Abstract
Objective: Hepatocellular carcinoma (HCC) is still one of the most common death-related malignancies worldwide. Because the way onset and progression are hidden most, HCC diagnoses are made at an advanced stage, when they are unsuitable for surgical resection. MicroRNAs are a class of small non-coding RNAs, participating in many aspects of cancers. In this study, we tried to establish the role of microRNA-718 (miR-718) in the malignant phenotype of HCC cells and its possible role in HCC diagnosis. Methods: Here we first used a methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay, Transwell migration and invasion assays, and colony formation assay to evaluate the impact of miR-718 on the malignant phenotypes of HCC cells. Then, we used bioinformatic methods to predict the target gene of miR-718 and used green fluorescence protein (GFP) reporter assay, Western blot, and quantitative real-time polymerase chain reaction (qRT-PCR) to validate the regulation relationship. Finally, we determined the role of the target gene in the HCC phenotype. Results: We found that the expression of miR-718 was significantly reduced in various HCC cell lines and HCC tissues. Re-expression of miR-718 significantly reduced the cellular viability and colony formation ability as well as inhibited the migration and invasion abilities of HCC cell lines. Early growth response protein 3 (EGR3) is a direct target of miR-718 and is negatively regulated by miR-718. EGR3 could increase the viability and proliferation of HCC cells, and promot the migration and invasion of HCC cells. Conclusions: miR-718 acts as a tumor suppressive microRNA in HCC via regulating the expression of EGR3, which may provide a new diagnostic marker and treatment target for HCC.
Keywords: miR-718; MicroRNA; Early growth response protein 3 (EGR3); Hepatocellular carcinoma (HCC); Malignant phenotype
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent and aggressive human malignancies worldwide, and it has a very poor prognosis (Waller et al., 2015). Men are more susceptible to HCC than women and it is a leading cause of cancer-related death of men around the world (Fong and Tanabe, 2014; Kimhofer et al., 2015). The onset and progress of HCC are not evident, and thus most HCC cases are diagnosed at an advanced stage, which may mean that they are not then suitable for surgical resection. Additionally, the drugs for advanced HCC are limited and ineffective, which contributes to the poor prognosis (Bronte et al., 2015; Sugimachi et al., 2015). Thus, novel strategies for the early diagnosis and therapy of HCC need to be developed. MicroRNA, about 21–23 nucleotides in length, is one of the most important regulators of gene expression in eukaryocytes (Shukla et al., 2011; Ameres and Zamore, 2013). In most cases, microRNAs downregulate the expression of their target genes through repressing the translation of targeting mRNA or mediating the degradation of targeting mRNA via binding to the 3' untranslated regions (3' UTR) of their targets (Bartel, 2009; Shukla et al., 2011). Dysregulation of microRNAs has been reported in many kinds of human diseases, including cancers (Felekkis et al., 2010; Hu et al., 2010). MicroRNAs participate in many aspects of HCC, including cell proliferation, apoptosis, migration, invasion, and vasculogenic mimicry (Wan et al., 2014; Singh et al., 2015). Although the role of microRNAs in the pathogenesis of HCC has been extensively studied, the role of microRNA-718 (miR-718) in HCC onset and progression is still unknown.
Our present work demonstrated that miR-718 may function as a tumor suppressive microRNA through targeting early growth response protein 3 (EGR3) in HCC, but EGR3 has an opposite effect on HCC cell lines. This may shed new light on the mechanism of how microRNA affects the onset and progression of HCC.
2. Materials and methods
2.1. Materials
Five pairs of hepatic tissue, consisting of human HCC and matched normal hepatic tissue from the same patient, were used in this study. All samples were obtained with the patients’ informed consent. The samples were received from the Department of Oncology, Affiliated Hospital of Taishan Medical University (Tai’an, China). Total RNA was extracted from the tissue samples and purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions.
2.2. Cell culture and transfection
HepG2 was cultured in minimum essential medium (MEM) with 10% fetal bovine serum (Gibco, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Hyclone, China). SMMC-7721, LO2, and QGY-7703 were cultured in RPMI 1640 medium with 10% fetal bovine serum and 100 U/ml penicillin and 100 μg/ml streptomycin. HepG2, LO2, QGY-7703, and SMMC-7721 were maintained in a humidified incubator at 37 °C with 5% CO2. HepG2, LO2, and QGY-7703 were transfected with the target vectors, respectively, using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
2.3. Antibody
Anti-EGR3 antibody was purchased from Univ-Biology Company, Tianjin, China.
