Centromeres evolve rapidly and display a remarkable lack of sequence conservation (Henikoff et al., 2001). They can be small (125 bp in Saccharomyces cerevisiae) or large (9 million bp in maize) and show a relatively ordered arrangement of long repeats (Schizosaccharomyces pombe) or a wider array of small repeats interrupted by transposable elements (humans, Drosophila, and plants) (Choo, 1997). In addition, much of the DNA at centromeres appears to be unnecessary for function. There are many examples from both plants and animals to illustrate the impressive plasticity of centromeres, but few are as dramatic as human neocentromeres (Amor and Choo, 2002). Human neocentromeres have been found on 60 different chromosome fragments generated by rare breakage events. Such fragments have acquired new centromeres that lack any homology with the 23 normal centromeres in regions that generally would be considered typical euchromatin. Despite the adoption of novel centromeric DNA, human neocentromeres form fully functional kinetochores that are indistinguishable from those on true centromeres.
Discussions of the mechanisms that underlie centromere plasticity have focused on epigenetics in a general sense (Karpen and Allshire, 1997), structural codes that are not apparent in traditional sequence comparisons (Koch, 2000), or subtle DNA-protein binding preferences that are favored through evolutionary mechanisms such as meiotic drive (i.e., non-Mendelian segregation ratios; Henikoff et al., 2001). However, recent studies from the fission yeast S. pombe, particularly those by Volpe et al. (2002) and Hall et al. (2002), are likely to shift the focus of the discussion toward RNA and its wide-ranging effects in gene silencing and chromatin modification. At least for the regions surrounding the centromeres in this species, RNA appears to be a key epigenetic messenger for establishing the specialized chromatin organization required for chromosome segregation. The question now is whether these observations are broadly applicable to other eukaryotes and whether they can help explain the specification of the centromere itself.
RNA INTERFERENCE GENES ARE REQUIRED FOR S. POMBE PERICENTROMERE ORGANIZATION
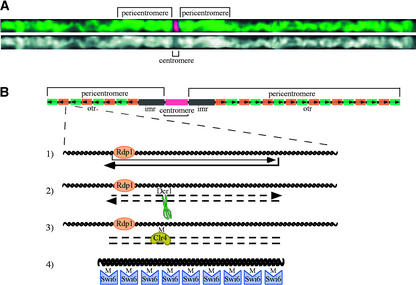
The centromeres of S. pombe are arranged in a manner similar to what is observed in metazoans, with a centromere core that binds the key kinetochore protein CENH3 flanked by repetitive regions analogous to the pericentromeric DNA of plants and animals (Figure 1). The centromeric domain usually is defined as those sequences that organize the kinetochore, the large multiprotein complex that orchestrates chromosome movement (Yu et al., 2000). By contrast, pericentromeres can be viewed as anchor zones that hold sister centromeres together and provide a boundary that isolates the centromeres from the chromosome arms, where most of the genes are found (Henikoff et al., 2001). In S. pombe, the pericentromeric sequences are indispensable and recruit the Rad21 subunit of cohesin that maintains chromatid cohesion until disjunction at anaphase (Bernard et al., 2001). Pericentromeric DNA in S. pombe can be subdivided into an innermost region (imr) composed of tRNA genes and a larger outermost region (otr). The otr contains alternating copies of related ∼4- to 5-kb repeats known as dg and dh that flank the imr in an inverted manner (Wood et al., 2002). The chromatin within S. pombe pericentromeres is characteristically heterochromatic and causes reporter genes to be silenced. By screening for mutations that allow the expression of pericentromere-inserted reporter genes, proteins required for chromatin modification and accurate chromosome segregation have been identified (Ekwall et al., 1999). The Heterochromatin Protein1 (HP1) homolog Swi6 and the histone methylase Clr4 are examples of proteins that meet the criteria of both repressing genes inserted in the centromere and providing functions that are required for accurate chromosome segregation.
Figure 1.
Centromere and Pericentromere Structures in Maize and S. pombe.
