Abstract
Earlier reported small interfering RNA (siRNA) high-throughput screens, identified seven-transmembrane superfamily member 3 (TM7SF3) as a novel inhibitor of pancreatic β-cell death. Here we show that TM7SF3 maintains protein homeostasis and promotes cell survival through attenuation of ER stress. Overexpression of TM7SF3 inhibits caspase 3/7 activation. In contrast, siRNA-mediated silencing of TM7SF3 accelerates ER stress and activation of the unfolded protein response (UPR). This involves inhibitory phosphorylation of eukaryotic translation initiation factor 2α activity and increased expression of activating transcription factor-3 (ATF3), ATF4 and C/EBP homologous protein, followed by induction of apoptosis. This process is observed both in human pancreatic islets and in a number of cell lines. Some of the effects of TM7SF3 silencing are evident both under basal conditions, in otherwise untreated cells, as well as under different stress conditions induced by thapsigargin, tunicamycin or a mixture of pro-inflammatory cytokines (tumor necrosis factor alpha, interleukin-1 beta and interferon gamma). Notably, TM7SF3 is a downstream target of p53: activation of p53 by Nutlin increases TM7SF3 expression in a time-dependent manner, although silencing of p53 abrogates this effect. Furthermore, p53 is found in physical association with the TM7SF3 promoter. Interestingly, silencing of TM7SF3 promotes p53 activity, suggesting the existence of a negative-feedback loop, whereby p53 promotes expression of TM7SF3 that acts to restrict p53 activity. Our findings implicate TM7SF3 as a novel p53-regulated pro-survival homeostatic factor that attenuates the development of cellular stress and the subsequent induction of the UPR.
Proper functionality and robustness of protein homeostasis (proteostasis) is regulated by several defense mechanisms.1, 2, 3 These include the heat-shock response (HSR)4 and the unfolded protein responses (UPRs).5 The UPR is an elaborate adaptive response that evolved to restore protein-folding homeostasis under conditions that challenge ER function and induce ER stress.5, 6 It involves dissociation of BiP/GRP78 from the three principal ER stress sensors: PKR-like ER kinase (PERK), inositol-requiring kinase-1 (IRE1) and activating transcription factor-6 (ATF6), and activation of the signal transduction pathways emanating from these stress sensors.5, 7 The main role of the UPR is to restore ER homeostasis by reducing protein load, and increasing ER-folding capacity and misfolded protein degradation. Attenuation of protein translation is executed by PERK through phosphorylation of the eukaryotic translation initiation factor 2α (eIF2α), whereas the two other branches of the UPR increase ER function by upregulating the expression of ER chaperones and the ER-associated protein degradation machinery. The combined action of these cascades serves to reduce ER stress. However, sustained ER stress induces apoptotic cell death through increased expression of ATF4, ATF3 and the C/EBP homologous protein (CHOP).8, 9, 10
The tumor suppressor p53 is activated in response to numerous cellular insults, including ER stress, and may lead to the repair of the cellular damage, cell–cycle arrest, apoptosis or senescence.11 p53 attenuates the UPR caused by ER stress as evidenced by augmented alternative splicing of X-box binding protein-1 (XBP1), the transcription factor that executes the UPR, in p53-deficient mice.12 Accordingly, reduced expression of BiP/GRP78 is observed in liver and pancreas of p53 knockout mice subjected to ER stress in vivo, but not in spleen and lung.13 These findings indicate that p53 deficiency may confer hypersensitivity to ER stress and the UPR in selected tissues. In addition, p53 plays a more general role in apoptosis induced by sustained and unresolved UPR.14, 15, 16 Under such conditions, elevation in p53 expression in certain cell types, including MEF, MCF-7 and HCT116 cells, is accompanied by upregulation of apoptosis modulators such as p53-upregulated modulator of apoptosis (PUMA) and NOXA, a BH3-Only Member of the Bcl-2 Family.15 Of interest, PUMA has been implicated as a key player in apoptosis of pancreatic β-cells as a result of palmitate-induced ER stress.17
Pancreatic β-cells are particularly susceptible to cellular stress due to their high rate of protein synthesis.18, 19 Pro-inflammatory cytokines are prominent inducers of ER stress and pancreatic β-cells death, through a number of pathways, including induction of the transcription factors nuclear factor kappa B (NF-κΒ), and signal transducer and activator of transcription 1 (STAT-1).20 STAT-1 activates Jun N-terminal kinase, whereas NF-κΒ promotes the production of nitric oxide (NO), as well as depletion of calcium from the ER that results in ER stress and subsequent apoptosis.8, 21 In search for novel players involved in stress-induced death of pancreatic β-cells, we conducted high-throughput screens (HTSs) for the effect of specific small interfering RNAs (siRNAs) on the survival of the MIN6 β-cell line22 and human pancreatic islets treated with pro-inflammatory cytokines. These studies revealed several genes that had not been previously associated with cytokine-induced β-cell death.22, 23 Among them were Otubain-2,23 Nedd4 family-interacting protein-1 (ref. 24) and the seven-transmembrane superfamily member 3 protein (TM7SF3).25
In a previous study,22 we have shown that TM7SF3 is a potential new player in the inhibition of cytokine-induced death, and in the promotion of insulin secretion from pancreatic β-cells. Its main action was associated with maintaining cellular reducing power within physiological levels and reducing the cellular content of pro-apoptotic proteins such as FAS, Fas-associated via death domain and caspase-8.22 Here we show that TM7SF3 maintains protein homeostasis and promotes survival of different cell types through attenuation of ER stress and the subsequent induction of the UPR. We further show that TM7SF3 is a downstream transcriptional target of p53. Our findings implicate TM7SF3 as a novel pro-survival homeostatic factor, regulated by p53, that attenuates the development of cellular stress.
Results
TM7SF3 inhibits cell death
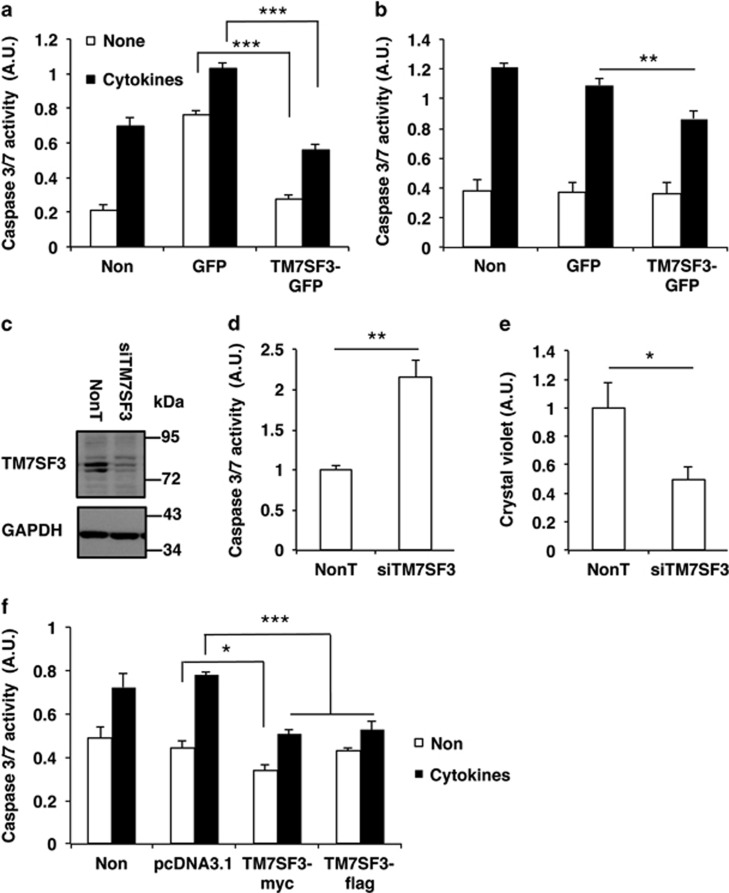
Earlier reported siRNA HTSs22, 23 identified novel genes involved in cytokine-induced death of the MIN6 β-cell line and human pancreatic islets. One of the top ‘hits' was TM7SF3, a protein of unknown function, whose silencing inhibited insulin secretion and promoted cytokine-induced death of pancreatic β-cells.22 To study its underlying mode of action, TM7SF3 was overexpressed in dispersed human islets treated with cytokines. Overexpression of GFP (control) plasmid significantly increased basal caspase activity in dispersed human islets, presumably owing to the stress induced by the transfection procedure in these hypersensitive cells; however, overexpression of TM7SF3 decreased by ~65%, this elevated activity (Figure 1a). A similar ~45% decrease in caspase 3/7 activity was observed in cells treated with cytokines. Smaller effects were observed in MIN6 cells overexpressing TM7SF3 that were treated with cytokines (Figure 1b). We have previously shown22 that the increase in caspase 3/7 activity induced by cytokines and other stress inducers reflects increased cellular apoptosis, at least in MIN6 cells, as evident by their altered cellular morphology, reduced cell viability and cellular reducing power, and the increase in the fraction of apoptotic (Annexin V-positive) cells, determined by FACS analysis.22
Figure 1.