2.4. Plasmid construction
For the miR-718 expression vector (pri-miR-718), the region containing miR-718 precursor was amplified from genomic DNA and cloned into the pcDNA3 vector. Pri-miR-718-S-BI: CGCGGATCC ACCCAGGGCGGCACCTCGTA; Pri-miR-718-AS-EI: CCGGAATTCGCCGTTCGTCCTCCAGAGCC. The 2'-O-methyl-modified miR-718 antisense oligonucleotide (ASO-miR-718) was commercially synthesized as an inhibitor of miR-718. ASO-miR-718: CGACGCCCGGCGGGGCGGAAG; ASO-NC: CA GUACUUUUGUGUAGUACAA. The segment of 3' UTR of EGR3 containing miR-718 was acquired by polymerase chain reaction (PCR) with gene specific primers, and then cloned into pcDNA3/luciferase following the stop codon of luciferase with BamHI and HindIII sites. EGR3-UTR-BI-S: GATCCCGTGGGGGCGGAAAGGTGGCGTCAAGCTTG; EGR3-UTR-EcoRI-AS: AATTCAAGCTTGACGCCACCTTTCCGCCCCCACGG.
To construct EGR3 over-expression plasmid, PCR products, acquired with EGR3 specific primers, were cloned into pcDNA3, with BamHI and EcoRI sites. EGR3-S-BI: CGCGGATCCATGACCGGCAAACTCGCCGAGAAGC; EGR3-AS-EI: CCGGAATTCGGCGCAGGTGGTGACCACGGGGGCC.
2.5. RNA isolation and detection
Total RNA was isolated with TRIzol reagent (Sigma, Shanghai, China) as per the manufacturer’s instructions. The quality and integrity of the RNA were evaluated by Nanodrop and gel electrophoresis, respectively. For reverse transcription, 1 μg of total RNA was reverse-transcripted with miR-718, U6 RT primers, or Oligo (dT) with Moloney murine leukemia virus (M-MLV) revertase. Real time PCR was performed with kits.
2.6. MTT assay
Cells, post-transfected with pri-miR-718, ASO-miR-718, pEGR3, or shR-EGR3, or in the control group, were seeded in a 96-well plate at 5000 cells/well, and cell viabilities were detected by methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay at the indicated time.
2.7. Colony formation assay
Cells transfected with pri-miR-718 or EGR3 expression plasmid or respective controls were seeded in a 24-well plate in 300 cells/well, and then cultured for about 14 d before staining with crystal violet, and any colony with more than 50 cells was counted.
2.8. Target prediction
The potential targets of miR-718 were predicted by Targetscan 7.1 (http://www.targetscan.org) and MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5 ), and the predicted targets were selected in accordance with the phenotype of miR-718.
2.9. Luciferase reporter assay
The enhanced green fluorescence protein (EGFP) reporter plasmid with EGR3 3' UTR containing miR-718 targeting sites was transfected into SMMC-7721 and HepG2 cells with Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA, USA), and red fluorescent protein (RFP) expressing plasmid was integrated as the transfection efficiency control. Cells were lysised 48 h post transfection, and the intensities of EGFP and RFP fluorescence were determined with a spectrophotometer.
2.10. Statistical analysis
Two-tailed Student’s t tests were used for comparisons, and each experiment was performed at least three times. A P-value less than 0.05 was considered significant (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, ns = not significant).
