Abstract
Microdeletions in the chromosomal region 17p13.3 are associated with neuronal migration disorders, and PAFAB1H1 is the main gene involved. The largest genomic imbalances, including the YWHAE and CRK genes, cause more severe structural abnormalities of the brain and other associated dysmorphic features. Here, we describe a 3-year-old boy with a microdeletion in 17p13.3 presenting with minor facial dysmorphisms, a cleft palate, neurodevelopmental delay, and behavioral disorder with no structural malformation of the brain. The patient was evaluated by a clinician using a standard protocol. Laboratory investigation included GTG-banding, whole-genome AGH, and array-CGH. Whole-genome AGH and array-CGH analysis identified an estimated 2.1-Mb deletion in the 17p13.3 region showing haploinsufficiency of the YWHAE, CRK, H1C1, and OVCA1 genes and no deletion of PAFAH1B1. The complex gene interaction on brain development and function is illustrated in the genotype-phenotype correlation described here. This report reinforces the importance of the 17p13.3 region in developmental abnormalities and highlights the weak implication of the HIC1 and OVCA1 genes in palatogenesis.
Key Words: Behavioral disorder, Deletion 17p13.3, Microarray, Neurodevelopmental delay, Palatogenesis
Genomic imbalances in 17p13.3 are mainly associated with morphological and/or functional defects of the brain such as neuronal migration disorders [Cardoso et al., 2003; Nagamani et al., 2009; Enomoto et al., 2012]. The malformation spectrum ranges from an isolated lissencephaly sequence to Miller-Dieker syndrome (MDS), and the phenotype is usually associated with the genomic size of the imbalance involving the 17p13.3 region [Cardoso et al., 2003; Toyo-oka et al., 2003; Chen et al., 2013]. Isolated lissencephaly sequence and MDS are characterized by deletions in the PAFAH1B1 gene, which is the main gene involved in brain malformations observed in both conditions. However, when deletions also include the YWHAE and CRK genes, other associated defects are present, and brain malformations are usually more severe, leading to a phenotype characterized by a smooth brain with absence of sulci or gyra [Cardoso et al., 2003; Toyo-oka et al., 2003; Mignon-Ravix et al., 2010; Enomoto et al., 2012].
Microdeletion 17p13.3 syndrome is a recognized disorder usually associated with telomeric deletions affecting the YWHAE and CRK genes preserving PAFAH1B1. This syndrome is associated with postnatal growth retardation, mild to moderate mental retardation, facial dymorphisms, and mild structural abnormalities of the brain identified on MRI [Nagamani et al., 2009; Bruno et al., 2010; Schiff et al., 2010; Østergaard et al., 2011; Enomoto et al., 2012].
There are 19 microdeletion 17p13.3 cases already reported [Nagamani et al., 2009; Bruno et al., 2010; Schiff et al., 2010; Shimojima et al., 2010; Tenney et al., 2011; Enomoto et al., 2012]. Up to now, a correlation between size of the deletion and clinical severity has not been established [Bruno et al., 2010; Enomoto et al., 2012]. The smallest deletion reported has approximately 258 kb and is located within the critical region of MDS, which includes YWHAE and CRK as well as 4 other genes: TUSC5, MYO1C, SKIP, and PITPNA [Bruno et al., 2010]. YWHAE and CRK are associated with major phenotypic abnormalities, including craniofacial dysmorphisms, while TUSC5 appears to have a minor contribution to the phenotype [Nagamani et al., 2009; Bruno et al., 2010; Østergaard et al., 2011; Enomoto et al., 2012].
The YWHAE gene (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon; OMIM 605066; http://omim.org/) encodes 14-3-3ε, a member of the 14-3-3 protein family. They are ubiquitous proteins highly conserved with several molecular and cellular functions that act as regulatory molecules by binding to phosphoproteins and playing important roles in neuronal migration [Cardoso et al., 2003; Toyo-oka et al., 2003; Mignon-Ravix et al., 2010; Enomoto et al., 2012]. In addition, YWHAE and TUSC5 also seem to contribute to craniofacial dysmorphism [Bruno et al., 2010].
The CRK gene (v-crk avian sarcoma virus CT10 oncogene homolog; OMIM 164762) codes for a signaling protein that plays a critical role in cell proliferation, differentiation, migration, and axonal growth. Deletions in CRK are strongly associated with physical growth deficit observed in these cases [Cardoso et al., 2003; Nagamani et al., 2009; Bruno et al., 2010; Østergaard et al., 2011]. Furthermore, CRK seems to be related to limb abnormalities, since CRK interacts with PLNA - a gene that is known to be involved in the development of limbs [Østergaard et al., 2011].