Effects of TM7SF3 on caspase activity in human islets and other cell types. Dispersed human islets (a), MIN6 (b), U2-OS (c–e) and HEK cells (f) were transfected with the indicated plasmids (harboring the cDNA of TM7SF3 or GFP alone) or siRNAs for 48 h (a, b and f) or for 8 days (by triple transfections) (c–e). Cells remained untreated (open bars) or were treated with Cytomix ‘3X' for 16 h (black bars), followed by assay of caspase activity (a, b and d) and cell number (Crystal Violet) (e). Alternatively, total cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (c). Data are the mean±S.E.M. of at least three independent experiments in four replicates. *P<0.05; **P<0.01; ***P<0.001
The protective effects of TM7SF3 on cell viability were not restricted to β-cells. Silencing of TM7SF3 in U2-OS cells almost completely abolished the expression of the protein (Figure 1c), and was accompanied by a marked twofold increase in basal caspase 3/7 activity (Figure 1d) and a significant 50% reduction in cell number (Figure 1e). Accordingly, overexpression of TM7SF3-Myc or TM7SF3-Flag in HEK293 cells significantly reduced caspase activity mainly following treatment with cytokines, although transfection of an ‘empty' (control) vector was without effect (Figure 1f). Collectively, these results suggest that inhibition of apoptosis and increased cellular viability mediated by TM7SF3 is a general phenomenon, taking place in a variety of cell types.
Silencing of TM7SF3 increases iNOS mRNA levels and nitric oxide production
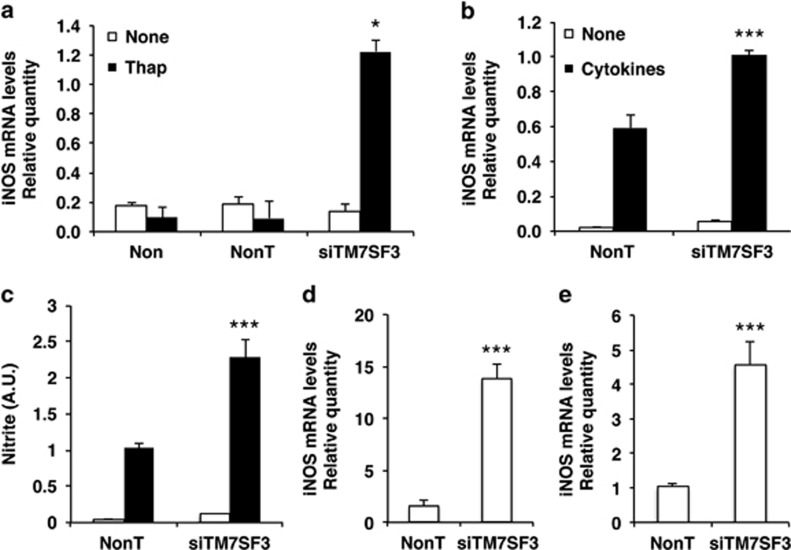
To determine whether silencing of TM7SF3 promotes apoptosis through the induction of endoplasmic reticulum (ER) stress, we studied the effects of silencing of TM7SF3 on activation of iNOS and production of NO that are key contributors to ER stress and apoptosis in pancreatic β-cells26 and other cell types.27, 28 Human pancreatic islets were treated with 100 nM of thapsigargin – an inhibitor of the sarco/ER Ca2+ ATPase, which depletes the ER Ca2+ stores.29 Such treatment did not increase iNOS transcription on its own; however, when it was combined with silencing of TM7SF3, a marked ~10-fold increase in the transcription of iNOS could be observed (Figure 2a). Treatment of human islets with cytokines for 24 h significantly increased iNOS mRNA levels, but silencing of TM7SF3 did not exert any further effect (Supplementary Figure 1S). On the other hand, silencing of TM7SF3 in MIN6 cells in the presence of cytokines, resulted in a significant increase of ~1.7-fold in iNOS mRNA (Figure 2b) that was accompanied by a significant ~2-fold increase in NO production compared with control (Figure 2c). The ability of TM7SF3 to suppress iNOS expression seems to be a general phenomenon. Thus, silencing of TM7SF3 in HepG2 and U2-OS cells caused a marked 4–12-fold increase in iNOS mRNA levels (Figures 2d and e). These findings suggest that TM7SF3 can inhibit iNOS expression and NO production both under basal and stress conditions.
Figure 2.
Effects of TM7SF3-siRNA on iNOS mRNA levels and NO production. Dispersed human islets (a), MIN6 (b and c), HepG2 (d) and U2-OS (e) cells were transfected for 48 h (a–d) or 6 days (e) with TM7SF3-siRNA or with a non-targeting sequence (control). Human islets (a) were then treated with thapsigargin (Thap; 100 nM) for 16 h or were left untreated. MIN6 cells (b and c) were treated with Cytomix ‘1X' for 12 h. HepG2 (d) and U2-OS (e) cells remained untreated. RNA was extracted from all cells and quantified by qRT-PCR (a, b, d and e). Alternatively, the medium was collected, and the concentration of nitrite was measured by Griess assay (c). Bar graphs are the mean±S.E.M. of three independent experiments in duplicates. *P<0.05; ***P<0.001 versus non-targeting siRNA assayed under the same conditions. Results in a were obtained in three (out of four batches) of human islets
TM7SF3 inhibits ER stress and the unfolded protein response
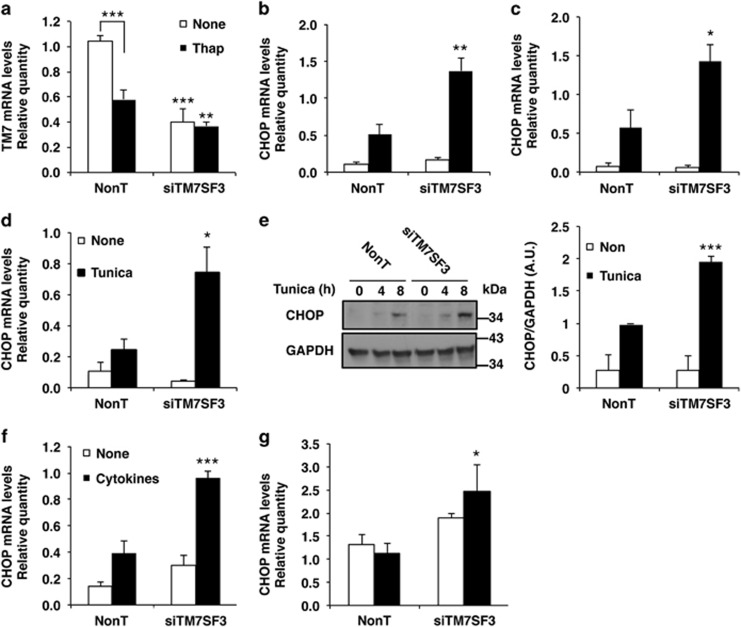
A number of cell types, including pancreatic β-cells, are highly sensitive to the induction endoplasmic reticulum (ER) stress.21 Given that TM7SF3 acted as an anti-apoptotic gene and an inhibitor of nitric oxide production that promotes ER stress, we studied its effects on CHOP, a key inducer of apoptosis as a result sustained activation of the UPR. As shown in Figure 3a, treatment of MIN6 cells for 16 h with thapsigargin reduced the mRNA levels of TM7SF3. The mRNA levels of TM7SF3 were further reduced (by ~60% and 40% for basal and thapsigargin treatment, respectively) in cells transfected with TM7SF3-siRNA. Silencing of TM7SF3 was accompanied by ~2.5-fold increase in the transcription of CHOP in thapsigargin-treated MIN6 (Figure 3b) or βTC (Figure 3c) cells. Similar effects could be observed in cells treated with tunicamycin, an inhibitor of GlcNAc phosphotransferase and protein glycosylation30 that leads to accumulation of unfolded proteins in the ER and triggers the UPR (Figure 3d). The effects of TM7SF3 on CHOP expression were also confirmed at the protein level. TM7SF3-siRNA significantly increased (approximately twofold) the levels of CHOP protein in tunicamycin-treated MIN6 cells (Figure 3e). TM7SF3 silencing also augmented CHOP transcription in MIN6 cells treated with cytokines (Figure 3f); a similar, albeit less pronounced effect was seen in human pancreatic islets (Figure 3g). These findings implicate TM7SF3 as an inhibitor of CHOP expression following ER stress in different cell types.
