Abstract
Latent‐transforming growth factor beta‐binding protein 3 (LTBP‐3) is important for craniofacial morphogenesis and hard tissue mineralization, as it is essential for activation of transforming growth factor‐β (TGF‐β). To investigate the role of LTBP‐3 in tooth formation we performed micro‐computed tomography (micro‐CT), histology, and scanning electron microscopy analyses of adult Ltbp3‐/‐ mice. The Ltbp3‐/‐ mutants presented with unique craniofacial malformations and reductions in enamel formation that began at the matrix formation stage. Organization of maturation‐stage ameloblasts was severely disrupted. The lateral side of the incisor was affected most. Reduced enamel mineralization, modification of the enamel prism pattern, and enamel nodules were observed throughout the incisors, as revealed by scanning electron microscopy. Molar roots had internal irregular bulbous‐like formations. The cementum thickness was reduced, and microscopic dentinal tubules showed minor nanostructural changes. Thus, LTBP‐3 is required for ameloblast differentiation and for the formation of decussating enamel prisms, to prevent enamel nodule formation, and for proper root morphogenesis. Also, and consistent with the role of TGF‐β signaling during mineralization, almost all craniofacial bone components were affected in Ltbp3‐/‐ mice, especially those involving the upper jaw and snout. This mouse model demonstrates phenotypic overlap with Verloes Bourguignon syndrome, also caused by mutation of LTBP3, which is hallmarked by craniofacial anomalies and amelogenesis imperfecta phenotypes.
Keywords: amelogenesis, enamel, Ltbp3, mouse, scanning electron microscopy
Enamel is the body's strongest tissue because of its dense mineralized content, which forms in a unique stage‐specific process. Enamel formation is a sensitive process susceptible to environmental effects, as well as to the consequences of gene mutations, sometimes encountered in rare diseases 1. Enamel‐formation defects observed in rodent genetic models are excellent systems for using to study the basis of analogous human malformations 2. Enamel formation begins when an organic protein‐enriched matrix is deposited by ameloblasts during the secretory stage. This protein matrix is then modified through the transition and maturation stages 3. During the secretory stage, polarized columnar‐shaped ameloblasts, adjacent to forming enamel, secrete enamel proteins, such as amelogenin, ameloblastin, and enamelin. Tomes’ processes on the distal surface of secretory ameloblasts organize the direction of enamel deposition into rods 4. During the maturation and mineralization stages, enamel matrix proteins are degraded in a stepwise manner by matrix metalloproteinase‐20 (MMP‐20) and kallikrein‐4 (KLK‐4) to form unique enamel prisms 5, 6. Murine mutations of enamel processing and degradation proteins produce enamel defects resembling human amelogenesis imperfecta (AI) phenotypes 5, 6, 7, 8.
Transforming growth factor beta (TGF‐β) is part of a superfamily of growth factors that regulate a broad range of cell growth, differentiation, and extracellular morphogenic events 9. Inhibited TGF‐β signaling (by mutation of TGF‐β ligands, TGF‐β receptors, or intracellular SMADs) leads to reduced enamel formation and detachment of ameloblast cells from the dentin surface, causing the secretion of bubble‐like masses that form cystic structures 10, 11. For example, mice with a conditional knockout mutation for TGF‐β receptor II display enamel attrition with thinner crystals 12. Both Smad3‐/‐ and Smad7‐/‐ mutations reduce enamel mineralization 13, 14. Hence, disrupted TGF‐β signaling has stage‐specific consequences for amelogenesis in a variety of rodent models.
Transforming growth factor‐β family proteins are secreted in the form of high‐molecular‐mass latency complexes that contain other proteins, including latent‐transforming growth factor beta‐binding proteins (LTBPs) 15. To date, four members of the LTBP family (LTBP‐1, LTBP‐2, LTBP‐3, and LTBP‐4) are known. Through their interactions with other extracellular proteins, LTBPs are important regulators of the bioavailability and action of TGF‐β 16. Immunoprecipitation studies using mouse pre‐osteoblast MC3T3‐E1 cells revealed that the LTBP polypeptide forms a complex with the TGF‐β1 precursor 16. Moreover, LTBP‐3 allows latent TGF‐β complexes to be targeted to connective tissue matrices and cells 17, 18. Ltbp3‐/‐ mice show severe skull deformities 19, 20 and an osteopetrosis‐like phenotype 21, while other LTBP members have broad functions in many systems 16. Exactly how craniofacial hard‐tissue defects occur under Ltbp3 deficiency is still unclear, especially those concerning tooth and enamel malformation.
Mutations in human LTBP3 were first observed in a consanguineous Pakistani family, in which all affected members presented with short stature, vertebral and skull bone alterations, and oligodontia 22. In another family, two sisters with homozygous‐recessive truncating mutations in LTBP3 also had oligodontia, short stature, and mitral valve prolapse 23. Our published report identified recessive hypomorphic LTBP3 mutations (including deletion, nonsense, and aberrant splice mutations) in patients with dental anomalies and short stature (MIM; 601216) 24 or Verloes Bourguignon syndrome 25. Using the adult Ltbp3‐/‐ mouse model to characterize all dental hard‐tissue components, we previously described very thin or absent enamel in both incisors and molars 24. More complete understanding of Ltbp3‐/‐ mouse dental phenotypes will allow us to address how mutations of this gene in humans produce tooth abnormalities 19, 20, and to clarify the role of LTBP‐3 in modulating TGF‐β bioavailability 19.
Here we explore dental and cranial morphological differences caused by Ltbp3 deficiency using a three‐dimensional (3D) imaging system [X‐ray micro‐computed tomography (micro‐CT)], classical histology, and scanning electron microscopy. We observed (i) alterations in enamel formation and deposition of enamel nodules, (ii) maturation‐stage ameloblast disruptions, (iii) small bulbous‐like formations inside molar roots, and (iv) enamel prism pattern malformations and reduced cementum thickness, collectively providing a framework for investigating the genetic basis of TGF‐β signaling defects.