In this report, we describe a child without structural malformation of the brain associated with neurodevelopmental delay, behavioral disorder, failure to thrive, cleft palate (CP) and other dysmorphisms, presenting with haploinsufficiency of the YWHAE, CRK, HIC1, and OVCA1 genes and no deletion of PAFAH1B1.
Patient and Methods
Clinical Report
The propositus, a 3-year-old boy at first evaluation, was referred for genetic assessment because of syndromic CP and failure to thrive. He is the only child of a nonconsanguineous couple; the mother is 24 years old. He has a healthy half-sister, and his father's history is unknown.
Despite an apparently uneventful pregnancy, the boy was born at term (41 weeks) with intrauterine growth retardation, weight 2,440 g (-2.5 SD), length 46 cm (-2.5 SD), and a head circumference of 30.5 cm (-2.5 SD). Apgar scores of 8 and 10 at 1 and 5 min, respectively. He evolved with poor suction and dehydration.
Developmental milestones were delayed, and he attended supportive therapy up to 2.5 years of age. Surgical history included gastrostomy, fundoplication for gastroesophageal reflux disease, palatoplasty, and orchiopexy.
At 3 years and 6 months of age, he had dolichocephaly with a prominent front and occiput, bitemporal contraction, low-set ears, sparse and arched eyebrows, broad and high nasal root, smooth philtrum, and thick lips (Fig. 1a, b). The penis had chordee and a length of 4.5 cm (-1.0 SD), hypoplastic scrotum with testicles of 1-2 mL bilaterally (-1.0 SD), clinodactyly of the 5th fingers (Fig. 1c), and flat feet. On physical examination at the age of 7 years, his height, weight, and head circumference were 105 cm (-3.5 SD), 17 kg (-1.7 SD), and 50 cm (-2.0 SD).
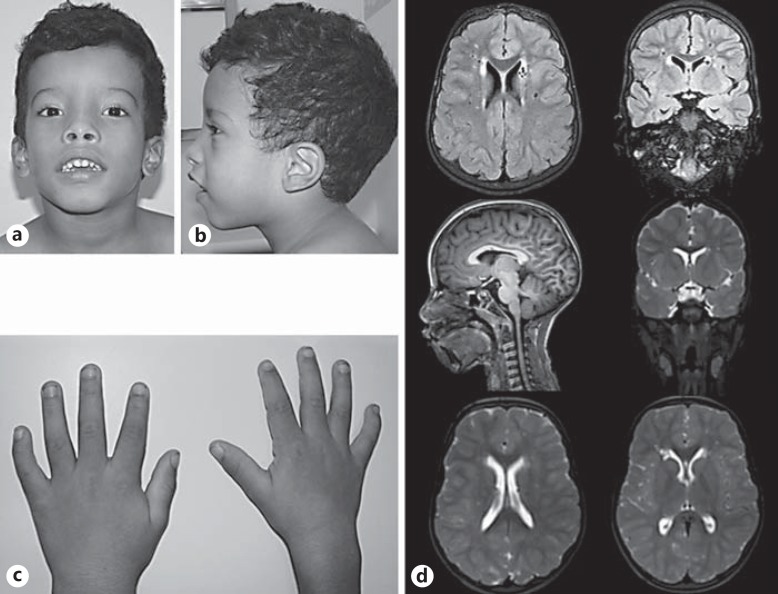
Fig. 1.
Patient at 7 years of age. a, b Mild dolichocephaly with bitemporal contraction, a prominent front and occiput, sparse and arched eyebrows, broad nasal bridge, thick lips with smooth philtrum, and low-set ears. c Clinodactyly of the 5th fingers. d Brain MRI showing no structural abnormalities.
Follow-up revealed failure to thrive and attention and intellectual deficits, with an inability for self-care. There is no history of epilepsy or seizures.
Biochemical analysis revealed normal levels of basal growth hormone. Before genetic evaluation, the boy received growth hormone treatment which was not effective to achieve catch-up growth.
Ultrasound evaluation of the abdomen and heart was normal. EEG performed during sleep showed disorganized background activity. MRI of the brain showed no structural abnormalities (Fig. 1d).
Cytogenetics
Chromosome analysis by GTG-banding from cultured peripheral blood lymphocytes from the patient and his mother were performed at the 500-band level. The father was not available for analysis.