Figure 3.
Effects of TM7SF3-siRNA on CHOP expression in cells subjected to ER stress. MIN6 cells (a, b, d–f), βTC cells (c) and dispersed human pancreatic islets (g) were transfected for 48 h with TM7SF3-siRNA or with a non-targeting sequence. Cells remained untreated or were treated with thapsigargin (Thap; 100 nM, 16 h) (a–c), tunicamycin (2 μg/ml, 8 h) (d and e), tunicamycin (2 μg/ml, 4 and 8 h) (e), Cytomix ‘1X', 12 h (f) or Cytomix ‘3X', 4 h (g). RNA (a–d, f–g) was extracted and quantified by qRT-PCR. Total extracts (e) of cells treated with tunicamycin were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Bar graphs are the mean±S.E.M. of at least three (a, b, d–g) or two (c) independent experiments in duplicates. *P<0.05; **P<0.01; ***P<0.001 versus non-targeting siRNA assayed under the same conditions
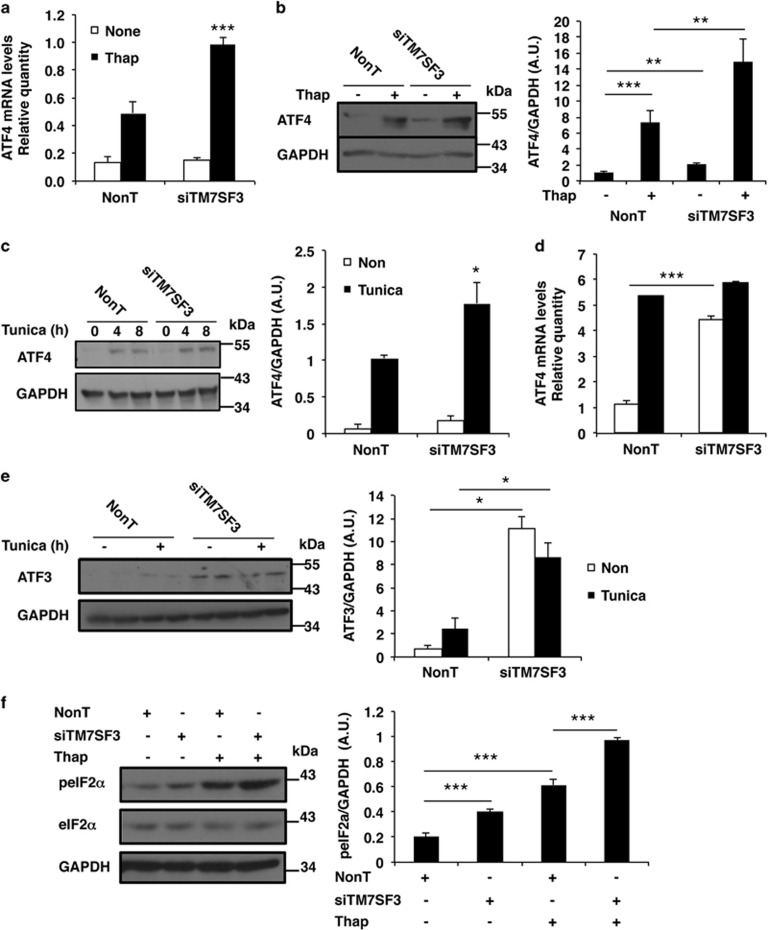
ATF4 is the upstream activator of CHOP expression.31 Therefore, we studied whether TM7SF3 affects ATF4 expression. Silencing of TM7SF3 stimulated ~1.5–2-fold ATF4 mRNA and protein levels in MIN6 cells treated with thapsigargin (Figures 4a and b) or tunicamycin (Figure 4c). These effects were not limited to MIN6 cells: silencing of TM7SF3 in untreated U2-OS cells increased about fivefold the mRNA levels of ATF4, similar to the levels induced by treatment with tunicamycin, but no further increase was observed when silencing of TM7SF3 was combined with addition of tunicamycin (Figure 4d). TM7SF3 also inhibited the expression of ATF3, a downstream target of ATF4 and an upstream regulator of CHOP. Silencing of TM7SF3 significantly increased the protein levels of ATF3 (Figure 4e), although addition of tunicamycin did not exert an additional effect. Of note, silencing of TM7SF3 did not affect other aspects of the UPR: it did not promote splicing of XBP1,32 nor did it affect the cleavage of ATF6 (ref. 33) (Supplementary Figures 2S a and b).
Figure 4.
Effects of TM7SF3-siRNA on ATF3, ATF4 and eIF2α in stress-induced MIN6 and U2-OS cells. MIN6 cells (a–c and f) and U2-OS (d and e) were transfected for 48 h (a–c and f) or for 6 days (d and e) with TM7SF3-siRNA or with a non-targeting sequence. Cells were remained untreated or were treated with thapsigargin (Thap 100 nM) for 16 h (a, b and f); tunicamycin (2 μg/ml) for 4 and 8 h (c); or tunicamycin (2 μg/ml) for 8 h (d and e). RNA (a and d) was extracted and quantified by qRT-PCR. Total cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (b, c, e and f, left panels). Quantitation of ATF4, ATF3 and phospho-eIF2α intensities (control versus treatment with tunicamycin (8 h) or thapsigargin (16 h)) is shown as bar graphs (b, c, e and f, right panels). Bar graphs are the mean±S.E.M. of at least three independent experiments in duplicates. *P<0.05; **P<0.01; ***P<0.001 versus non-targeting siRNA assayed under the same conditions
ER stress and the induction of the UPR are accompanied by attenuation of global protein translation through phosphorylation and inhibition of the eIF2α.34 To determine whether TM7SF3 affects this aspect of the UPR, we studied the state of phosphorylation of eIF2α. As shown in Figure 4f, silencing of TM7SF3 in MIN6 cells promoted the phosphorylation (inhibition) of eIF2α already at the basal state, and this effect was further potentiated in the presence of thapsigargin, suggesting that TM7SF3 is required for maintenance of eIF2α in its dephosphorylated active state.