Material and methods
Animals
Ltbp3‐/‐ mice (C57BL/6;Sv129;SW mixed background) were produced and genotyped as previously described 19, 20. All procedures with animals were performed according to the standards approved by New York University School of Medicine Institutional Animal Care and Use Committee.
Micro‐CT imaging
The heads of five adult male Ltbp3‐/‐ mutant mice and of five corresponding age/sex‐matched wild‐type (WT) littermates were examined at 3.5 months of age. To investigate malformations at slightly later stages, two male 5.5‐month‐old Ltbp3‐/‐ mutants and two WT matching controls were also examined. All samples were scanned using the Quantum FX micro‐CT pre‐clinical in vivo imaging system (Caliper Life Sciences, Hopkinton, MA, USA), which operates at an energy of 80 kV and current intensity of 160 μA. We found no obvious skull morphological changes with aging. Micro‐CT data acquisitions (with resolution at 80, 40, 20, and 10 μm pixel sizes for skulls, mandibles, incisors, and molars, respectively) were reconstructed using Analyze11.0 (Biomedical Imaging Resource, Mayo Clinic, Rochester, MN, USA). Bony dento‐cranio‐facial anatomy was sorted in coronal, transverse, and sagittal planes at skull, mandible, incisor, and molar levels. The segmented voxels were compiled to represent bony tissue and then were separated according to density to quantitate incisor and molar enamel. Cranial anatomical landmarks were analyzed using Euclidean distance matrix analysis (EDMA) for morphometric analysis 26, 27.
Histology
Samples (heads of 3.5‐month‐old WT and Ltbp3‐/‐ mice) were fixed in 10% formalin for 30 d or longer and then transferred into 70% ethanol, washed in water, and demineralized in 10% ethylenediaminetetraacetic acid (EDTA) at 37°C for 10 d (the demineralizing solution was changed every day for the first 3 d and then every other day). After thoroughly washing in water, the heads were dehydrated in graded ethanol, cleared in Histo‐Clear (National Diagnostics, Atlanta, GA, USA), and embedded in paraffin at 60°C. Ten‐micrometer‐thick transverse sections were collected, deparaffinized, and stained with hematoxylin and eosin (H&E). A detailed histology protocol can be found at http://www.empress.har.mrc.ac.uk.
Scanning electron microscopy
The upper and lower murine incisors of 3.5‐month‐old Ltbp3‐/‐ and WT mice were dissected out of the alveolar bone. After rinsing with distilled water, the teeth were dehydrated in a graded series of ethanol, transferred in a solution of propylene oxide/epon resin (1:1, vol/vol), and embedded in Epon 812 (Euromedex, Souffelweyersheim, France). The teeth were sectioned into two halves along their sagittal axes using a water‐cooled diamond circular saw (Bronwill Scientific, Rochester, NY, USA), and both surfaces were polished with diamond paste (Escil, Chassieu, France). One‐half was etched with a 20% (wt/vol) citric acid solution for 2 min, rinsed with distilled water, dehydrated in a graded series of ethanol solutions and left to dry at room temperature. The samples were coated with a gold‐palladium alloy using a Hummer Jr sputtering device (Technics, Union City, CA, USA). Scanning electron microscopy assessments were performed using a Quanta 250 ESEM (FEI Company, Eindhoven, the Netherlands) operating with an accelerating voltage of electrons of 5 kV.
Statistical analysis
Non‐parametric statistical testing (using the Mann–Whitney U‐test) was used to compare the enamel volume of incisor and molar enamel segmentations. The level of statistical significance was set at P < 0.05. For EDMA analysis, a parametric bootstrap approach was performed to validate the statistical significance, as described previously 27. All significance points were then recalculated as length in millimeters, aiding data interpretation.
Results
Alterations in enamel formation and deposition of enamel nodules
Macroscopically, the upper and lower incisors of normal teeth (from WT mice) are naturally yellow in color and the enamel is smooth and present only on the labial side (Fig. 1A). In Ltbp3‐/‐ mutants, reduced coloration and irregular enamel surfaces were observed, with the lateral zones having less enamel than the central zones (Fig. 1B). The 3D rendered micro‐CT images showed disrupted enamel patterns at the junction between the alveolar bone and the crown area, where numerous enamel nodules had formed laterally (Fig. 1C,D; white arrowheads). Early enamel formed at the distal part of the lower incisor showed obvious changes, as seen in two‐dimensional (2D) sagittal plane reconstructions from micro‐CT images (Fig. 1E,F; red arrowheads). This impaired enamel formation was confirmed by histological analysis (Fig. 1G,H). The enamel matrix in Ltbp3‐/‐ mutants was irregular and failed to form a steady pattern (Fig. 1H). Changes in enamel volume were not uniform throughout the tooth crown, as enamel was completely missing from the lateral side of the labial portion of the incisor. However, a band of enamel of similar thickness as in WT mice was present in the central portion of the labial crown of the incisors.
Figure 1.