Array-GH and Array-CGH Investigation
High-resolution AGH was performed using the GeneChip CytoScan® 750 Array Kit (Affymetrix Inc., Santa Clara, CA, USA). Data were analyzed by the Chromosome Analysis Suite (ChAS) Software (Affymetrix). Standard analysis adopted the following parameters: markers, >25 for deletion and >50 for duplication. Confirmation of the results was through the SurePrint G3 Human CGH Microarray Kit, 4×180K (Agilent Technologies Inc., Santa Clara, CA, USA). The results were analyzed employing Agilent Genomic Workbench software using the default analysis method CHG+SNP v2 and considering a minimum of 3 probes for deletion or duplication.
Fluorescence in situ Hybridization
FISH analysis from the boy's mother was performed using BAC clone RP11-96B11 mapped at 17p13.3.
Results
GTG-banding revealed that the karyotypes of our patient and his mother were normal: 46,XY, and 46,XX, respectively.
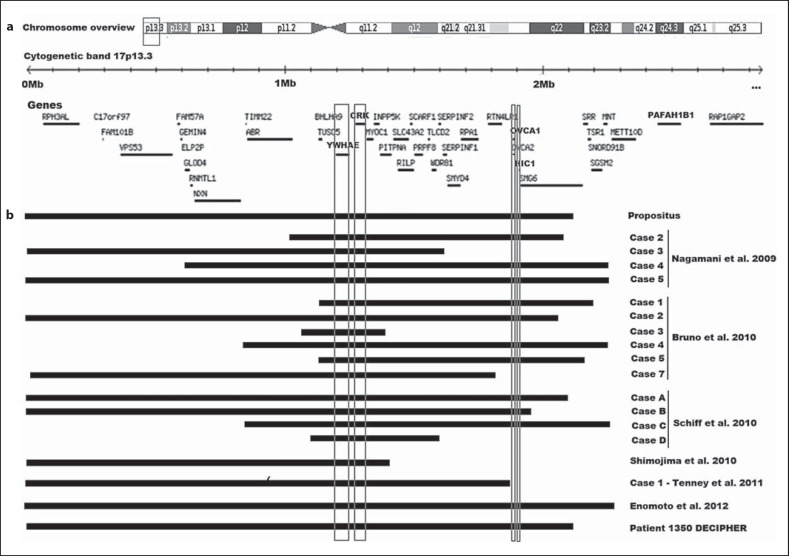
Both whole-genome AGH and aCGH analysis of the patient identified an estimated 2.1-Mb deletion in the 17p13.3 region (525-2,101,504 bp) [hg19-build37NCBI] covering 896 markers on AGH and 156 probes on aCGH. Microarray nomenclature: arr[hg19] 17p13.3(525-2,101,504)×1. The deleted region encompasses 42 RefSeq genes including 30 OMIM genes. Comparison between genomics features of this patient with other published cases with 17p13.3 deletion is shown in Figure 2.
Fig. 2.
Schematic representation of chromosome 17 and the 17p13.3 region. a The ideogram of chromosome 17 shows the region of interest as well as the gene content (box). b Size, extent, and CNV content of the deletion found in our patient and in other reported cases.
A deletion, approximately 163 kb in size, in the 14q24.1 region (68,394,217-68,556,878 bp) [hg19-build37NCBI], which encompasses one RefSeq and OMIM gene, also was detected. The AGH analysis, as well as FISH investigation, of the proband's mother did not show any pathogenic structural variation.
Discussion
In this study, we describe a patient with a telomeric microdeletion of 17p13.3 involving the YWHAE and CRK genes, but not PAFAH1B1 and compared the results with 19 cases previously reported that present the same genomic rearrangement (Table 1). The breakpoint of the microdeletion identified in our patient is distal to PAFAH1B1, about 2.1 Mb from 17pter.
Table 1.