p53 is an upstream regulator of TM7SF3
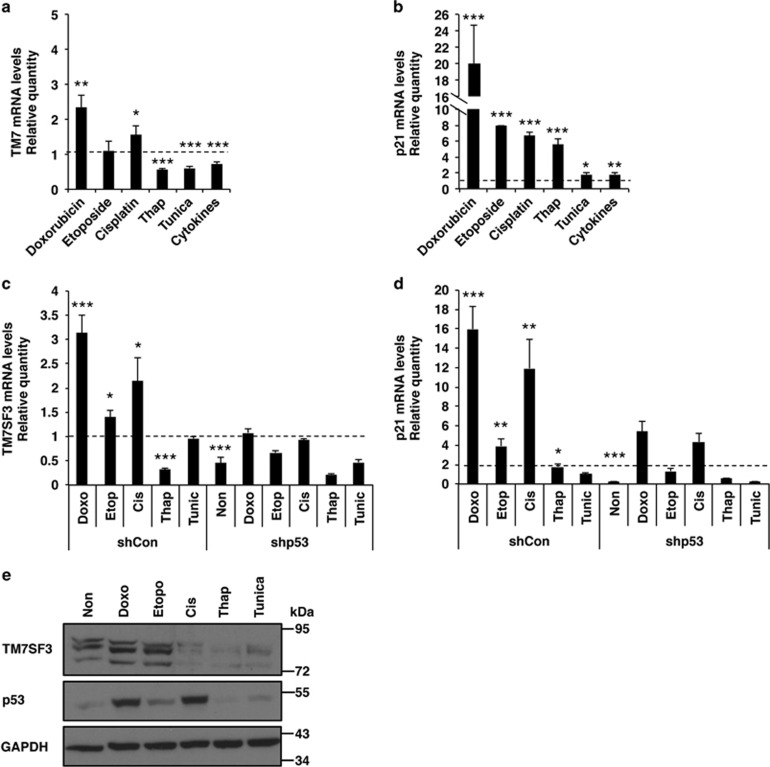
The above findings suggest that TM7SF3 dampens ER stress and the subsequent activation of the UPR. We therefore sought to unravel the mechanisms that regulate the expression of TM7SF3. As shown in Figure 5a, treatment of MIN6 cells with cisplatin35 or doxorubicin36 increased TM7SF3 mRNA levels, suggesting that it could be upregulated also under certain types of genotoxic stress. Interestingly, thapsigargin, tunicamycin, cytokines and etoposide,37 failed to increase or even slightly decreased the mRNA levels of TM7SF3 (Figure 5a), indicating that only a selected set of stress inducers affect TM7SF3 expression. Of note, a number of the agents that promoted expression of TM7SF3, also increased transcription of p21 (Figure 5b), a known downstream target of p53.38 Indeed, silencing of p53 in HepG2 and other cells, that abolished p53 protein expression (Supplementary Figure 3S), significantly reduced the mRNA levels of both TM7SF3 and p21, stimulated by the different stress inducers (Figures 5c and d). The same general trend was also observed at the protein level (Figure 5e), where doxorubicin and etoposide, but not thapsigargin or tunicamycin increased the protein level of both TM7SF3 and p53. Of note, there was no complete correlation between the effects of the different stress inducers on mRNA and protein levels of TM7SF3, p21 and p53. For example, etoposide was a strong inducer of TM7SF3 expression, mainly at the protein level, although having a rather modest effect on the expression of p53, while the opposite was true for cis-platinum. These findings suggest that somewhat different pathways regulate the expression of TM7SF3 and p53. Given that certain inducers of p53 promote transcription and expression of TM7SF3, and given that ER and genotoxic stress up-regulates p53,15, 16, 39 we set out to explore the possible involvement of p53 in the regulation of TM7SF3 transcription.
Figure 5.
Effects of stress inducers on mRNA levels of TM7SF3 and p21. MIN6 cells (a and b), HepG2sh–con or HepG2sh-p53 cells (c and d), as well as naive HepG2 cells (e) were treated with the indicated stress inducers: cis-platinum −6 μg/ml (24 h); etoposide –2 μM (24 h); doxorubicin −1 μM (24 h); thapsigargin 100 nM (8 h); tunicamycin (Tu) 2 μg/ml (8 h) and Cytomix ‘1X' (5 h) or were left untreated. mRNA levels of TM7SF3 (a and c) and p21 (b and d) were quantified by qRT-PCR. Total cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (e). Bar graphs are the mean±S.E.M. of three independent experiments in duplicates. *P<0.05; **P<0.01; ***P<0.001 versus non-treated cells assayed under the same conditions
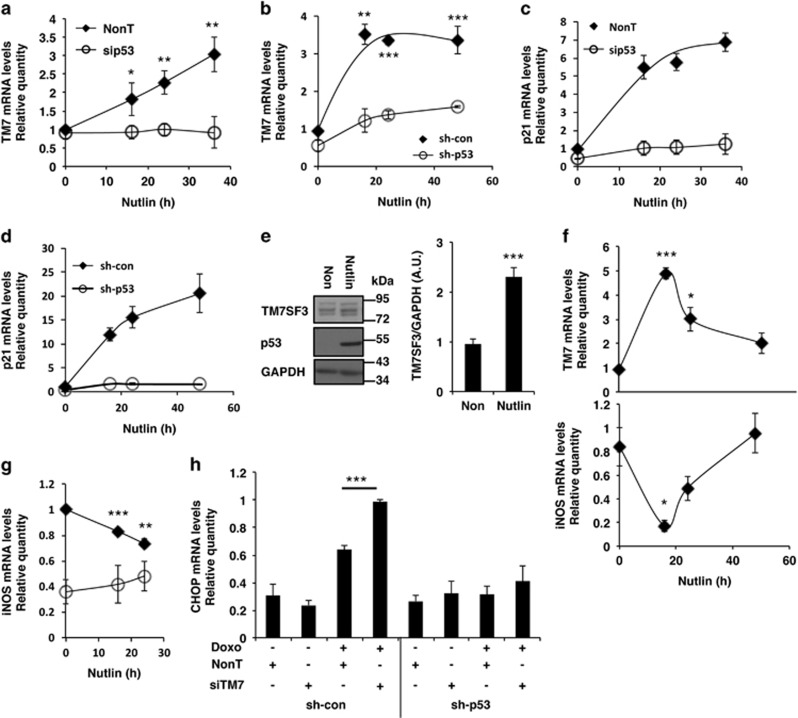
We found that Nutlin-3a, an inhibitor of MDM2, and a potent inducer of p53,40 increased the amounts of TM7SF3 mRNA and protein. Treatment of human islets or HepG2 cells with Nutlin increased approximately threefold TM7SF3 mRNA in a time-dependent manner (Figures 6a and b). A marked increase in p21 mRNA was observed as well (Figures 6c and d). Silencing of p53 markedly reduced the effects of Nutlin (Figures 6a and b), indicating that indeed they were mediated by p53. Similar results were observed in HCT116 (Supplementary Figures 4Sa and b) and MCF10A cells (Supplementary Figures 4Sc and d). Nutlin also increased twofold the protein levels of TM7SF3, in association with a marked increase in expression of p53 itself (Figure 6e). Given that silencing of TM7SF3 promoted iNOS transcription (Figure 2) and given that suppression of iNOS transcription is linked to p53 activity,41 we could show that indeed Nutlin, induced TM7SF3 mRNA and inhibited transcription of NOS2 (Figure 6f), an effect that was abolished upon silencing of p53 (Figure 6g). Similarly, silencing of TM7SF3 increased CHOP mRNA levels in HepG2 cells treated with doxorubicin, and this effect was also abolished upon silencing of p53 (Figure 6h).
Figure 6.
Effects of Nutlin or silencing of p53 on mRNA levels of TM7SF3, p21, iNOS and CHOP. Human islets (a and c) were transfected for 24 h with 25 nM of p53-siRNA or with a non-targeting siRNA (control). Cells were then treated with nutlin (10 μM) for the indicated periods of time. Alternatively, HepG2sh–con or HepG2sh-p53 cells (b, d and g), naive HepG2 (e) or HCT116 cells (f) were treated with Nutlin (10 μM) for 24 h (e) or for the indicated periods of time (a–d, f and g). In parallel, HepG2sh–control or HepG2sh-p53 cells were transfected for 48 h with 25 nM of TM7SF3-siRNA or with a non-targeting control sequence. Then the cells were treated with doxorubicin (2 μM; 24 h) (h). RNA was extracted from all cells and quantification of the indicated mRNAs was carried out by qRT-PCR (a–d, f–h). Alternatively, total cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (e). Bar graphs are the mean±S.E.M. of at least three independent experiments. *P<0.05, **P<0.01, ***P<0.001 versus non-targeting siRNA assayed under the same conditions
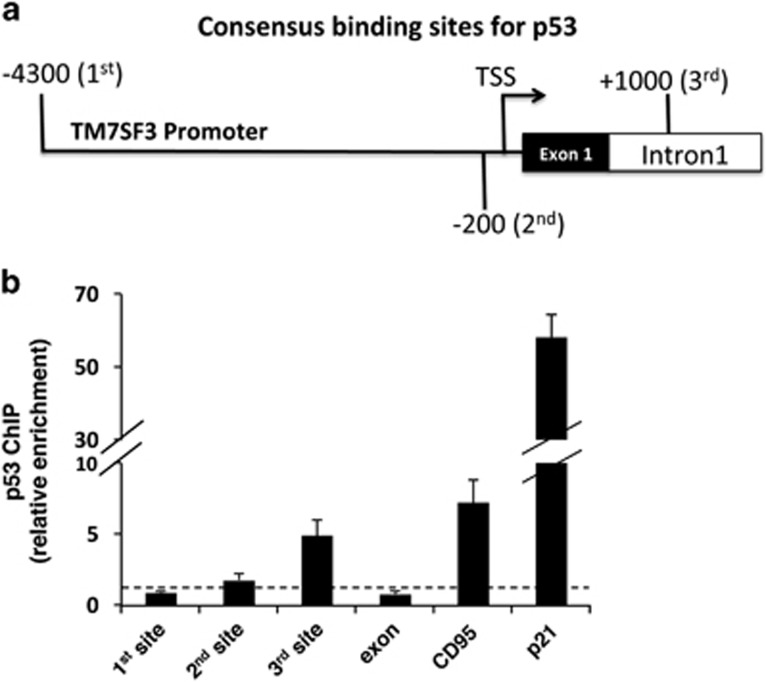
Analysis of the TM7SF3 gene identified three sites that could function as potential p53-binding sites, located respectively, 4300 and 180 bp upstream or 1000 bp downstream of the transcription start site (TSS) of TM7SF3 (site-3 is within the first intron, Figure 7a). To determine whether p53 directly regulates TM7SF3 transcription, MCF10A cells were subjected to chromatin immunoprecipitation (ChIP)42 with anti-p53 or control antibodies. The ChIP analysis (Figure 7b) revealed ~5-fold enrichment in p53 binding to the third site of the TM7SF3 gene, versus ~7- and 60-fold enrichment in p53 binding to the promoter regions of CD95 (ref. 43) and p21,38 respectively, both being known targets of p53. These finding suggest that TM7SF3 may be a direct target of p53 that binds to site-3 of the TM7SF3 gene.