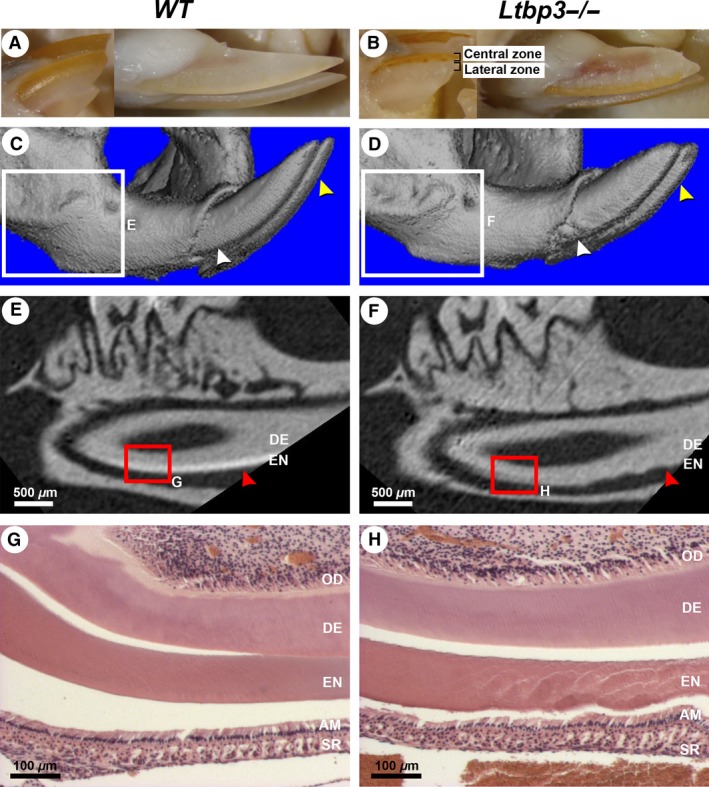
External morphology, micro‐computed tomography (micro‐CT), and histology analysis of lower incisors of a 3.5‐month‐old wild‐type (WT) mouse (left column of panels) or a latent transforming growth factor beta binding protein mutant (Ltbp3‐/‐) mouse (right column of panels). The same WT or Ltbp3‐/‐ mutant is shown in all respective panels. (A, B) In external views of lower incisors, the incisors of the mutant are shorter, laterally white, and centrally orange. (C, D) Three‐dimensional rendered micro‐CT images of the distal part of the mandible. The surface of enamel from the Ltbp3‐/‐ mouse appears irregular (yellow arrows), and enamel nodules are seen near the alveolar bone crest area of the incisor (white arrows). (E, F) Two‐dimensional micro‐CT sagittal sections of the first lower molar and the underlying incisor (obtained from the respective boxed region in panels C and D), showing the enamel density. In the mutant the enamel layer is clearly reduced and has a rough appearance (red arrows). (G, H) Histological sections (hematoxylin and eosin staining) of the lower incisor (respective red boxed area in panels E and F) showing an irregular enamel layer (EN) in the Ltbp3‐/‐ mutant. The odontoblast (OD) and dentin (DE) layers show no obvious changes. AM, ameloblasts; SR, stellate reticulum.
Maturation‐stage ameloblast disruptions
Histological analysis of secretory, transition, maturation, and postmaturation stage ameloblasts of the lower incisors revealed striking alterations in differentiation in Ltbp3‐/‐ mutants (Fig. 2). At the secretory and transition stages, ameloblasts from Ltbp3‐/‐ mice showed no obvious morphological changes compared with ameloblasts from WT mice, although the papillary layer underneath ameloblasts appeared impaired (Fig. 2A–H). Then, during the maturation stage, ameloblasts displayed a clearly disrupted morphology in mutants, especially within the lateral region of the incisor where the most severe ameloblast differentiation defects were observed (Fig. 2L). In addition, the ameloblast columnar epithelium was reduced in thickness within the central region (Fig. 2K). All these changes were more severe at postmaturation stages (Fig. 2M–P; compare WT with Ltbp3‐/‐).
Figure 2.
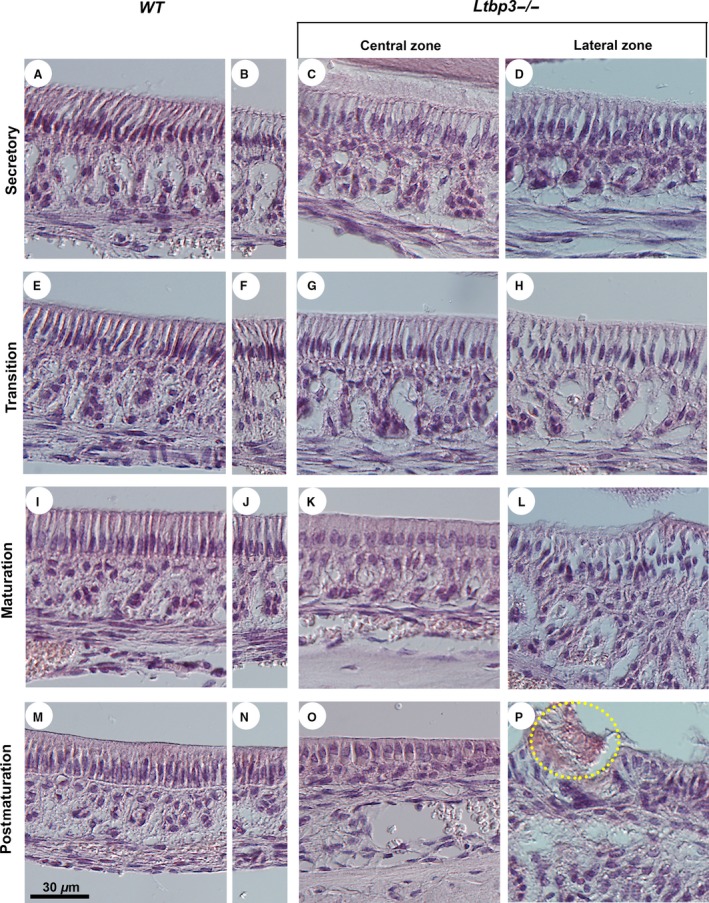
Histological analysis (hematoxylin and eosin staining) of the lower incisor ameloblasts. Normal ameloblast development starts at the cervical loop region of the incisor and involves differentiation in consecutive secretory, transition, maturation, and postmaturation stages toward the incisal edge. These regions are illustrated both in central and lateral zones of the incisor in latent transforming growth factor beta binding protein (Ltbp3‐/‐) mutants (right‐side panels) and respective wild‐type (WT) controls (left‐side panels). The central (A, E, I, M) and lateral (B, F, J, N) zones of WT mice show virtually identical morphology. (A‐H) Ameloblasts of Ltbp3‐/‐ mice show no obvious changes at the secretory and transition stages. However, the papillae underlying the ameloblasts have an irregular organization, with an increased space between each papilla. (I–L) At the maturation stage, the ameloblasts of Ltbp3‐/‐ mice differentiate as shorter columnar cells (K). Ameloblasts appear highly abnormal in the lateral zone (L). (M–P) At the postmaturation stage, the ameloblast layer is disorganized in the mutants (O, P), with cell disruptions localizing mainly to the lateral zone, including the detachment of enamel (circled in P).