Comparison between features of our patient and reported patients with 17p13.3 deletion
| Deletion size, Mb | DD/ID | Structural brain abnormalitiesa | IUGR/growth retardation | Craniofacial dysmorphism | CP | Deletion HIC1 | Deletion OVCA1 | |
|---|---|---|---|---|---|---|---|---|
| Present case | 2.12 | + | – | + | + | + | + | + |
| Nagamani et al., 2009 | ||||||||
| Case 2 | 1.06 | + | + | + | + | – | + | + |
| Case 3 | 1.62 | + | – | + | + | – | – | – |
| Case 4 | 1.64 | + | + | + | + | – | + | + |
| Case 5 | 2.26 | + | + | + | + | + | + | + |
| Bruno et al., 2010 | ||||||||
| Case 1 | 1.07 | + | + | + | + | – | + | + |
| Case 2 | 2.07 | + | + | + | + | + | + | + |
| Case 3 | 0.33 | + | ND | + | – | – | – | – |
| Case 4 | 1.42 | + | + | + | – | – | + | + |
| Case 5 | 1.03 | + | ND | – | – | – | + | + |
| Case 7 | 1.80 | + | ND | + | – | – | – | – |
| Schiff et al., 2010 | ||||||||
| Case A | 2.18 | + | + | + | + | – | + | + |
| Case B | 1.99 | + | + | + | + | – | + | + |
| Case C | 1.46 | + | + | + | + | – | + | + |
| Case D | 0.53 | + | + | + | + | – | – | – |
| Shimojima et al., 2010 | 1.28 | + | + | + | + | – | – | – |
| Tenney et al., 2011 | ||||||||
| Case 1 | 1.90 | + | + | + | + | – | ND | ND |
| Case 2 | ND | + | + | + | + | – | ND | ND |
| Enomoto et al., 2010 | 2.30 | + | – | + | + | – | + | + |
| DECIPHER patient 1350 | 2.14 | + | ND | + | + | + | + | + |
| Total | – | 20/20 | 13/16 | 19/20 | 16/20 | 4/20 | 13/18 | 13/18 |
DD, developmental delay; ID, intellectual disability; CP, cleft palate; ND, no data available.
Identified on MRI.
CNV del14q24.1 (approximately 163 kb) encompasses the RAD51B gene (OMIM 602948). The protein encoded by this gene is a member of the RAD51 protein family, evolutionarily conserved proteins, and essential for DNA repair by homologous recombination. Up to now, there is no evidence of its involvement in the features presented by patients with microdeletion 17p13.3.
Bruno et al. [2010] was the first to suggest that deletions involving only YWHAE and CRK, sparing PAFAH1B1, are associated with neurodevelopmental deficit, growth retardation, craniofacial dysmorphisms, but no severe brain malformations such as lissencephaly. The patient reported here presents a similar clinical picture observed in 19 cases already published [Nagamani et al., 2009; Bruno et al., 2010; Schiff et al., 2010; Shimojima et al., 2010; Tenney et al., 2011; Enomoto et al., 2012].
Growth retardation is a common feature in 17p13.3 microdeletion syndrome and is observed in the present case. It has been related to CRK gene haploinsufficiency [Nagamani et al., 2009; Bruno et al., 2010; Mignon-Ravix et al., 2010; Schiff et al., 2010; Østergaard et al., 2011]. Only one case among all the previous reported does not present this abnormality [Bruno et al., 2010].
Craniofacial dysmorphisms seem to be related to haploinsufficiency of the CRK gene. [Nagamani et al., 2009; Bruno et al., 2010; Mignon-Ravix et al., 2010; Schiff et al., 2010; Østergaard et al., 2011]. According to Bruno et al. [2010], the most common facial features are laterally extended eyebrows, infraorbital folds, broad nasal tip, maxillary prominence, and a prominent upper and/or lower lip. The craniofacial phenotype of our patient is quite similar.
CP was described in 4 cases including the present one. All have deletions of approximately 2 Mb that include HIC1 and OVCA1 [Nagamani et al., 2009; Bruno et al., 2010]. Among them, patient 1350 of the browser DECIPHER is the only one who also has another CNV (approximately 1.18 Mb) leading to a duplication of the PAFAH1B1 gene.
An association between HIC1 and orofacial clefts was previously described by Jugessur et al. [2010]. More recently, Yu et al. [2014] investigated whether OVCA1 plays a role in the craniofacial defects exhibited by individuals with MDS. They showed that Ovca1 plays an important role in the neural crest cell-derived nasal bone and mandible development in mice. Therefore, the inactivation of OVCA1 in human could be involved in craniofacial dysmorphisms such as a CP phenotype.
Haploinsufficiency of HIC1 and OVCA1 is reported in 13 out of 19 patients with 17p13.3 microdeletion; in 2 cases [Tenney et al., 2011], this information is unavailable. Craniofacial dysmorphism is described in 11 patients; however, CP is reported in only 4 of them. These results support the involvement of the HIC1 and OVCA1 genes with craniofacial dysmorphism but suggest a weak correlation with CP.
Among 15 cases that performed MRI, only 2 have no structural abnormalities [Nagamani et al., 2009; Enomoto et al.; 2012]. Our patient seems to be the third case of 17p13.3 microdeletion syndrome with no severe brain malformation on MRI despite the presence of a significant mental handicap.