Figure 7.
Chromatin immunoprecipitation (ChIP) of p53 in MCF10A cells. MCF10A cells were subjected to ChIP with p53-specific polyclonal antibodies (CM1) or no antibody control (mock). Quantification of the fragment containing the predicted p53-binding site in the TM7SF3 (site 1 – 4300 bp and site 2 – 180 bp upstream to the TSS; site 3 – 1000 bp downstream (first intron) to the TSS); CD95, or the p21 promoter regions was done by qRT-PCR; dashed line – Mock. Bar graphs are the mean±S.E.M. of three experiments
TM7SF3 regulates p53 activity
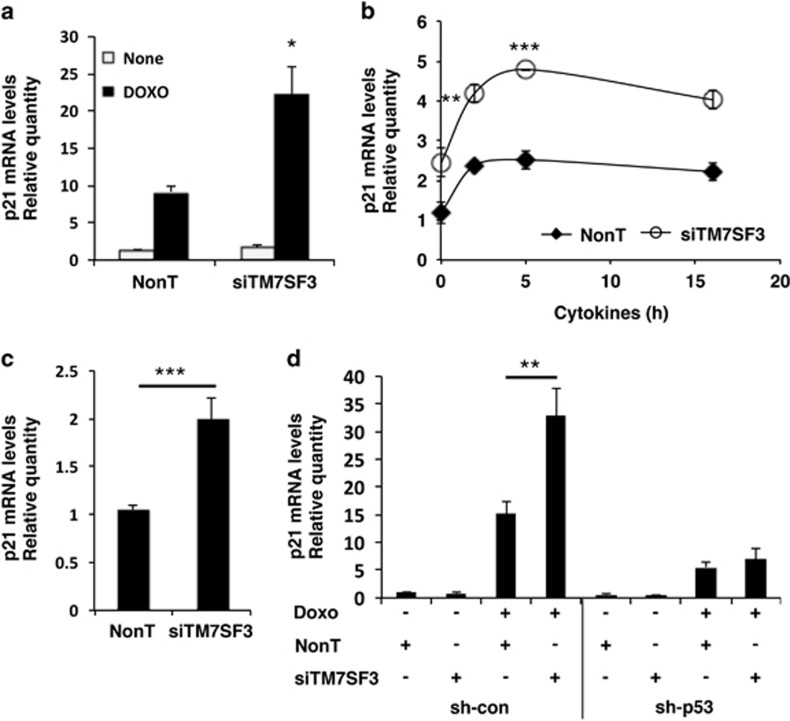
Feedback regulatory loops represent a common paradigm in biological systems. Therefore, we asked whether TM7SF3 could modulate p53 activity. Indeed, silencing of TM7SF3 markedly potentiated the stimulatory effect of stress inducers on p53 activity, as determined by increased p21 mRNA levels (Figure 8). This effect was observed both in HepG2 (Figure 8a), MIN6 (Figure 8b) and U2-OS cells (Figure 8c), under basal conditions (Figure 8c), as well as in cells treated with doxorubicin (Figure 8a) or with cytokines (Figure 8b). As expected, silencing of p53 eliminated the increase in p21 mRNA (Figure 8d). Collectively, these findings suggest that p53 and TM7SF3 generate a negative regulatory feedback loop, whereby p53 promotes the expression of TM7SF3, that acts as an inhibitor of p53 activity.
Figure 8.
Effects of TM7SF3-siRNA on p21 mRNA levels. Nave HepG2 (a), MIN6 (b), U2-OS (c) and HepG2sh–control or HepG2sh-p53 cells (d) were transfected for 48 h (a, b and d) or for 6 days (c) with TM7SF3-siRNA or with a non-targeting sequence. Cells were treated with doxorubicin (2 μM for 24 h) (a and d); with Cytomix ‘1X' for the indicated periods of time (b); or were left untreated (c). RNA was extracted and quantified by qRT-PCR. Bar graphs are the mean±S.E.M. of at least three independent experiments. *P<0.05; **P<0.01; ***P<0.001 versus non-targeting siRNA assayed under the same conditions
Discussion
The present work highlights the role of TM7SF3 as a novel regulator of ER stress and the UPR in pancreatic β-cells and other cell types. The results indicate that silencing of TM7SF3 promotes the induction of ER stress under basal conditions, and potentiates the effects of known ER stress inducers such as cytokines, thapsigargin and tunicamycin. Lack of TM7SF3 activates NO production and the PERK arm of the UPR that involves inhibition of eIF2α activity, enhanced expression of ATF4 and ATF3, and induction of CHOP that culminates in apoptosis. These observations implicate TM7SF3 itself as an inhibitor of ER stress and a promoter of cell survival. Furthermore, TM7SF3 seems to be both a positive downstream direct target and a negative regulator of p53, implicating the existence of a regulatory network that controls expression of both proteins.
Little is known about TM7SF3 and its biological functions. Our interest in this protein emerged when our earlier siRNA screen revealed that TM7SF3 acts as an anti-apoptotic gene in pancreatic β-cells.22 This was evident by the increase in caspase 3/7 activity upon silencing of TM7SF3 both at the basal level and under stress induced by cytokines.22 We could corroborate these findings in the present work by showing that overexpression of TM7SF3 in human pancreatic islets and in the MIN6 β-cell line inhibits caspase 3/7 activity, both under basal conditions and following induction of ER stress. We could further show that this is a general phenomenon, taking place in a number of diverse cell types, including HEK and U2-OS cells. Two microarray analyses of luminal breast tumors44 and testicular germ cell tumors45 revealed upregulation of TM7SF3 transcription. These studies suggest, though not proven, that TM7SF3 might function as a pro-survival protein and support our data that TM7SF3 could exert anti-apoptotic functions.
Silencing of TM7SF3 promotes ER stress stimulated by different stress inducers, as evident by the activation of iNOS and the increased production of NO observed in a number of cell lines, as well as in human pancreatic islets. Induction of the UPR is a natural outcome of ER stress. Among the three main branches that execute the UPR, silencing of TM7SF3 only affects the PERK pathway, although having little or no effect on the ATF6 and the IRE1α arms. Preferential activation of a selected branch of the UPR network is a known phenomenon. For example, the dominant effect of the UPR in photoreceptors of P23H rhodopsin knock-in mice involves the preferential use of IRE1-mediated induction of XBP1.46 Similarly, there is a direct role for the PERK pathway in prion disease pathogenesis, although the IRE1 arm is dispensable in this process.47, 48
Silencing of TM7SF3 significantly activates the PERK pathway, resulting in the inhibition of eIF2α and activation of ATF4, ATF3 and CHOP, already at basal state, in different cell types. These results suggest that even without external induction of ER stress, TM7SF3 helps in maintaining the cells in unstressed conditions, suggesting that it plays a significant role in keeping cellular homeostasis. These effects of TM7SF3 are accentuated in the presence of stress inducers. Three such inducers were used to promote the UPR: thapsigargin,29 tunicamycin49 and cytokines.21 Our data demonstrate that silencing of TM7SF3 had different effects on elements along the PERK pathway, depending on the nature of the stress being inflicted upon the cells. Although silencing of TM7SF3 potentiated the effects of all stress inducers on CHOP expression, analysis of ATF4, the upstream regulator of CHOP, revealed a somewhat different picture. Thus, when cells were subjected to a strong ER stress (e.g., upon treatment with tunicamycin), the increase in ATF4 transcription was already maximal and silencing of TM7SF3 had little additional contribution. However, when the ER stress was mild (e.g., upon treatment with thapsigargin), then silencing of TM7SF3 had a potentiating effect. These findings suggest that lack of TM7SF3 supports the induction of ER stress and the UPR, although it is not a strong stress inducer on its own.