(A) Computationally straightened chromosome 4 from maize, derived from a three-dimensional rendering of a cell in the pachytene substage of meiosis I. The chromosome was stained for the CENH3 protein using indirect immunofluorescence (Zhong et al., 2002). Chromatin is shown in green and CENH3 is shown in red. At bottom is the chromatin-only image illustrating the euchromatic appearance of the centromere and the heterochromatic appearance of the pericentromere.
(B) Illustration of the centromere on S. pombe chromosome 3 and an interpretation of the data provided by Volpe et al. (2002). The dh and dg repeats are shown in orange and green, respectively; arrows indicate the directionality of the repeats. A small region in which transcripts were detected by Volpe et al. (2002) is shown in larger detail. Reverse transcripts were detected in wild-type cells, but forward transcripts were detected only in ago1−, dcr1−, rdp1−, and swi6− mutant cells. The forward transcript may be initiated at a low level from the otr and/or through the action of Rdp1 on the reverse transcript. The Ago1 and Dcr1 products may generate small RNA cleavage products that serve as cofactors for Clr4, the histone H3 methyltransferase in S. pombe. Once histone H3 is methylated at Lys-9, Swi6 (the S. pombe HP1 homolog) binds to the histone and establishes/reinforces the heterochromatic state.
Volpe et al. (2002) exploited the silencing-based assay for centromere function and the fact that S. pombe has only one copy of each of three genes known to be required for RNA interference (RNAi): one that encodes a helicase RNASEIII gene commonly known as dicer (dcr1+), another that encodes an RNA-dependent RNA polymerase (rdp1+), and a third that encodes ARGONAUTE (ago1+), a part of the RNAi silencing complex that cleaves and destroys mRNAs that are bound to small interfering RNAs (siRNAs). Mutations in these genes abolish RNAi in plants, animals, and fungi. The surprising finding was that ura4+ selectable marker transgenes inserted into either the imr or the otr region of the centromere were derepressed by mutations in ago1−, dcr1−, and rdp1−. The ago1− mutation is an allele of csp9 (Ekwall et al., 1999), which causes defects in chromosome segregation (cited by Volpe et al. [2002] as a personal communication with R.C. Allshire). Furthermore, forward and reverse transcripts corresponding to the dg-dh repeats were detected in each of the mutant strains. Nuclear run-on experiments indicated that the reverse transcript was always transcribed but presumably destroyed by RNAi in wild-type cells. A forward transcript, which is not present in wild-type strains but is detected in ago1−, dcr1−, rdp1−, and swi6− mutants, likely initiates RNAi. Strong support for this interpretation came from an accompanying article (Reinhart and Bartel, 2002), which demonstrated that a large percentage of the small RNAs cloned from S. pombe, which were thought to be dicer cleavage products, showed perfect homology with the portions of the centromere affected by the RNAi pathway (Volpe et al., 2002).
Derepression of ura4+ in the centromere also was correlated with a reduction in the levels of Lys-9–methylated histone H3 (H3 Lys-9) at dh repeats as well as the inserted ura4+ genes. H3 Lys-9 is a well-documented characteristic of heterochromatin that is required for the recruitment of HP1/Swi6, a protein that helps to establish, reinforce, and extend the heterochromatic state along the chromosome (Richards and Elgin, 2002). As expected, each of the RNAi mutations caused the delocalization of Swi6 from the centromere. A final experiment supporting a functional relationship between RNAi and chromatin struc-ture was an experiment showing that the Rdp1 protein interacts with the dg repeat by coimmunoprecipitation, providing preliminary evidence that the RNA processing events may occur in cis (i.e., in close proximity to the chromatin being modified). The inverted repeat structure of the otr is similar to the inverted repeats of class-II transposons, suggesting that dg and dh are rem-nants of ancient transposable elements (Volpe et al., 2002). Because genetic evidence from several organisms indicates that transposable elements are regulated by RNAi (Waterhouse et al., 2001), the elaborate RNAi-mediated chromatin modification system may be the evolutionary outcome of a primitive transposon repression system (Volpe et al., 2002).