Small bulbous‐like formations inside molar roots
Micro‐CT analysis allows us to segment each tissue according to its density, using different density thresholds. Using this method, we performed morphometric analysis of the skull and facial bones of Ltbp3‐/‐ mice (Figs S1, S2). We were also able to segment the enamel to calculate the enamel volume (Fig. 3A,B; Fig. S3). The enamel volume of Ltbp3‐/‐ mice was significantly reduced in all teeth (Fig. S3). In molars, even in the case of complete absence of enamel, the molar cusp pattern was maintained (Fig. 3C,D). As seen in Fig. 3C,D, the overall root pattern and molar surfaces, as revealed by 3D micro‐CT, appear well preserved. However, irregular circles (bulbous‐like structures) were seen, with an apparent random distribution on the mesial and/or distal molar roots, especially for M1 (Fig. 3H; yellow arrowhead). From histological analysis, small circle‐like growths were observed within Ltbp3‐/‐ roots, in contrast to the smooth mineralization pattern in roots from WT mice (Fig. 3I,J; red arrows). These findings indicate that LTBP‐3 is not critical for proper root formation, but has important actions in ameloblasts in regulating proper differentiation of enamel.
Figure 3.
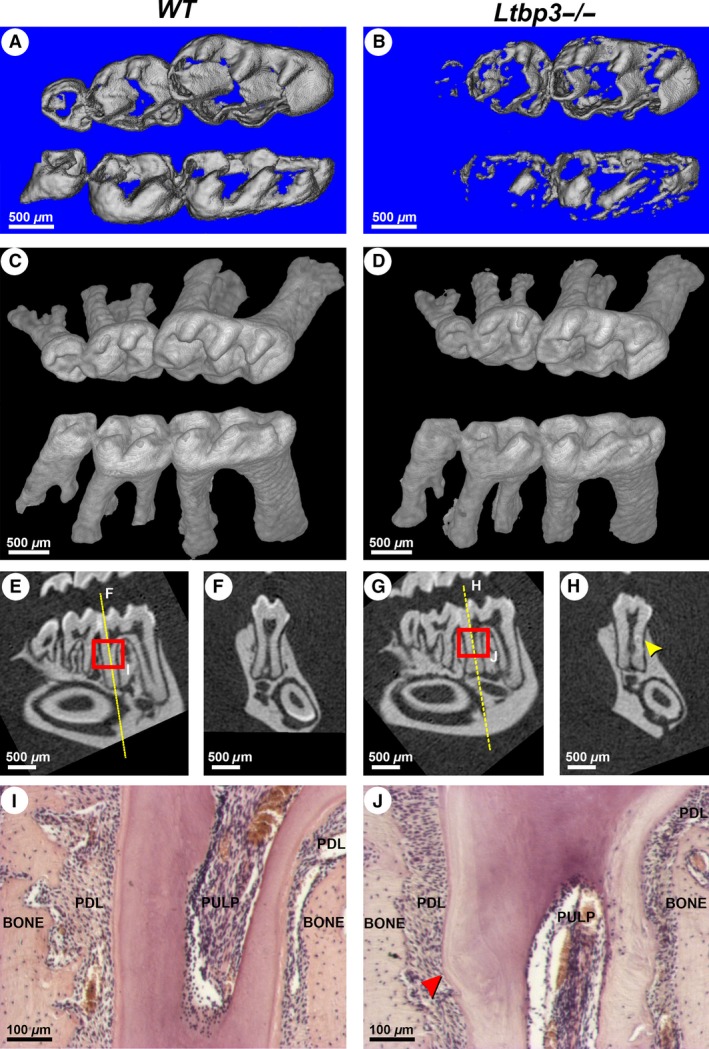
Alterations of enamel in molars and the molar root phenotype in latent transforming growth factor beta binding protein mutant (Ltbp3‐/‐) mice. (A, B) High‐density renderings of micro‐computed tomography (micro‐CT) data from wild‐type (WT) and Ltbp3‐/‐ molars, highlighting enamel structural deficits. The enamel volume of the Ltbp3‐/‐ molar is significantly reduced, especially at the third molar (also see Fig. S3). (C, D) Three‐dimensional rendering of micro‐CT images showing less density in molar crowns of Ltbp3‐/‐ mice. The root pattern of Ltbp3‐/‐ mice is similar to that of WT mice. (E–H) Two‐dimensional sections confirming enamel reduction in both molars and incisors of Ltbp3‐/‐ mutants. Section planes (F, H) are generated at the level of the yellow line. (H) A bulbous‐like structure can be found in some areas of Ltbp3‐/‐ molar roots (yellow arrowhead). (I, J) Histology (hematoxylin and eosin staining; regions highlighted by red boxes in E and G) showing a correctly organized root dentin in the lower molar of WT mouse, whereas irregular dentin staining is seen in the Ltbp3‐/‐ mutant mouse. A bulbous‐like structure (red arrowhead) is also observed. The periodontium, cementum, alveolar bone, and dental pulp tissues have no obvious morphological changes at the histological scale. BONE, alveolar bone; PDL, periodontium; PULP, dental pulp tissue.