Some inducers of ER stress, such as thapsigargin, tunicamycin or cytokines, reduce the mRNA levels of TM7SF3, yet other stress inducers such as doxorubicin50 had no effect or even upregulated TM7SF3 mRNA and protein levels. Given that all these inducers activate the UPR,21, 29, 49, 51 we conclude that TM7SF3 does not belong to the set of genes that are activated as part of the UPR (e.g., certain chaperons). We could show, however, that TM7SF3 transcription is directly regulated by p53. This is based on our findings that inducers of p53, such as Nutlin, promoted the accumulation of TM7SF3 mRNA and protein, whereas silencing of p53 reduced TM7SF3 mRNA. Furthermore, ChIP analysis revealed a direct interaction of p53 with site-3 of the TM7SF3 gene, localized 1000 bp downstream of the TSS of TM7SF3, within the first intron. As p53 supports TM7SF3 transcription at basal state, it suggests that TM7SF3 belongs to the set of genes that help maintain proteostasis and are p53 targets under basal conditions. The fact that p53 promotes expression of TM7SF3, that attenuates the UPR, is in line with the role of p53 as a suppressor of the UPR.12, 13 For example, E2F1, whose activity is repressed upon activation of p21 by p53,52, 53 inhibits transcription of GRP78/BiP,54 thus stimulating the propagation of the UPR. This suggests a molecular link between activation of the p53/p21-signaling pathway and attenuation of the UPR, as a result of E2F1 inhibition and GRP78/BiP accumulation. Still, our findings do not implicate p53 as the sole inducer of TM7SF3 expression because some inducers of p53 fail to promote TM7SF3 expression, indicating that this gene is subject to regulation by other transcription factors as well.
Of interest, TM7SF3 itself presumably acts to restrict p53 activity, implicating the existence of a negative-feedback loop whereby the pro-apoptotic p53 promotes expression of the pro-survival gene TM7SF3 that acts to inhibit p53 expression. The existence of such regulatory loop could serve to maintain the levels of TM7SF3 within a confined range and prevent overproduction of the protein under stress conditions.
In summary, the present study highlights the role of TM7SF3, as a regulator of cellular homeostasis that helps in inhibiting the development of ER stress and the subsequent activation of the UPR both under basal conditions and following the induction of cellular stress. It implicates this protein as being a downstream target of p53, thus revealing the existence of a novel link between p53, ER stress and the UPR, with TM7SF3 playing a central role in this network.
Materials and Methods
Materials
siRNAs and Dharmafect transfection reagents were provided by Dharmacon Inc. (Lafayette, CO, USA). Interleukin-1 beta (IL-1β) and interferon gamma (IFNγ) were purchased from MD Biosciences (Ness Ziona, Israel) Ltd. Tumor necrosis factor alpha (TNFα) was provided by Prospec-Tany Technogene (Rehovot, Israel). Collagenase (type XI), protease inhibitor mixture and gentamycin were purchased from Sigma Chemicals Co (St Louise, MO, USA); Lipofectamine 2000 was from GIBCO-BRL (Grand Island, NY, USA); concentrated T4 ligase, CellTiter-Blue viability assay, Greiss kit, gel-extraction and miniprep DNA purification systems were from Promega (Madison, WI, USA); PerfectPure Cell & Tissue RNA extraction kit was from 5Prime (Hamburg, Germany); cDNA reverse transcription kit was purchased from ABI (Applied Biosystems, Foster City, CA, USA); Real-time PCR kit (SYBR green PCR master mix) was from Invitrogen (Carlsbad, CA, USA). Enzolyte-caspaseTM RH110 caspase 3/7 assay kit was from AnaSpec, Ltd. (San Jose, CA, USA). The cytokines mixture (′1X' Cytomix) consisted of 3 nM TNFα, 3 nM IFNγ and 1.5 nM IL-1β. Their biological activities were 10 units per ng (TNFα and IFNγ) and 200 units per ng (IL-1β).
Antibodies
Polyclonal CHOP (SC-793) and ATF3 (SC-188) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). ATF4 (#11815), eIF2α and phospho-eIF2αSer51 (#339) antibodies were from Cell signaling (Beverly, MA, USA). Monoclonal GAPDH (MAB374) antibodies were from Millipore (Billerica, MA, USA). Rabbit polyclonal TM7SF3 antibodies were generated by Sigma (Rehovot, Israel) against a peptide (CIKELFQKEQPAGERTPL) at the C-terminal end of TM7SF3. Peroxidase-conjugated affinity purified goat anti-mouse and goat anti-rabbit were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Cells
MIN6 cells; naive HepG2 cells; HepG2 cells stably expressing control shRNA (HepG2sh–con) or sh-p53 construct (HepG2sh-p53);55 naive HCT116cells and their p53-null derivative (HCTp53−/−);56 HEK293; U2-OS; and MCF10A cells were maintained at 37 °C. Cells were grown in Dulbecco's Modified Eagle's medium or McCoy's 5A medium (for HCT116 cells), supplemented with 10% FBS. Human islets (~90% purity) were provided by the European Consortium for Islets Transplantation (Islet for Basic Research program) through a Juvenile Diabetes Research Foundation Award 31-2008-413. Islets were cultured at 37 °C in a 5% CO2 humidified atmosphere in CMRL 1066 medium containing 10% (v/v) FBS, 2 mM l-glutamine, 100 units per ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin and 40 μg/ml gentamycin. The medium was changed every other day. Human islets studies received the WIS Ethics Committee approval. Intact human islets were dispersed by a 4 min incubation at 37 °C with 1 mg/ml trypsin/EDTA and by passing the cells twice through a 21 G needle. Trypsinized islets were washed with CMRL 1066 medium containing 10% FBS, and were resuspended in CMRL 1066 containing 10% FBS.
Cloning of mouse TM7SF3
The DNA encoding mouse TM7SF3 (GI:119964708) was cloned from total mouse pancreatic islets cDNA by PCR using the following primers: 5′-ACGAATTCATGTGGCGGCTGCGCCTGCTGGTCCTAGCGGTGCTGGCGGCGGG CAGCGCCGAGGCCCAGGCGAACTCCAGCGACGGTT-3′ and 5′-GTCGACGCAGAAGCAAGGGTGTCCT-3′. PCR products were gel-purified, by the Gel-purification kit (Promega) and cut by EcoRI and SalI restriction enzymes. The restricted PCR products were ligated into pEGFP-N1 to generate TM7SF3-GFP. Constructs of TM7SF3 tagged with Flag or Myc at the C terminus were generated as follows: TM7SF3-GFP was digested using SalI and NotI restriction enzymes to remove the DNA encoding GFP. The GFP was replaced by a double strand synthetic oligonucleotide, containing matching overhangs of SalI and NotI, coding for immunogenic sequence derived from the c-Myc protein (underlined): S 5′-TCGACATGGAACAAAAGCTCATCTCAGAAGAAGATCTGAATACGC-3′AS 5′-GGCCGCGTATTCAGATCTTCTTCTGAGATGAGCTTTTGTTCCATG-3′. The construct of TM7SF3 tagged with Flag at the C terminus was generated by the proteomics unit at Weizmann institute, using ‘restriction site-free cloning'.57 In brief, primers were designed to contain a tag (Flag or Myc) and overlapping sequences to the TM7SF3 plasmid. These primers were used for whole-plasmid amplification of the insert and the plasmid. The primers used for replacing the c-Myc with Flag in into pEGEF-TM7SF3-Myc are: S 5′- GAGGACACCCTTGCTTCTGGGTCGACATGGATTACAAGGACGCCGCGACTCTAGATCATAATCAGCCATAC-3′ AS 5′- GTATGGCTGATTATGATCTAGAGTCGCGGCGTCCTTGTAATCCATGTCGACCCAGAAGCAAGGGTGTCCTC-3′.