3. Results
3.1. Tumor suppressive function of miR-718 in HCC
miR-718 has been reported to be downregulated in many kinds of cancers, but its role in HCC is not well studied. In order to find out the possible function of miR-718 in HCC, we determined the expression of miR-718 in several HCC cell lines and the normal hepatocellular cell L02 by quantitative real-time PCR (qRT-PCR). In contrast with the L02 cell, the expressions of miR-718 in SMMC-7721, QGY-7703, and HepG2 HCC cell lines were significantly reduced (Fig. 1a), implying a tumor suppressive function of miR-718 in HCC. To validate this speculation, we used serial cellular experiments to investigate the function of miR-718 in HCC cells. The expressing plasmids of miR-718 and specific ASO-miR-718 could efficiently increase or reduce the mature miR-718 level in HCC cell lines (Fig. 1b), and miR-718 significantly reduced cellular viability (Figs. 1c and 1d) and colony formation ability (Figs. 1e and 1f) of HepG2 and SMMC-7721 cells as detected by MTT assay and colony formation assay, respectively. We further used Transwell migration and invasion assays to measure the influence of miR-718 on the mobility ability of HCC cells. In accordance with the above results, miR-718 inhibited the migration and invasion of HepG2 and SMMC-7721 cells (Figs. 1g–1j), further supporting the tumor inhibitory function of miR-718 in HCC cells.
Fig. 1.
Function of miR-718 in HCC cells
(a) Expressions of miR-718 in L02 and HCC cell lines were determined by qRT-PCR. (b) Efficiency of pri-miR-718 expression plasmid or ASO-miR-718 was confirmed by qRT-PCR. (c, d) Impacts of miR-718 on HepG2 and SMMC-7721 cellular viabilities were determined by MTT assay. (e, f) Colony formation abilities of HepG2 and SMMC-7721 cells with indicated treatment were determined by colony formation assay. Impacts of miR-718 on the migration (g, h) and invasion (i, j) abilities of HepG2 and SMMC-7721 cells were evaluated by Transwell migration and invasion assays, respectively. Data were expressed as mean±standard deviation (SD). All experiments were repeated three times. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001 vs. the control group
3.2. Regulation of miR-718 in the expression of EGR3 in HCC
Most microRNAs elicit their bio-functions through regulating the expression of their target genes. In an attempt to find out how miR-718 performs its tumor suppressive function, we first predicted the potential target gene of miR-718. As shown in Fig. 2a, there is a predicted target site of miR-718 in EGR3 mRNA 3' UTR in mammals, indicating that miR-718 might regulate the expression of EGR3. To experimentally explore whether EGR3 is a bona fide miR-718 target, we detected the expression of EGR3 in L02 and several other HCC cell lines. Interestingly, compared with the L02 cell, the protein level of EGR3 was significantly increased in SMMC-7721, QGY-7703, and HepG2 HCC cell lines (Fig. 2b), while the expression of miR-718 in these cells had the opposite trends (Fig. 1a), which demonstrated that EGR3 might be regulated by miR-718 in HCC cells. We then constructed an EGFP reporter plasmid containing miR-718 targeting sites in EGR3 3' UTR. The plasmid was co-transfected with miR-718 overexpressing plasmid or ASO-miR-718 into HepG2 or SMMC-7721 cells. As indicated in Fig. 2c, the expression of EGFP was significantly affected by miR-718 in both cell lines, and mutations of miR-718 seed targeting sites abolished the impact of miR-718 on the expression of EGFP, indicating that miR-718 is directly targeting EGR3 3' UTR. To further investigate the relationship between miR-718 and EGR3, we detected the impacts of miR-718 on endogenous EGR3 mRNA and protein levels. In accordance with the results of EGFP reporter assay, miR-718 significantly reduced EGR3 mRNA and protein in both HepG2 and SMMC-7721 cells (Figs. 2d and 2e). All the above results demonstrated that EGR3 is a bona fide target gene of miR-718, and is negatively regulated by miR-718 in HCC cells.
Fig. 2.