Enamel prism pattern malformations and reduced cementum thickness – scanning electron microscopy analysis
Figure 4 shows enamel prism patterns on sagittal views of the lower incisor, which were obtained by high‐resolution scanning electron microscopy. Mineralization starts by forming an enamel ribbon, with ameloblasts in WT incisors secreting a rod‐like structure. This typical pattern was not observed in the Ltbp3‐/‐ incisors (Fig. 4A,B). Figure 4C shows normal decussating enamel rods, with adjacent rows that criss‐cross near the dentinoenamel junction. In contrast, enamel from Ltbp3‐/‐ mice was poorly organized, with a clearly impaired prism pattern (Fig. 4D) and almost no rod pattern at the incisal edge (Fig. 4F). At its thickest area, the enamel from Ltbp3‐/‐ teeth appears to be more porous (Fig. 4G,H). The whole outer enamel pattern, oriented in the opposite direction of the inner enamel, exhibited a disturbed structure (Fig. 4L; arrowhead). These observations suggest that ameloblasts from Ltbp3‐/‐ mice fail to complete enamel formation.
Figure 4.
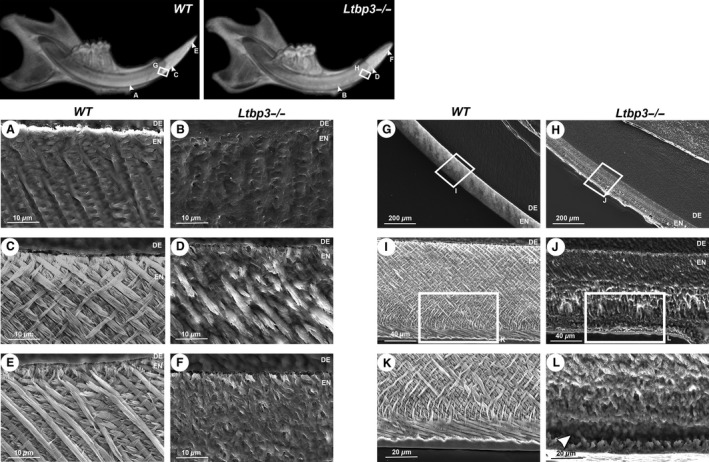
Comparative scanning electron microscopy images of midsagittal sections of lower incisor enamel at various distances from the incisor extremity (the approximate location of the areas shown is given in the upper, low‐magnification views), comparing teeth from wild‐type (WT) (A, C, E, G, I, K) and latent transforming growth factor beta binding protein mutant (Ltbp3‐/‐) (B, D, F, H, J, L) mice. (A, B) Early maturation stage of enamel crystal deposition showing enamel ribbon patterns in WT mouse, whereas no clear pattern is seen in Ltbp3‐/‐ mouse. (C, D) Junction area between the alveolar bone crest and crown analog of incisor, showing a criss‐cross pattern in WT mouse, whereas a poor organization of enamel rods is observed in Ltpb3‐/‐ mouse. (E, F) Incisal (or tip) area of incisor showing a typical enamel prism pattern in WT mouse, whereas no such pattern is observed in mutant mouse. (G, H) Fully mineralized enamel showing a rough external surface in Ltbp3‐/‐ mouse. The enamel thickness in this zone is similar in WT and Ltbp3‐/‐ mice. Boxed areas indicate areas shown under higher magnification in the next panels. (I, J) The enamel layer appears porous and non‐decussated in Ltbp3‐/‐ mutant mouse compared with WT mouse. (K, L) Under higher magnification, the outer enamel shows poorly organized, non‐structured prism patterns (arrowhead), while an increased roughness at the outer enamel surface is also observed in the Ltbp3‐/‐ mutant. DE, dentin; EN, enamel.
Similar alterations in enamel formation were also found in molars, consistent with the observations made for incisors (Fig. 5). Root regions in Ltbp3‐/‐ mutants showed a more condensed, unsteady pattern, according to the presence of bulbous‐like formations (Fig. 5B; oval area shown by a broken line), and an irregular distribution of dentinal tubules beneath a reduction of cementum thickness (Fig. 5A,B, white arrowheads; Fig. 5C,D). These findings suggest that LTBP‐3 is required for ameloblasts to secrete a proper enamel matrix, to establish a typical enamel rod structure, and for formation of normal dentinal tissue.
Figure 5.
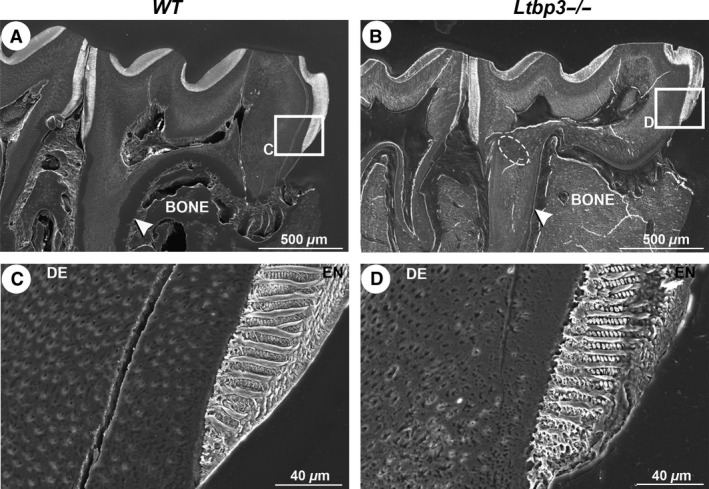
Scanning electron microscopy analysis of sagittal sections of lower molars. (A, B) In this section of the mid‐sagittal plane, the enamel thickness observed in the molar of latent transforming growth factor beta binding protein mutant (Ltbp3‐/‐) mouse is nearly identical to that of wild‐type (WT) mouse. The white arrows (A, B) indicate the cementum thickness, and the dashed oval (B) indicates a bulbous‐like formation. (C, D) Higher‐magnification views (boxed areas in A, B), showing an irregular organization of the enamel rods and a rough outer surface in the molar from Ltbp3‐/‐ mouse compared with that from WT mouse. The dentinal tubule distribution is also impaired. BONE, alveolar bone; DE, dentin.