Treatment of cells, transfection and preparation of cell extracts
Cells were seeded in 96-well/24-well/12-well plates (30 × 103/150 × 103/500 × 103 cells per well, respectively). Twenty -four hours later MIN6 cells were transfected with siRNA SMARTpools using Dharmafect-4 transfection reagent (Dharmacon), according to the manufacturer's instructions. Human Islets, HepG2, U2-OS and HEK293 cells were transfected with siRNA SMARTpools using Dharmafect-1 transfection reagent. Cells were transfected with DNA plasmids using Lipofectamine 2000 transfection reagent (GIBCO-BRL Grand Island, NY, USA) according to the manufacturer's instructions. Cells were then treated as indicated, washed three times with PBS and collected in extraction buffer (25 mM Tris-HCl, 2 mM sodium orthovanadate, 0.5 mM EGTA, 10 mM NaF, 10 mM sodium pyrophosphate, 80 mM β-glycerophosphate, 25 mM NaCl, 1% Triton X-100 and protease inhibitor mixture (diluted 1:1000, pH 7.4)). Cell extracts were centrifuged at 20 000 × g for 15 min at 4 °C, and the supernatants were collected. Samples (10–40 μg) were mixed with 5 × Laemmli sample buffer, boiled and resolved by 8% SDS-PAGE under reducing conditions. The proteins were transferred to nitrocellulose membrane for western blotting with the different antibodies.
Assay of caspase 3/7 activity and cellular reducing power
Apoptosis was determined by caspase 3/7 activity kit (Enzolyte-caspase 3-RH110, AnaSpec, Ltd.) using fluorescent microplate reader excitation/emission=496/520 nm. Cellular reducing power was determined using the CellTiter-Blue assay kit (Promega) using fluorescent microplate reader excitation/emission=480/530 nm.
RNA analysis
Cells were grown in 24-well plates. After treatment, cells were collected and total RNA was extracted using the PerfectPure RNA kit (5Prime, Hamburg, Germany). First-strand cDNA was generated by cDNA Reverse Transcription kit (Applied Biosystems, CA, USA). Quantitive detection of specific mRNA transcripts was carried out by real-time PCR using ABI-Prism 7300 instrument (Applied Biosystems, CA, USA); SYBR Green PCR mix (Invitrogen) and the following oligonucleotide primers: inducible NO synthase (mouse), 5′-GCCCTGCTTTGTGCGAAGTG-3′ and 5′-AGCCCTTTGTGCTGGGAGTC-3′ ATF4(mouse), 5′-CCTTCGACCAGTCGGGTTTG-3′ and 5′-GTCGCTGGAGAACCCATGAG-3′ CHOP (mouse), 5′-GCAGGAGAACGAGCGGAAAG-3′ and 5′-CTGACCATGCGGTCGATCAG-3′ HPRT (mouse), 5′-GCAGTACAGCCCCAAAATGG-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′ p21 (mouse), 5′-CCAGGCCAAGATGGTGTCTT-3′ and 5′-TGAGAAAGGATCAGCCATTGC-3′ TM7SF3 (mouse), 5′-GCCGTCCTGCGCCATATTTC-3′ and 5′- CCCTCTGTCGTTGCCTGTTC-3′ inducible NO synthase (human), 5′-GGTGGAAGCGGTAACAAAGG-3′ and 5′-TGCTTGGTGGCGAAGATGA-3′ ATF4 (human), 5′-CTTACGTTGCCATGATCCCT-3′ and 5′-CTTCTGGCGGTAGTGG-3′ p21 (mouse), 5′-GGCGGGCTGCATCCA-3′ and 5′-AGTGGTGTCTCGGTGACAAAGTC-3′ TM7SF3 (mouse), 5′-AGTGCTCTCAAGGTGGTTAC-3′ and 5′-GAAACGGGTCCCAAACAATG-3′. Data were normalized for the content of actin or HPRT mRNA levels.
Chromatin Immunoprecipitation
ChIP was performed as described.58 Immunoprecipitated chromatin was subjected to qPCR analysis using gene-specific primer pairs as follows: TM7SF3 site-1 (4300 bp upstream to the TSS) 5′-GCCACCTGTGTATCTAGGACTACAG-3′ and 5′-CACACAACCAGAAAAAAAAATTAGC -3′ TM7SF3 site-2 (180 bp upstream to the TSS) 5′-TTGCCTACTTGGTGAGATAAAAGTG-3′ and 5′-GGGAGCCCCCAGGAAGTC-3′ TM7SF3 site-3 (1000 bp downstream (first intron) of the TSS) 5′-TGTGGGAGCCAAGAAGCA-3′ and 5′-AATCATCAAAAATGGTGTACTCATAGG-3′ TM7SF3 exon, 5′-CATTGTTTGGGACCCGTTTC-3′ and 5′-AAGCGTATGTGTGAGCAGGAA-3′. PCR of the p21 promoter 100 bp upstream of the 5′ p53-responsive element was performed on immunoprecipitated chromatin using the following pair of oligonucleotide primers: 5′-AGCAGGCTGTGGCTCTGATT-3′ and 5′-CAAAATAGCCACCAGCCTCTTCT-3′. PCR of the CD95 promoter 610 bp downstream of the TSS was performed on immunoprecipitated chromatin, using the following pair of oligonucleotide primers: 5′-AAGCGGAAGTCTGGGAAGCT-3′ and 5′-CTTGTCCAGGAGTTCCGCTC-3′. PCR of a sequence from GAPDH (exon 4) was performed on immunoprecipitated chromatin, using the following pair of oligonucleotide primers: 5′-GTATTCCCCCAGGTTTACAT-3′ and 5′-TTCTGTCTTCCACTCACTCC-3′.
Densitometry and statistical analysis
The intensity of bands in autoradiograms was determined by densitometry that was carried out on exposures within the linear range. Graphic analysis was performed with NIH image software. Results are presented as means±S.E.M. For comparison between two groups, a two-sample equal variance Student's t-test (two tailed) was used when appropriate; P<0.05 were considered significant. *, ** and *** correspond to P<0.05, 0.01 and 0.001, respectively.
Acknowledgments
We thank Dr Yoav Peleg (Weizmann Institute) for production of different TM7SF3 constructs; Dr JI Miazaki (Osaka University, Japan) for providing MIN6 cells. Insightful discussions with Dr Sanford Sampson (Bar Ilan University) are greatly acknowledged. This work was supported by The Juvenile Diabetes Research Foundation International (YZ); The Israel Science Foundation (YZ), a Center of Excellence grant (1779/11) from the Israel Science Foundation (VR and MO) and the Minerva Foundation with funding from the Federal German Ministry for Education and Research (MO).
Author contributions
RI, IG, NF, SBH, EE, VR, MO and YZ conceived and designed the experiments. RI, IG, NF, SS and SBH performed the experiments and statistical analysis. EE, VR, MO and YZ wrote the manuscript.
Glossary
- ATF3/4/6
activating transcription factor-3/4/6
- ChIP
chromatin immunoprecipitation
- CHOP
C/EBP homologous protein
- eIF2α
eukaryotic translation initiation factor 2α
- ER
Endoplasmic reticulum
- HTS
high-throughput screen
- IFNγ
interferon gamma
- IL-1β
interleukin-1 beta
- IRE1
inositol-requiring kinase-1
- NF-kB
Nuclear factor kappa b.
