51.Dalleau S, Baradat M, Gueraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- 53.Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 54.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang X, et al. Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. J. Am. Chem. Soc. 2013;135:14496–14499. doi: 10.1021/ja405400k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin HY, Haegele JA, Disare MT, Lin Q, Aye Y. A generalizable platform for interrogating target- and signal-specific consequences of electrophilic modifications in redox-dependent cell signaling. J. Am. Chem. Soc. 2015;137:6232–6244. doi: 10.1021/ja5132648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parvez S, et al. Substoichiometric hydroxynonenylation of a single protein recapitulates whole-cell-stimulated antioxidant response. J. Am. Chem. Soc. 2015;137:10–13. doi: 10.1021/ja5084249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janssen DB. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 2004;8:150–159. doi: 10.1016/j.cbpa.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Los GV, Wood K. A novel technology for cell imaging and protein analysis. Methods Mol. Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 60.Los GV, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 61.Ohana RF, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 2009;68:110–120. doi: 10.1016/j.pep.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 62.Long MJC, Poganik JR, Aye Y. On-demand targeting: investigating biology with proximity-directed chemistry. J. Am. Chem. Soc. 2016;138:3610–3622. doi: 10.1021/jacs.5b12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 64.Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tjalkens RB, Cook LW, Petersen DR. Formation and export of the glutathione conjugate of 4-hydroxy-2, 3-E-nonenal (4-HNE) in hepatoma cells. Arch. Biochem. Biophys. 1999;361:113–119. doi: 10.1006/abbi.1998.0946. [DOI] [PubMed] [Google Scholar]
- 66.Volkel W, et al. Glutathione conjugates of 4-hydroxy-2(E)-nonenal as biomarkers of hepatic oxidative stress-induced lipid peroxidation in rats. Free Radic. Biol. Med. 2005;38:1526–1536. doi: 10.1016/j.freeradbiomed.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 67.Banerjee R. Redox Biochemistry. Wiley; 2007. [Google Scholar]
- 68.Cao Z, Hardej D, Trombetta LD, Li Y. The role of chemically induced glutathione and glutathione S-transferase in protecting against 4-hydroxy-2-nonenal-mediated cytotoxicity in vascular smooth muscle cells. Cardiovasc. Toxicol. 2003;3:165–177. doi: 10.1385/ct:3:2:165. [DOI] [PubMed] [Google Scholar]
- 69.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 70.Aye Y, Li M, Long MJC, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 71.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 72.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krief S, et al. Identification and characterization of cvHsp: a novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J. Biol. Chem. 1999;274:36592–36600. doi: 10.1074/jbc.274.51.36592. [DOI] [PubMed] [Google Scholar]
- 74.Rosenfeld GE, Mercer EJ, Mason CE, Evans T. Small heat shock proteins Hspb7 and Hspb12 regulate early steps of cardiac morphogenesis. Dev. Biol. 2013;381:389–400. doi: 10.1016/j.ydbio.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marvin M, et al. Developmental expression patterns of the zebrafish small heat shock proteins. Dev. Dyn. 2008;237:454–463. doi: 10.1002/dvdy.21414. [DOI] [PubMed] [Google Scholar]
- 76.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garber K. Biochemistry: a radical treatment. Nature. 2012;489:S4–S6. doi: 10.1038/489S4a. [DOI] [PubMed] [Google Scholar]
- 79.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc. Natl. Acad. Sci. USA. 1992;89:4544–4548. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uchida K, Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J. Biol. Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- 81.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem. Res. Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 82.Papa A, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Covey TM, Edes K, Coombs GS, Virshup DM, Fitzpatrick FA. Alkylation of the tumor suppressor PTEN activates Akt and β-catenin signaling: a mechanism linking inflammation and oxidative stress with cancer. PLoS One. 2010;5:e13545. doi: 10.1371/journal.pone.0013545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shearn CT, et al. Increased carbonylation of the lipid phosphatase PTEN contributes to Akt2 activation in a murine model of early alcohol-induced steatosis. Free Radic. Biol. Med. 2013;65:680–692. doi: 10.1016/j.freeradbiomed.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shearn CT, et al. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) inhibition by 4-hydroxynonenal leads to increased Akt activation in hepatocytes. Mol. Pharmacol. 2011;79:941–952. doi: 10.1124/mol.110.069534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol. Sci. 2011;32:131–140. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 87.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Covey TM, Edes K, Fitzpatrick FA. Akt activation by arachidonic acid metabolism occurs via oxidation and inactivation of PTEN tumor suppressor. Oncogene. 2007;26:5784–5792. doi: 10.1038/sj.onc.1210391. [DOI] [PubMed] [Google Scholar]
- 89.Ananthanarayanan B, Ni Q, Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc. Natl. Acad. Sci. USA. 2005;102:15081–15086. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Antal CE, Newton AC. Spatiotemporal dynamics of phosphorylation in lipid second messenger signaling. Mol. Cell Proteomics. 2013;12:3498–3508. doi: 10.1074/mcp.R113.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 92.Fellmann CL, Scott W. Stable RNA interference rules for silencing. Nat. Cell Biol. 2014;16:10–18. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Assou S, et al. Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Hum. Reprod. Update. 2011;17:272–290. doi: 10.1093/humupd/dmq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.García-Santamarina S, et al. Monitoring in vivo reversible cysteine oxidation in proteins using ICAT and mass spectrometry. Nat. Protoc. 2014;9:1131–1145. doi: 10.1038/nprot.2014.065. [DOI] [PubMed] [Google Scholar]
- 95.Lo Conte M, Lin J, Wilson MA, Carroll KS. A chemical approach for the detection of protein sulfinylation. ACS Chem. Biol. 2015;10:1825–1830. doi: 10.1021/acschembio.5b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang J, et al. Global in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 2015;10:1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 98.Grune T, Siems WG, Zollner H, Esterbauer H. Metabolism of 4-hydroxynonenal, a cytotoxic lipid peroxidation product, in Ehrlich mouse ascites cells at different proliferation stages. Cancer Res. 1994;54:5231–5235. [PubMed] [Google Scholar]
- 99.Srivastava S, et al. Metabolism of the lipid peroxidation product, 4-hydroxy-trans-2-nonenal, in isolated perfused rat heart. J. Biol. Chem. 1998;273:10893–10900. doi: 10.1074/jbc.273.18.10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blankespoor RL, et al. Photochemistry of 1-alkoxy- and 1-(benzyloxy)-9,10-anthraquinones in methanol: a facile process for the preparation of aldehydes and ketones. J. Org. Chem. 1995;60:6852–6859. [Google Scholar]
- 101.Wood KV. Luciferase Assay Method. 5,283,179. US patent. 1994
- 102.Sherf BA, Wood KV, Schenborn ET. Quenching Reagents and Assays for Enzyme-mediated Luminescence. 5,744,320. US patent. 1998
- 103.Grune T, Davies KJA. The proteasomal system and HNE-modified proteins. Mol. Aspects Med. 2003;24:195–204. doi: 10.1016/s0098-2997(03)00014-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.