Discussion
Transforming growth factor‐β proteins are thought to play a major role in the morphogenesis of developing teeth 14. Both TGF‐β1 and TGF‐β3 are produced by the enamel organ and are activated by components of the basement membrane, including those induced by odontoblast differentiation 28. Inactivation of the Tgfbr2 gene (which encodes TGF‐β receptor II) increases the proliferation of odontogenic epithelial cells 29. Both Tgfbr2 conditional knockout mice and Tgfß1 over‐expressing transgenic mice exhibit an abnormal tooth phenotype at early stages of enamel formation 11, 12. As TGF‐β proteins are present in the extracellular matrix as inactive forms, the absence of the targeting LTBP probably results in reduced TGF‐β activity, mirroring other Tgfβ mouse mutant models 15. The incisors of Ltbp3‐/‐ mice have a partial white color, with some yellow–orange bands centrally, indicative of loss of enamel, as confirmed by micro‐CT, histology, and scanning electron microscopy analyses. In accordance with this result, TGF‐β‐activating SMAD2 and SMAD3, and TGF‐inhibiting SMAD7, are found both in the enamel epithelium and in the dental mesenchyme, with alterations in these SMAD signaling components producing a variety of tooth phenotypes 14, 30, 31, 32.
Both histological and micro‐CT analyses confirmed alterations in enamel formation in the mutants, with obvious reductions in incisor and molar mineralized enamel volume. A gradient of reduced enamel mineralization seen from central to lateral zones of incisors, and from the first to the third molars, suggests site‐specific or temporally defined roles of LTBP‐3. In addition, some incisor enamel areas also showed enamel nodules, especially at the lateral zones near the alveolar bone crest, suggesting that enamel formation and ameloblast differentiation are not maintained throughout tooth development. Normal fully formed murine incisor enamel has a decussating mineralized rod pattern. Each rod is presumably formed from a single ameloblast, preserving a complete record of its migratory path during its formation 3. Our scanning electron microscopy analysis showed enamel defects throughout the lower and upper incisors, as the prism patterns always failed to form. Consistently, SMAD3‐signaling mutants showed alterations in enamel mineralization 13. In addition, Klk4 is down‐regulated in 7‐d‐old Tgfbr2 conditional mouse mutants 12. The phenotypic changes in our Ltbp3‐/‐ mice suggest that active TGF‐β signaling complexes may regulate enamel matrix protein levels and/or enamel proteases, such as KLK‐4, thereby regulating dental hard‐tissue mineralization.
Severe ameloblast morphological alterations are observed at the maturation stage, revealed as both shortening and severe morphological alterations of these cells. Transforming growth factor‐β family members are known to regulate blood‐vessel formation and hematopoiesis 33, and in our histological analysis we observed some vessel enlargements and an impaired papillary layer below the ameloblasts. This suggests that the ameloblast alterations may potentially be caused by a vascularization defect, although this hypothesis will need further analysis to be substantiated. It has been reported that transgenic over‐expression of TGF‐β1 in early secretory stage ameloblasts triggers dentin adhesion and the formation of cyst‐like structures and mineralized globules 11. The loss of enamel ribbon formation observed in Ltbp3‐/‐ mice confirms that enamel defects must start at the beginning of enamel deposition, suggesting that LTBP‐3 signaling regulates enamel mineralization at early stages, with the effects continuing through amelogenesis.
Dental‐targeted over‐expression of TGF‐β1 (using a Dspp–Tgfβ1 transgene) results in a phenotype similar to dentin dysplasia 34, and over‐expression of TGF‐β2 in the dentin matrix results in the reduction of dentin hardness and elasticity in male mice 35. Our histological analysis shows no obvious changes in dentin or odontoblasts in Ltbp3‐/‐ mutants. Osteodentin, which was found in teeth from Dspp–Tgfβ1 transgenic mice 11, was not found in Ltbp3‐/‐ mice. Even so, scanning electron microscopy analysis reveals that Ltbp3‐/‐ mice have an impairment of dentinal tubular patterns, suggesting an indirect effect through alterations of enamel differentiation.
Some regions of the Ltbp3‐/‐ molar roots have irregular bulbous‐like formations under the cementum. Despite this, the overall root pattern is consistently similar to that of WT mice. In vivo assays of TGF‐β signaling effects in ameloblasts using Ki67, a cellular marker for proliferation, showed marked proliferation in the Hertwig's epithelial root sheath (HERS) at postnatal day 7 12. Likewise, the presence of LTBP‐3 in outer dental epithelium could regulate molar proliferation 24, explaining the reduction of cementum thickness.
Ltbp3‐/‐ mice also had striking craniofacial defects, such as reductions of viscerocranium and dome‐shape neurocranium, which suggests premature ossification of some cranial sutures 19. Our results are consistent with previous work on Ltbp3‐/‐ mice, which described craniofacial malformations 20 and a high bone‐mass phenotype 21. Our measurements show that the reduction of the upper jaw is greater than that of the mandible (see molar relationship classification in Fig. S4).
Our previous study reported that four unrelated families had mutations in LTBP3, which probably produced AI and brachyolmia phenotypes. This highlights the role of LTBP‐3 and TGF‐β signaling in amelogenesis 24. Here, our findings of morphological changes within all tooth components, including enamel and ameloblasts, of Ltbp3‐knockout mice supports multiple roles of LTBP‐3 in odontogenesis. Understanding how LTBP‐3 regulates hard‐tissue mineralization and tooth development might provide treatment strategies for patients with a variety of craniofacial anomalies.
The Ltbp3‐/‐ mouse phenotype described herein is characterized by severe enamel defects and overlaps with those of human LTBP3 mutations and mouse mutants for other genes of the TGF‐β pathway. This study demonstrates that LTBP‐3 is necessary for initiating and maintaining an enamel matrix, for regulating ameloblast differentiation, for complete enamel prism formation, and for establishing root mineralization.