- TM7SF3
seven-transmembrane superfamily member 3
- TNFα
Tumor necrosis factor alpha
- PERK
PKR-like ER kinase
- PUMA
p53-upregulated modulator of apoptosis
- STAT-1
signal transducer and activator of transcription 1
- UPR
Unfolded Protein Response
- XBP1
X-box binding protein-1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by KH Vousden
Supplementary Material
References
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell 2010; 40: 253–266. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J 2011; 30: 2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P, Morimoto RI. Organismal proteostasis: role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev 2014; 28: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichko AK, Markova EN, Petrova NV, Razin SV, Kantidze OL. Mechanisms of heat shock response in mammals. Cell Mol Life Sci 2013; 70: 4229–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–529. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 2014; 14: 581–597. [DOI] [PubMed] [Google Scholar]
- Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta 2013; 1833: 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DL, Miani M, Cardozo AK. Signalling danger: endoplasmic reticulum stress and the unfolded protein response in pancreatic islet inflammation. Diabetologia 2013; 56: 234–241. [DOI] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol 2015; 17: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011; 13: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Marcel V, Olivier M, Oren M, Rotter V, Hainaut P. Understanding wild-type and mutant p53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Ther 2011; 18: 2–11. [DOI] [PubMed] [Google Scholar]
- Dioufa N, Chatzistamou I, Farmaki E, Papavassiliou AG, Kiaris H. p53 antagonizes the unfolded protein response and inhibits ground glass hepatocyte development during endoplasmic reticulum stress. Exp Biol Med (Maywood) 2012; 237: 1173–1180. [DOI] [PubMed] [Google Scholar]
- Mihailidou C, Chatzistamou I, Papavassiliou AG, Kiaris H. Improvement of chemotherapeutic drug efficacy by endoplasmic reticulum stress. Endocr Relat Cancer 2015; 22: 229–238. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 2013; 5: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem 2006; 281: 7260–7270. [DOI] [PubMed] [Google Scholar]
- Lin WC, Chuang YC, Chang YS, Lai MD, Teng YN, Su IJ et al. Endoplasmic reticulum stress stimulates p53 expression through NF-kappaB activation. PLoS One 2012; 7: e39120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha DA, Igoillo-Esteve M, Gurzov EN, Germano CM, Naamane N, Marhfour I et al. Death protein 5 and p53-upregulated modulator of apoptosis mediate the endoplasmic reticulum stress-mitochondrial dialog triggering lipotoxic rodent and human beta-cell apoptosis. Diabetes 2012; 61: 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I et al. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci USA 2001; 98: 10845–10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 2008; 29: 42–61. [DOI] [PubMed] [Google Scholar]
- Donath MY, Storling J, Berchtold LA, Billestrup N, Mandrup-Poulsen T. Cytokines and beta-cell biology: from concept to clinical translation. Endocr Rev 2008; 29: 334–350. [DOI] [PubMed] [Google Scholar]
- Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL. Mechanisms of Pancreatic {beta}-Cell Death in Type 1 and Type 2 Diabetes: Many Differences, Few Similarities. Diabetes 2005; 54(Suppl 2): S97–S107. [DOI] [PubMed] [Google Scholar]
- Beck A, Isaac R, Lavelin I, Hart Y, Volberg T, Shatz-Azoulay H et al. An siRNA screen identifies transmembrane 7 superfamily member 3 (TM7SF3), a seven transmembrane orphan receptor, as an inhibitor of cytokine-induced death of pancreatic beta cells. Diabetologia 2011; 54: 2845–2855. [DOI] [PubMed] [Google Scholar]
- Beck A, Vinik Y, Shatz-Azoulay H, Isaac R, Streim S, Jona G et al. Otubain 2 is a novel promoter of beta cell survival as revealed by siRNA high-throughput screens of human pancreatic islets. Diabetologia 2013; 56: 1317–1326. [DOI] [PubMed] [Google Scholar]
- Oliver PM, Cao X, Worthen GS, Shi P, Briones N, MacLeod M et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 2006; 25: 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Han HJ, Iizaka M, Nakajima Y, Furukawa Y, Sugano S et al. Isolation and characterization of a novel gene encoding a putative seven-span transmembrane protein, TM7SF3. Cytogenet Cell Genet 2000; 88: 305–309. [DOI] [PubMed] [Google Scholar]
- Zumsteg U, Frigerio S, Hollander GA. Nitric oxide production and Fas surface expression mediate two independent pathways of cytokine-induced murine beta-cell damage. Diabetes 2000; 49: 39–47. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Mori M. Nitric oxide and endoplasmic reticulum stress. Arterioscler Thromb Vasc Biol 2006; 26: 1439–1446. [DOI] [PubMed] [Google Scholar]
- Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol 2008; 10: 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T, Hara H, Adachi T. Effect of endoplasmic reticulum (ER) stress inducer thapsigargin on the expression of extracellular-superoxide dismutase in mouse 3T3-L1 adipocytes. J Clin Biochem Nutr 2013; 52: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu H, Zhang W, Wu Q, Liu W, Liu Y et al. N-acetylglucosaminyltransferase V confers hepatoma cells with resistance to anoikis through EGFR/PAK1 activation. Glycobiology 2013; 23: 1097–1109. [DOI] [PubMed] [Google Scholar]
- Pirot P, Ortis F, Cnop M, Ma Y, Hendershot LM, Eizirik DL et al. Transcriptional regulation of the endoplasmic reticulum stress gene chop in pancreatic insulin-producing cells. Diabetes 2007; 56: 1069–1077. [DOI] [PubMed] [Google Scholar]
- He Y, Sun S, Sha H, Liu Z, Yang L, Xue Z et al. Emerging roles for XBP1, a sUPeR transcription factor. Gene expression 2010; 15: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Prywes R. ER stress signaling by regulated proteolysis of ATF6. Methods 2005; 35: 382–389. [DOI] [PubMed] [Google Scholar]
- Chambers KT, Unverferth JA, Weber SM, Wek RC, Urano F, Corbett JA. The role of nitric oxide and the unfolded protein response in cytokine-induced beta-cell death. Diabetes 2008; 57: 124–132. [DOI] [PubMed] [Google Scholar]
- Fine JH, Chen P, Mesci A, Allan DS, Gasser S, Raulet DH et al. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res 2010; 70: 7102–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Rojas JM, Taya Y, Gutkind JS. A role for the p38 mitogen-acitvated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res 2000; 60: 2464–2472. [PubMed] [Google Scholar]
- Kweon KH, Lee CR, Jung SJ, Ban EJ, Kang SW, Jeong JJ et al. Sirt1 induction confers resistance to etoposide-induced genotoxic apoptosis in thyroid cancers. Int J Oncol 2014; 45: 2065–2075. [DOI] [PubMed] [Google Scholar]
- Garner E, Raj K. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle 2008; 7: 277–282. [DOI] [PubMed] [Google Scholar]
- Kumari R, Kohli S, Das S. p53 regulation upon genotoxic stress: intricacies and complexities. Mol Cell Oncol 2014; 1: e969653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem 2006; 281: 33030–33035. [DOI] [PubMed] [Google Scholar]
- Wada S, Matsushita Y, Tazawa H, Aoi W, Naito Y, Higashi A et al. Loss of p53 in stromal fibroblasts enhances tumor cell proliferation through nitric-oxide-mediated cyclooxygenase 2 activation. Free Radic Res 2015; 49: 269–278. [DOI] [PubMed] [Google Scholar]
- Ren B, Dynlacht BD. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol 2004; 376: 304–315. [DOI] [PubMed] [Google Scholar]
- Muller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med 1998; 188: 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics 2007; 8: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Jafer O, Goker H, Summersgill BM, Zafarana G, Gillis AJ et al. Expression profile of genes from 12p in testicular germ cell tumors of adolescents and adults associated with i(12p) and amplification at 12p11.2-p12.1. Oncogene 2003; 22: 1880–1891. [DOI] [PubMed] [Google Scholar]
- Chiang WC, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT et al. Robust Endoplasmic Reticulum-Associated Degradation of Rhodopsin Precedes Retinal Degeneration. Mol Neurobiol 2015; 52: 679–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C et al. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci USA 2008; 105: 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu N, Chiang WC, Kurt TD, Sigurdson CJ, Lin JH. Multiple mechanisms of unfolded protein response-induced cell death. Am J Pathol 2015; 185: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012; 2012: 857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones 2013; 18: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiter A, Yerushalmi R, Hardy B. Pharmacological induction of cell surface GRP78 contributes to apoptosis in triple negative breast cancer cells. Oncotarget 2014; 5: 11452–11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayakumar T, Shareef MM, Diaz DA, Ahmed MM, Pollack A. The E2F1/Rb and p53/MDM2 pathways in DNA repair and apoptosis: understanding the crosstalk to develop novel strategies for prostate cancer radiotherapy. Semin Radiat Oncol 2010; 20: 258–266. [DOI] [PubMed] [Google Scholar]
- Laine A, Westermarck J. Molecular pathways: harnessing E2F1 regulation for prosenescence therapy in p53-defective cancer cells. Clin Cancer Res 2014; 20: 3644–3650. [DOI] [PubMed] [Google Scholar]
- Racek T, Buhlmann S, Rust F, Knoll S, Alla V, Putzer BM. Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1. J Biol Chem 2008; 283: 34305–34314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N et al. p53, a novel regulator of lipid metabolism pathways. J Hepatol 2012; 56: 656–662. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Gershoni A, Rotkopf R, Biton IE, Porat Z, Koh AP et al. The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes Dev 2016; 30: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol 2010; 172: 34–44. [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 2004; 16: 631–639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.