Regulation of miR-718 in the expression of EGR3 in HCC cells
(a) Targeting sites of miR-718 in human and other mammal EGR3 mRNA 3' UTR are shown. (b) Expressions of EGR3 in normal hepatocellular cell L02 and SMMC-7721, QGY-7703, and HepG2 HCC cells were detected by Western blot. (c) EGFP intensities of HepG2 and SMMC-7721 cells with indicated treatment were determined by spectrophotometer, and the value of the control group (pcDNA3 or ASO-NC) was set to one. (d) EGR3 mRNA levels in HepG2 and SMMC-7721 cells with indicated treatment were measured by qRT-PCR. (e) EGR3 protein levels in HepG2 and SMMC-7721 cells transfected with pri-miR-718 or ASO-miR-718 and respective controls were determined by Western blot. Data were expressed as mean±standard deviation (SD). All experiments were repeated three times. * P<0.05, ** P<0.01 vs. the control group. ns: not significant
3.3. Tumor promoting function of EGR3 in HCC
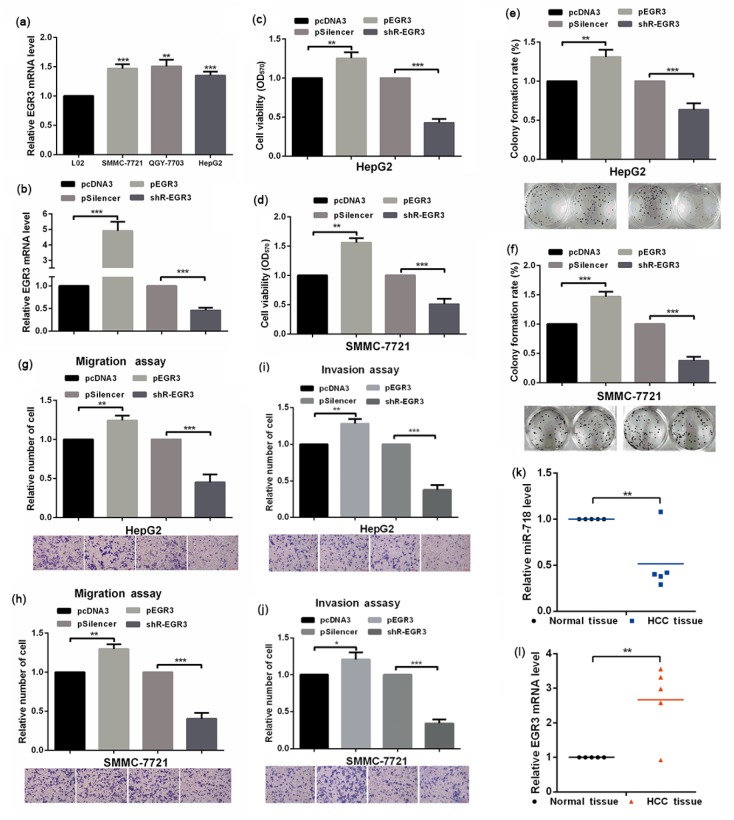
The above results showed that miR-718 is downregulated in HCC cells and functions as a tumor inhibitory microRNA, and EGR3 is negatively regulated by miR-718. We sought to find out whether miR-718 performs its tumor inhibitory role through EGR3. To answer this question, we tested the role of EGR3 in the malignancy phenotype of HCC cell lines. The expression of EGR3 was significantly increased in HCC cell lines (Fig. 3a), and the overexpression or knockdown plasmid of EGR3 was effective in HCC cell lines (Fig. 3b). Overexpression of EGR3 in HepG2 and SMMC-7721 cells increased cellular viability, while knockdown of EGR3 reduced it (Figs. 3c and 3d). The colony formation ability of HepG2 and SMMC-7721 cells was also increased by EGR3 (Figs. 3e and 3f), indicating that EGR3 functions as the tumor promoting factor in HCC cell lines. The increase of migration and invasion ability of HepG2 and SMMC-7721 cells by EGR3 further supported its tumor promoting function in HCC cells (Figs. 3g–3j). We also examined the levels of miR-718 and EGR3 in hepatocellular cancer and normal tissue by qRT-PCR, and the results showed that miR-718 had lower expression in hepatocellular cancer tissue than in normal tissue, but the level of EGR3 was highly expressed in hepatocellular cancer tissue (Figs. 3k and 3l). In all, these results demonstrated that EGR3 functions in the opposite direction to miR-718, and promotes the malignancy phenotype of HCC cells.
Fig. 3.