The general model described by Volpe et al. (2002) is supported by a number of observations from different organisms. In S. pombe, supporting data were published 2 weeks later in a study of the mating-type (mat) locus (Hall et al., 2002). Mating-type switching depends on a 7.5-kb sequence known as the K region, which contains a transcribed 4.3-kb sequence known as CenH that is 96% homologous with the dg-dh region of the centromeric otr. Mating-type interconversion and silencing at the mat locus are strongly correlated, and gene silencing can be initiated at an ectopic site by CenH. Hall et al. (2002) used high-resolution chromatin immunoprecipitation analysis to show that CenH functions as a nucleation center for H3 Lys-9 methylation, recruiting the Swi6 protein, which then spreads in cis along the chromosome. When CenH was moved to an ectopic site, the same phenomena were observed. Mutations in the RNAi genes ago1−, dcr1−, and rdp1− abolished the silencing effect of CenH at an ectopic site, strongly supporting the results of Volpe et al. (2002). Interestingly, RNAi mutations had no effect on an already-silenced mating-type locus; however, when the silenced state was erased by treatment with trichostatin A (a histone deacetylase inhibitor that causes H3 Lys-9 to be acetylated, not methylated) or by passage through a clr4− background (Clr4 methylates H3 at Lys-9), the silenced state could not be reestablished unless functional alleles of the RNAi genes were present. These data suggest that RNAi, at least when acting in concert with dg-dh homologous sequences in S. pombe, has a more important role in establishing the silenced state than in maintaining it.
The new literature based on experiments with S. pombe is consistent in many ways with extensive data from plants (Matzke et al., 2001) and more recent information from animals (Pal-Bhadra et al., 2002), which suggest a similar link between RNAi and transcriptional silencing of specific genes. For instance, in plants, a construct that forms double-stranded RNA homologous with the nopaline synthase promoter causes methylation of the promoter in trans (Aufsatz et al., 2002). The DNA methylation and histone modification pathways are known to interact and reinforce each other (Richards and Elgin, 2002), and the H3 Lys-9–HP1 association is highly conserved, suggesting that the link between RNAi and HP1 recruitment also may be conserved. Perhaps the strongest supporting evidence comes from studies in mouse cell lines, in which it was shown that RNase disrupts the spatial ordering of methylated histone H3 and HP1 in pericentromeres, as assayed by antisera that preferentially recognize higher order chromatin structure (Maison et al., 2002).
Together, the available data suggest that the RNAi pathway, presumably working in concert with the resulting small RNA products, can modify the chromatin state of the DNA that encodes the RNAs, effectively “converting nonspecific sequence information into distinct chromatin states” (Jenuwein, 2002). In the S. pombe pericentromere, distinct chromatin states are required for accurate chromosome segregation. How RNAi affects the organization of chromatin is not yet known, but it seems likely that small RNAs or RNA intermediates are involved in directing the chromatin-modifying enzymes to specific DNA sequences. One idea is that siRNAs bind transiently to the DNA by base pairing (Matzke et al., 2001), creating an unusual DNA structure that could attract these proteins. Another possibility is that a high local concentration of siRNAs may be all that is necessary (Volpe et al., 2002). The S. pombe RNA-dependent RNA polymerase (Rdp1) associates with the otr, where it is presumed to polymerize double-stranded RNAs from the reverse otr transcripts (Figure 1). During polymerization, the double-stranded RNAs would be bound to the chromatin as well; the double-stranded RNAs or siRNAs produced by the RNAi pathway could rapidly recruit chromatin-modifying enzymes in a less sequence-specific manner. Supporting both models is the fact that the Su(var)3-9 class of histone H3 Lys-9 methyltransferases (e.g., Clr4 of S. pombe) and the chromomethylases of plants contain chromodomains (Matzke et al., 2001), which are sequence motifs that have RNA binding properties (Akhtar et al., 2000). In Drosophila, the chromodomain of MOF (Males absent On the First) histone acetyltransferase interacts with a cellular RNA that is required for its localization to chromatin (Akhtar et al., 2000; Meller and Rattner, 2002).