Supporting information
Fig. S1. Anatomical landmarks used for morphometric analysis.
Fig. S2. Form difference matrix analysis of skulls and mandibles.
Fig. S3. Bar graph of mean enamel volume of WT vs. Ltbp3‐/‐ mutants.
Fig. S4. Molar relationship classification: the wild‐type mice have molar class I occlusions (a normal occlusion), but Ltbp3‐/‐ mutants have a class III molar bite (otherwise known as an anterior bite).
Acknowledgements
We would like to thank Brigitte Schuhbaur and Valérie Fraulob for their help in histological procedures, Isabelle Goncalves Da Cruz for her help in micro‐CT training, and Arnaud Duchon for his expertise in applying 3D landmark coordinates analysis. This work was supported by a training grant from the Centre Européen de Recherche en Biologie et en Médecine (CERBM GIE), institutional funds from the CNRS, Inserm, and University of Strasbourg, grants from the French Ministry of Health (National Program for Clinical Research, PHRC 2005 #4266 ‘Amelogenesis imperfecta’, HUS, API 2009‐2012 ‘Development of the oral cavity: from gene to clinical phenotype in Human’), and IFRO (Institut Français pour la Recherche Odontologique) to Agnès Bloch‐Zupan, grant CA034282 from the National Institutes of Health to Daniel B. Rifkin, and Franco‐Thai and Khon Kaen University scholarships to Supawich Morkmued. This initiative is supported by the project #1.7 ‘RARENET: a tri‐national network for education, research and management of complex and rare diseases in the Upper Rhine’ co‐financed by the European Regional Development Fund (ERDF) of the European Union in the framework of the INTERREG V Upper Rhine program. Agnès Bloch‐Zupan is a 2015 Fellow of the Institute of Advanced Studies (USIAS) of the University of Strasbourg.
Conflicts of interest
The authors have no conflict of interest to declare.
Morkmued S, Hemmerle J, Mathieu E, Laugel‐Haushalter V, Dabovic B, Rifkin DB, Dollé P, Niederreither K, Bloch‐Zupan A. Enamel and dental anomalies in latent‐transforming growth factor beta‐binding protein 3 mutant mice. Eur J Oral Sci 2017; 125: 8–17. © 2017 The Authors. Eur J Oral Sci published by John Wiley & Sons Ltd
References
- 1. Habelitz S. Materials engineering by ameloblasts. J Dent Res 2015; 94: 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pugach MK, Gibson CW. Analysis of enamel development using murine model systems: approaches and limitations. Front Physiol 2014; 5: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simmer JP, Richardson AS, Hu YY, Smith CE, Ching‐Chun Hu J. A post‐classical theory of enamel biomineralization.. and why we need one. Int. J Oral Sci 2012; 4: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res 2010; 89: 1024–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartlett JD, Simmer JP. Kallikrein‐related peptidase‐4 (KLK4): role in enamel formation and revelations from ablated mice. Front Physiol 2014; 5: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartlett JD, Skobe Z, Nanci A, Smith CE. Matrix metalloproteinase 20 promotes a smooth enamel surface, a strong dentino‐enamel junction, and a decussating enamel rod pattern. Eur J Oral Sci 2011; 119(Suppl 1): 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatakeyama J, Fukumoto S, Nakamura T, Haruyama N, Suzuki S, Hatakeyama Y, Shum L, Gibson CW, Yamada Y, Kulkarni AB. Synergistic roles of amelogenin and ameloblastin. J Dent Res 2009; 88: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, Papagerakis P, Hunter GK, Feng JQ, Yamakoshi F, Simmer JP. Enamel defects and ameloblast‐specific expression in Enam knock‐out/lacz knock‐in mice. J Biol Chem 2008; 283: 10858–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wan M, Cao X. BMP signaling in skeletal development. Biochem Biophys Res Commun 2005; 328: 651–657. [DOI] [PubMed] [Google Scholar]
- 10. Chen G, Deng C, Li YP. TGF‐beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci 2012; 8: 272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haruyama N, Thyagarajan T, Skobe Z, Wright JT, Septier D, Sreenath TL, Goldberg M, Kulkarni AB. Overexpression of transforming growth factor‐beta1 in teeth results in detachment of ameloblasts and enamel defects. Eur J Oral Sci 2006; 114(Suppl 1): 30–34; discussion 39‐41, 379. [DOI] [PubMed] [Google Scholar]
- 12. Cho A, Haruyama N, Hall B, Danton MJ, Zhang L, Arany P, Mooney DJ, Harichane Y, Goldberg M, Gibson CW, Kulkarni AB. TGF‐ss regulates enamel mineralization and maturation through KLK4 expression. PLoS ONE 2013; 8: e82267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yokozeki M, Afanador E, Nishi M, Kaneko K, Shimokawa H, Yokote K, Deng C, Tsuchida K, Sugino H, Moriyama K. Smad3 is required for enamel biomineralization. Biochem Biophys Res Commun 2003; 305: 684–690. [DOI] [PubMed] [Google Scholar]
- 14. Klopcic B, Maass T, Meyer E, Lehr HA, Metzger D, Chambon P, Mann A, Blessing M. TGF‐beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur J Cell Biol 2007; 86: 781–799. [DOI] [PubMed] [Google Scholar]