Effect of EGR3 on the malignancy phenotype in HCC cells
(a) Expressions of EGR3 in L02, SMMC-7721, QGY-7703, and HepG2 cells were determined by qRT-PCR. (b) Efficiency of EGR3 overexpression plasmid or knockdown plasmid was confirmed by qRT-PCR. (c, d) Impacts of EGR3 on HepG2 and SMMC-7721 cellular viabilities were determined by MTT assay. (e, f) Colony formation abilities of HepG2 and SMMC-7721 cells with indicated treatment were determined by colony formation assay. (g–j) Impacts of EGR3 on the migration and invasion abilities of HepG2 and SMMC-7721 cells were evaluated by Transwell migration and invasion assays. (k, l) Levels of miR-718 and EGR3 in HCC and normal tissues were examined by qRT-PCR. Data were expressed as mean±standard deviation (SD). All experiments were repeated three times. * P<0.05, ** P<0.01, *** P<0.001 vs. the control group
3.4. Rescue assay of the functional target of miR-718 in HCC cells
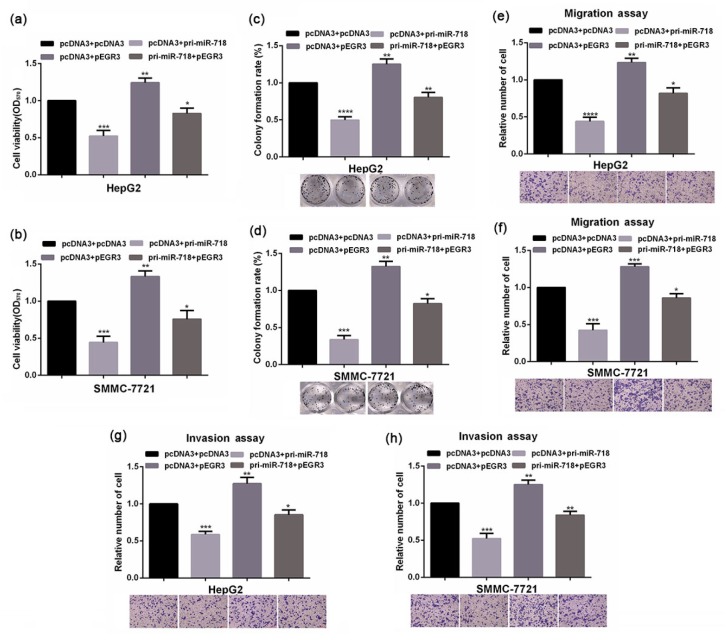
A microRNA can have many targets to elicit its bio-function, and in a given cell line microRNA may perform its function via a specific target gene. To find out whether EGR3 is a functional target of miR-718 in HCC cells, we conducted a rescue assay. Simultaneously transfected HepG2 and SMMC-7721 cells with an EGR3 expression plasmid were refractory to microRNA regulation, and miR-718 could restore the expression of EGR3 in these cells. Enhanced expression of EGR3 significantly increased the cellular viability, colony formation ability, and migration and invasion abilities of HCC cells, and the inhibitory effects of miR-718 on the malignancy phenotype of HCC cells were partially rescued by EGR3 (Fig. 4). Taken together all the above results indicated that miR-718 performs its tumor inhibitory function via downregulating the expression of EGR3 in HCC cells.
Fig. 4.
Rescue assay of the functional target of miR-718 in HCC cells
(a, b) Viabilities of HepG2 and SMMC-7721 cells with indicated treatment were tested by MTT assay. (c, d) Colony formation abilities of HepG2 and SMMC-7721 cells with indicated treatment were measured by colony formation assay. (e–h) Transwell migration and invasion assays were performed in HepG2 and SMMC-7721 cells with indicated treatments. Data were expressed as mean±standard deviation (SD). All experiments were repeated three times. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001 vs. the control group
4. Discussion
miRNAs are important regulators of many genes at the post-transcriptional level, and much more evidence has demonstrated that abnormal expression of miRNAs is closely related to the formation of cancer in human beings. Furthermore, increasing number of cancer-specific miRNAs have been considered as potential tumor biomarkers in various types of cancers (Hu et al., 2010; Lin et al., 2011). However, the functions of many miRNAs have not been elucidated so far.