WHAT ABOUT THE CENTROMERE?
Although the developments of recent months may provide the beginnings of a general model for the specification and organization of the pericentromeric domain, they also raise the question of whether RNAi is involved in centromere organization. Unlike in S. pombe, in which the major repeats are found in pericentromeres, the most highly repetitive sequences in metazoans are centromeric. The tandemly arrayed human α-satellites and maize CentC repeats (Ananiev et al., 1998) interact strongly with the conserved CENH3 protein that is thought to initiate kinetochore formation (Vafa and Sullivan, 1997; Zhong et al., 2002). The tandem repeats are located within the primary constrictions of chromosomes, which in plants often appear euchromatic (Figure 1A). This contrasts with the pericentromeres of metazoans, which stain densely in the manner typical of heterochromatin and are composed of degenerate repeats and a multitude of transposable elements.
Perhaps the most encouraging evidence in favor of the involvement of RNAi in the CENH3 binding centromere domain is the apparent involvement of transposable elements in both animal and plant centromere function. Centromere protein B (CENP-B) was one of the first kinetochore proteins identified in mammals, and it has been characterized extensively. Interestingly, CENP-B shows a high degree of similarity to the transposase of the TC1/mariner class of transposable elements and retains several of the key residues required for transposition (Kipling and Warburton, 1997). CENP-B binds to a specific sequence motif, the CENP-B box, which is present on a large fraction of the α-satellite in humans. Although mice homozygous for cenp-b mutations have no apparent segregation defects (Tomascik-Cheeseman et al., 2002), the CENP-B box contributes to the efficiency of artificial chromosome formation in humans, suggesting that CENP-B may help to establish centromeric chromatin (Ohzeki et al., 2002). CENP-B is a conserved protein, at least to the extent that three CENP-B homologs also exist in S. pombe. Two of the S. pombe CENP-B homologs contribute to the methylation of H3 Lys-9 at the centromere, and all three cooperate to recruit Swi6 to the otr (Nakagawa et al., 2002). It is possible that CENP-B is involved in the same RNAi-mediated pathway for heterochromatin assembly described by Volpe et al. (2002). Therefore, CENP-B provides not only a link between centromeres and transposable elements but a potential link between centromeres (mammals) and pericentromeres (S. pombe). Genes that have homology with CENP-B have been found in other organisms, but to date none has been shown to localize to centromeres or to function in chromosome segregation.
Additional evidence in favor of a role for transposons at the centromere comes from comparative and functional studies of the centromeres in cereal grains. Early studies in sorghum and Brachypodium identified small probes that in fluorescence in situ hybridization experiments identified centromeres with high specificity in a broad range of cereal grains, including rice, maize, wheat, barley, rye, and oat (Aragon-Alcaide et al., 1996; Jiang et al., 1996). Further studies established that the pan-cereal centromere probes are portions of sequence from a conserved clade of gypsy retrotransposons, known collectively as the centromeric retrotransposable (CR) elements. Sequence comparisons of the known full-length elements indicate that the CR elements are among the most conserved transposable elements known, with up to 80% sequence homology maintained over 60 million years (Zhong et al., 2002). BAC-scale sequencing and high-resolution fluorescence in situ hybridization have revealed that CR elements are interspersed with satellite repeats (Hudakova et al., 2001; Cheng et al., 2002). Most recently, it was shown that the CR elements from maize interact strongly with CENH3 in chromatin immunoprecipitation experiments, suggesting that they are components of the functional centromeric DNA (Zhong et al., 2002). To date, there are no small RNA libraries from cereal grains to suggest that CR elements are subject to RNAi. However, Athila retroelements found within and around Arabidopsis centromeres appear to be regulated by RNAi (Haupt et al., 2001; Llave et al., 2002).