Brain malformations are usually associated with haploinsufficiency of YWHAE. This gene participates in a complex pathway involving several genes that include PAFAB1H1 and NUDEL [Toyo-oka et al., 2003; Nagamani et al., 2009; Mignon-Ravix et al., 2010].
Enomoto et al. [2012] published the first evidence pointing to a dosage effect of the YWHAE gene in a patient with 17p13.3 microdeletion including YWHAE, presenting with developmental delay and intellectual disability without structural brain abnormalities. The authors suggest that YWHAE plays a crucial role in neuronal development along with other factors. However, a reduced expression level would not significantly impact the normal brain development. Features of our patient corroborate this hypothesis.
The complex gene interaction on brain development and function is illustrated in the genotype-phenotype correlation described here. The results reinforce the importance of the 17p13.3 region in embryonic development. The involvement of YWHAE, CRK, HIC1, and OVAC1 in brain and facial morphogenesis is clearly seen in cases reported. On the other hand, the presence of CP in a considerable number of cases is not yet known. Further studies are needed to strengthen the genotype-phenotype correlation.
Statement of Ethics
The patient was evaluated using the Brazil's CranioFacial Project Clefts Protocol (Ethics Committee 35316314.9.1001.5404). Clinical data and informed consent were obtained from the patient's mother.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Acknowledgments
The authors thank the family for their cooperation. This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (149600/2010-0) and the Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP (2012/51799-6). V.L.G.-d.-S.-L. is supported by CNPq (304455/2012-1).
References
- 1.Bruno DL, Anderlid BM, Lindstrand A, van Ravenswaaij-Arts C, Ganesamoorthy D, et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet. 2010;47:299–311. doi: 10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet. 2003;72:918–930. doi: 10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CP, Chang TY, Guo WY, Wu PC, Wang LK, et al. Chromosome 17p13.3 deletion syndrome: aCGH characterization, prenatal findings and diagnosis, and literature review. Gene. 2013;532:152–159. doi: 10.1016/j.gene.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 4.Enomoto K, Kishitani Y, Tominaga M, Ishikawa A, Furuya N, et al. Expression analysis of a 17p terminal deletion, including YWHAE, but not PAFAH1B1, associated with normal brain structure on MRI in a young girl. Am J Med Genet A. 2012;158A:2347–2012. doi: 10.1002/ajmg.a.35542. [DOI] [PubMed] [Google Scholar]
- 5.Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, et al. Maternal genes and facial clefts in offspring: a comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS One. 2010;5:e11493. doi: 10.1371/journal.pone.0011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mignon-Ravix C, Cacciagli P, El-Waly B, Moncla A, Milh M, et al. Deletion of YWHAE in a patient with periventricular heterotopias and pronounced corpus callosum hypoplasia. J Med Genet. 2010;47:132–136. doi: 10.1136/jmg.2009.069112. [DOI] [PubMed] [Google Scholar]
- 7.Nagamani SCS, Zhang F, Shchelochkov OA, Bi W, Ou Z, et al. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet. 2009;46:825–833. doi: 10.1136/jmg.2009.067637. [DOI] [PubMed] [Google Scholar]
- 8.Østergaard JR, Graakjær J, Brandt C, Birkebæk NH. Further delineation of 17p13.3 microdeletion involving CRK. The effect of growth hormone treatment. Eur J Med Genet. 2011;55:22–26. doi: 10.1016/j.ejmg.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Schiff M, Delahaye A, Andrieux J, Sanlaville D, Vincent-Delorme C, et al. Further delineation of the 17p13.3 microdeletion involving YWHAE but distal to PAFAH1B1: four additional patients. Eur J Med Genet. 2010;53:303–308. doi: 10.1016/j.ejmg.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Shimojima K, Sugiura C, Takahashi H, Ikegami M, Takahashi Y, et al. Genomic copy number variations at 17p13.3 and epileptogenesis. Epilepsy Res. 2010;89:303–309. doi: 10.1016/j.eplepsyres.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Tenney JR, Hopkin RJ, Schapiro MB. Deletion of 14-3-3ε and CRK: a clinical syndrome with macrocephaly, developmental delay, and generalized epilepsy. J Child Neurol. 2011;26:223–227. doi: 10.1177/0883073810379638. [DOI] [PubMed] [Google Scholar]
- 12.Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, et al. 14-3-3ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 13.Yu YR, You LR, Yan YT, Chen CM. Role of OVCA1/DPH1 in craniofacial abnormalities of Miller-Dieker syndrome. Hum Mol Genet. 2014;23:5579–5596. doi: 10.1093/hmg/ddu273. [DOI] [PubMed] [Google Scholar]