Prior reports have suggested that miR-718 is downregulated in ovarian cancer cells and HCC cells (Leng et al., 2014; Sugimachi et al., 2015). miR-718 can exhibit inhibitory effects against ovarian cancer through VEGF repression and it also can suppress cell proliferation in HCC cell lines (Huh7 and PLC/PRF/5) by targeting the HOXB8 gene. In the present study, qRT-PCR results validated that miR-718 expression was significantly downregulated in other HCC cell lines, such as SMMC-7721, QGY-7703, and HepG2. In addition, our results demonstrate the tumor inhibitory function of miR-718 in HepG2 and SMMC-7721 cells.
miRNAs can take effect by depending on the degree of complementarities with the 3' UTR of their target genes (Farazi et al., 2008). In this study, bioinformatics was used to predict target genes, and EGR3 might be a candidate target. Interestingly, we found that the expression of EGR3 was significantly increased in SMMC-7721, QGY-7703, and HepG2 HCC cell lines, which was inversely correlated with the miR-718 expression level. Furthermore, EGFP fluorescence reporter assay showed that EGR3 was a bona fide target gene of miR-718, and is negatively regulated by miR-718 in HCC cells.
EGR3 belongs to the EGR family of transcription factors that can regulate a wide range of biological processes (Fang et al., 2013), including central nervous system development, muscle stretch receptor function, angiogenesis, immunity, and cancer (Li et al., 2007; Gomez-Martin et al., 2010; Perez-Cadahia et al., 2011; Baron et al., 2015). Although evidence of EGR3 playing certain roles in cancer remains scant, it has been shown that EGR3 was relevant to the breast cancer cells, gastric cancer and prostate cancer cells (Suzuki et al., 2007; Liao et al., 2013; Pio et al., 2013). However, whether EGR3 displays correlation towards HCC remains unknown.
The current study indicated for the first time that EGR3 is highly expressed in HCC cells, enhancing HCC cell viability, colony formation, and migration and invasion abilities. Suzuki et al. (2007) found that overexpression of EGR3 in breast cancer cells increased cell invasion in vitro and in vivo. However, other evidence also showed that EGR3 expression was lower in gastric cancer tissue than in normal tissue (Liao et al., 2013). This indicated that EGR3 may exert its function in tissue and in a tumor-specific manner. In particular, our results demonstrated that EGR3 promotes the malignancy phenotype of HCC cells in the opposite direction to miR-718. The dysregulation of miR-718 performs its tumor inhibitory function via downregulating the expression of EGR3 in HCC cells. Although we have confirmed that EGR3 is another target of miR-718, the mechanism of EGR3 promoting the malignancy phenotype of HCC cells remains unclear. In addition, the mechanism that regulates the expression of miR-718 in HCC cells is not well understood. Therefore, the detail mechanism needs further investigation.
In conclusion, miR-718 functions as a tumor suppressive microRNA in HCC cells, and inhibits the growth of HCC in vitro through downregulating the expression of EGR3.
Footnotes
Project supported by the Science and Technology Project of Higher Education of Shandong Province (No. J12LK07), China
Compliance with ethics guidelines: Zhong-dong WANG, Fan-yong QU, Yuan-yuan CHEN, Zhang-shen RAN, Hai-yan LIU, and Hai-dong ZHANG declare that they have no conflict of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
References
- 1.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–488. doi: 10.1038/nrm3611. (Available from: http://dx.doi.org/10.1038/nrm3611) [DOI] [PubMed] [Google Scholar]
- 2.Baron VT, Pio R, Jia Z, et al. Early growth response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br J Cancer. 2015;112(4):755–764. doi: 10.1038/bjc.2014.622. (Available from: http://dx.doi.org/10.1038/bjc.2014.622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. (Available from: http://dx.doi.org/10.1016/j.cell.2009.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronte F, Bronte G, Fanale D, et al. Hepatomirnoma: the proposal of a new network of targets for diagnosis, prognosis and therapy in hepatocellular carcinoma. Crit Rev Oncol Hematol. 2015;97:312–321. doi: 10.1016/j.critrevonc.2015.09.007. (Available from: http://dx.doi.org/10.1016/j.critrevonc.2015.09.007) [DOI] [PubMed] [Google Scholar]