If RNAi is involved in CENH3-mediated centromere function, the mechanisms probably are quite different from what has been suggested for S. pombe. The CENH3s in animals are deposited in a replication-independent manner by means that are poorly understood (Ahmad and Henikoff, 2002). Once deposited, CENH3 necessarily removes any H3-specific modifications and associated proteins. As a group, the CENH3s have highly divergent N-terminal tails that bear little resemblance to the tail of histone H3 (Henikoff et al., 2001); the only post-translational modification known is a human Ser-7 phosphorylation, which seems to play little or no role in chromosome segregation (Zeitlin et al., 2001). In addition, the small RNAs that are anticipated from the S. pombe model for pericentromere formation have yet to be identified for the centromeric sequences that interact with CENH3. This fact does not exclude the involvement of RNAi but suggests that the active molecules may be RNAi processing intermediates, double-stranded RNAs, or siRNAs that are tightly associated with the centromere/kinetochore complex and difficult to clone. The open chromatin configuration typical of centromeres (Figure 1A) suggests that transcription may occur there. Future work is likely to take advantage of the already extensive collection of RNAi mutants in organisms such as Arabidopsis and more biochemically oriented strategies designed to detect RNAs associated with centromeric chromatin.
References
- Ahmad, K., and Henikoff, S. (2002). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99, 16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, A., Zink, D., and Becker, P.B. (2000). Chromodomains are protein-RNA interaction modules. Nature 407, 405–409. [DOI] [PubMed] [Google Scholar]
- Amor, D.J., and Choo, K.H. (2002). Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71, 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., Winden, J.V.D., Matzke, A.J., and Matzke, M. (2002). RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, C.R., Gu, M., Blattner, F.R., and Jiang, J. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. (1997). The Centromere. (New York: Oxford University Press).
- Ekwall, K., Cranston, G., and Allshire, R.C. (1999). Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153, 1153–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. (2002). Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P.F., and Torres-Ruiz, R.A. (2001). The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27, 285–296. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hudakova, S., Michalek, W., Presting, G.G., ten Hoopen, R., dos Santos, K., Jasencakova, Z., and Schubert, I. (2001). Sequence organization of barley centromeres. Nucleic Acids Res. 29, 5029–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T. (2002). An RNA-guided pathway for the epigenome. Science 297, 2215–2218. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Nasuda, A., Dong, F., Scherrer, C.W., Woo, S.-S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case of epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Kipling, D., and Warburton, P.E. (1997). Centromeres, CENP-B and Tigger too. Trends Genet. 13, 141–145. [DOI] [PubMed] [Google Scholar]
- Koch, J. (2000). Neocentromere and alpha satellite: A proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 9, 149–154. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A.H., Quivy, J.P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. (2002). Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Matzke, A.J., and Kooter, J.M. (2001). RNA: Guiding gene silencing. Science 293, 1080–1083. [DOI] [PubMed] [Google Scholar]
- Meller, V.H., and Rattner, B.P. (2002). The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, H., Lee, J.K., Hurwitz, J., Allshire, R.C., Nakayama, J., Grewal, S.I., Tanaka, K., and Murakami, Y. (2002). Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16, 1766–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki, J., Nakano, M., Okada, T., and Masumoto, H. (2002). CENP-B box is re-quired for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. (2002). RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9, 315–327. [DOI] [PubMed] [Google Scholar]
- Reinhart, R.J., and Bartel, D.P. (2002). Small RNAs correspond to centromere heterochromatic repeats. Science 297, 1831. [DOI] [PubMed] [Google Scholar]
- Richards, E.J., and Elgin, S.C. (2002). Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 108, 489–500. [DOI] [PubMed] [Google Scholar]
- Tomascik-Cheeseman, L., Marchetti, F., Lowe, X., Shamanski, F.L., Nath, J., Pedersen, R.A., and Wyrobek, A.J. (2002). CENP-B is not critical for meiotic chromosome segregation in male mice. Mutat. Res. 513, 197–203. [DOI] [PubMed] [Google Scholar]
- Vafa, O., and Sullivan, K.F. (1997). Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wood, V., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S.G., Shelby, R.D., and Sullivan, K.F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]