- 5.Fang F, Shangguan AJ, Kelly K, et al. Early growth response 3 (Egr-3) is induced by transforming growth factor-β and regulates fibrogenic responses. Am J Pathol. 2013;183(4):1197–1208. doi: 10.1016/j.ajpath.2013.06.016. (Available from: http://dx.doi.org/10.1016/j.ajpath.2013.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135(7):1201–1214. doi: 10.1242/dev.005629. (Available from: http://dx.doi.org/10.1242/dev.005629) [DOI] [PubMed] [Google Scholar]
- 7.Felekkis K, Touvana E, Stefanou C, et al. MicroRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 8.Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120(18):2824–2838. doi: 10.1002/cncr.28730. (Available from: http://dx.doi.org/10.1002/cncr.28730) [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Martin D, Diaz-Zamudio M, Galindo-Campos M, et al. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun Rev. 2010;9(6):454–458. doi: 10.1016/j.autrev.2009.12.006. (Available from: http://dx.doi.org/10.1016/j.autrev.2009.12.006) [DOI] [PubMed] [Google Scholar]
- 10.Hu X, Schwarz JK, Lewis JS, et al. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70(4):1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. (Available from: http://dx.doi.org/10.1158/0008-5472.can-09-3289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimhofer T, Fye H, Taylor-Robinson S, et al. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: a comprehensive review. Br J Cancer. 2015;112(7):1141–1156. doi: 10.1038/bjc.2015.38. (Available from: http://dx.doi.org/10.1038/bjc.2015.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leng R, Zha L, Tang L. MiR-718 represses VEGF and inhibits ovarian cancer cell progression. FEBS Lett. 2014;588(12):2078–2086. doi: 10.1016/j.febslet.2014.04.040. (Available from: http://dx.doi.org/10.1016/j.febslet.2014.04.040) [DOI] [PubMed] [Google Scholar]
- 13.Li L, Yun SH, Keblesh J, et al. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35(1):76–88. doi: 10.1016/j.mcn.2007.02.004. (Available from: http://dx.doi.org/10.1016/j.mcn.2007.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao F, Ji MY, Shen L, et al. Decreased Egr3 expression is related to poor prognosis in patients with gastric cancer. J Mol Histol. 2013;44(4):463–468. doi: 10.1007/s10735-013-9493-8. (Available from: http://dx.doi.org/10.1007/s10735-013-9493-8) [DOI] [PubMed] [Google Scholar]
- 15.Lin M, Chen W, Huang J, et al. MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol Rep. 2011;25(3):739–747. doi: 10.3892/or.2010.1112. (Available from: http://dx.doi.org/10.3892/or.2010.1112) [DOI] [PubMed] [Google Scholar]
- 16.Perez-Cadahia B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol. 2011;89(1):61–73. doi: 10.1139/O10-138. (Available from: http://dx.doi.org/10.1139/o10-138) [DOI] [PubMed] [Google Scholar]
- 17.Pio R, Jia Z, Baron VT, et al. Early growth response 3 (Egr3) is highly over-expressed in non-relapsing prostate cancer but not in relapsing prostate cancer. PLoS ONE. 2013;8(1):e54096. doi: 10.1371/journal.pone.0054096. (Available from: http://dx.doi.org/10.1371/journal.pone.0054096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, Yadav V, Kumar S, et al. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. 2015;5:17454. doi: 10.1038/srep17454. (Available from: http://dx.doi.org/10.1038/srep17454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimachi K, Matsumura T, Hirata H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–538. doi: 10.1038/bjc.2014.621. (Available from: http://dx.doi.org/10.1038/bjc.2014.621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T, Inoue A, Miki Y, et al. Early growth responsive gene 3 in human breast carcinoma: a regulator of estrogen-meditated invasion and a potent prognostic factor. Endocr Relat Cancer. 2007;14(2):279–292. doi: 10.1677/ERC-06-0005. (Available from: http://dx.doi.org/10.1677/erc-06-0005) [DOI] [PubMed] [Google Scholar]
- 22.Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: a comprehensive review. World J Hepatol. 2015;7(26):2648–2663. doi: 10.4254/wjh.v7.i26.2648. (Available from: http://dx.doi.org/10.4254/wjh.v7.i26.2648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan HY, Li QQ, Zhang Y, et al. MiR-124 represses vasculogenic mimicry and cell motility by targeting amotL1 in cervical cancer cells. Cancer Lett. 2014;355(1):148–158. doi: 10.1016/j.canlet.2014.09.005. (Available from: http://dx.doi.org/10.1016/j.canlet.2014.09.005) [DOI] [PubMed] [Google Scholar]