- 15. Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF‐beta ‐ an excellent servant but a bad master. J Transl Med 2012; 10: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Todorovic V, Rifkin DB. LTBPs, more than just an escort service. J Cell Biochem 2012; 113: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Yin W, Perez‐Jurado L, Bonadio J, Francke U. Mapping of human and murine genes for latent TGF‐beta binding protein‐2 (LTBP2). Mamm Genome 1995; 6: 42–45. [DOI] [PubMed] [Google Scholar]
- 18. Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor‐beta binding protein gene (LTBP‐3). J Biol Chem 1995; 270: 10147–10160. [DOI] [PubMed] [Google Scholar]
- 19. Dabovic B, Chen Y, Colarossi C, Obata H, Zambuto L, Perle MA, Rifkin DB. Bone abnormalities in latent TGF‐[beta] binding protein (Ltbp)‐3‐null mice indicate a role for Ltbp‐3 in modulating TGF‐[beta] bioavailability. J Cell Biol 2002; 156: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dabovic B, Chen Y, Colarossi C, Zambuto L, Obata H, Rifkin DB. Bone defects in latent TGF‐beta binding protein (Ltbp)‐3 null mice; a role for Ltbp in TGF‐beta presentation. J Endocrinol 2002; 175: 129–141. [DOI] [PubMed] [Google Scholar]
- 21. Dabovic B, Levasseur R, Zambuto L, Chen Y, Karsenty G, Rifkin DB. Osteopetrosis‐like phenotype in latent TGF‐beta binding protein 3 deficient mice. Bone 2005; 37: 25–31. [DOI] [PubMed] [Google Scholar]
- 22. Noor A, Windpassinger C, Vitcu I, Orlic M, Rafiq MA, Khalid M, Malik MN, Ayub M, Alman B, Vincent JB. Oligodontia is caused by mutation in LTBP3, the gene encoding latent TGF‐beta binding protein 3. Am J Hum Genet 2009; 84: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dugan SL, Temme RT, Olson RA, Mikhailov A, Law R, Mahmood H, Noor A, Vincent JB. New recessive truncating mutation in LTBP3 in a family with oligodontia, short stature, and mitral valve prolapse. Am J Med Genet A 2015; 167: 1396–1399. [DOI] [PubMed] [Google Scholar]
- 24. Huckert M, Stoetzel C, Morkmued S, Laugel‐Haushalter V, Geoffroy V, Muller J, Clauss F, Prasad MK, Obry F, Raymond JL, Switala M, Alembik Y, Soskin S, Mathieu E, Hemmerle J, Weickert JL, Dabovic BB, Rifkin DB, Dheedene A, Boudin E, Caluseriu O, Cholette MC, McLeod R, Antequera R, Gelle MP, Coeuriot JL, Jacquelin LF, Bailleul‐Forestier I, Maniere MC, Van Hul W, Bertola D, Dolle P, Verloes A, Mortier G, Dollfus H, Bloch‐Zupan A. Mutations in the latent TGF‐beta binding protein 3 (LTBP3) gene cause brachyolmia with amelogenesis imperfecta. Hum Mol Genet 2015; 24: 3038–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verloes A, Jamblin P, Koulischer L, Bourguignon JP. A new form of skeletal dysplasia with amelogenesis imperfecta and platyspondyly. Clin Genet 1996; 49: 2–5. [DOI] [PubMed] [Google Scholar]
- 26. Richtsmeier JT, Lele S. A coordinate‐free approach to the analysis of growth patterns: models and theoretical considerations. Biol Rev Camb Philos Soc 1993; 68: 381–411. [DOI] [PubMed] [Google Scholar]
- 27. Cole III TM, Lele S, Richtsmeier JT. A parametric bootstrap approach to the detection of phylogenetic signals in landmark data. In: Macleod N, Forey PL, eds. Morphology, shape and phylogeny. London: Taylor & Francis, 2002; 194–219. [Google Scholar]
- 28. Lesot H, Lisi S, Peterkova R, Peterka M, Mitolo V, Ruch JV. Epigenetic signals during odontoblast differentiation. Adv Dent Res 2001; 15: 8–13. [DOI] [PubMed] [Google Scholar]
- 29. Chai Y, Zhao J, Mogharei A, Xu B, Bringas P Jr, Shuler C, Warburton D. Inhibition of transforming growth factor‐beta type II receptor signaling accelerates tooth formation in mouse first branchial arch explants. Mech Dev 1999; 86: 63–74. [DOI] [PubMed] [Google Scholar]
- 30. Xu X, Jeong L, Han J, Ito Y, Bringas P Jr, Chai Y. Developmental expression of Smad1‐7 suggests critical function of TGF‐beta/BMP signaling in regulating epithelial‐mesenchymal interaction during tooth morphogenesis. Int J Dev Biol 2003; 47: 31–39. [PubMed] [Google Scholar]
- 31. Ito Y, Zhang YW. A RUNX2/PEBP2alphaA/CBFA1 mutation in cleidocranial dysplasia revealing the link between the gene and Smad. J Bone Miner Metab 2001; 19: 188–194. [DOI] [PubMed] [Google Scholar]
- 32. Ferguson CA, Tucker AS, Heikinheimo K, Nomura M, Oh P, Li E, Sharpe PT. The role of effectors of the activin signalling pathway, activin receptors IIA and IIB, and Smad2, in patterning of tooth development. Development 2001; 128: 4605–4613. [DOI] [PubMed] [Google Scholar]
- 33. Massague J. TGF[beta] signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thyagarajan T, Sreenath T, Cho A, Wright JT, Kulkarni AB. Reduced expression of dentin sialophosphoprotein is associated with dysplastic dentin in mice overexpressing transforming growth factor‐beta 1 in teeth. J Biol Chem 2001; 276: 11016–11020. [DOI] [PubMed] [Google Scholar]
- 35. Saeki K, Hilton JF, Alliston T, Habelitz S, Marshall SJ, Marshall GW, Denbesten P. Elevated TGF‐beta2 signaling in dentin results in sex related enamel defects. Arch Oral Biol 2007; 52: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Anatomical landmarks used for morphometric analysis.
Fig. S2. Form difference matrix analysis of skulls and mandibles.
Fig. S3. Bar graph of mean enamel volume of WT vs. Ltbp3‐/‐ mutants.
Fig. S4. Molar relationship classification: the wild‐type mice have molar class I occlusions (a normal occlusion), but Ltbp3‐/‐ mutants have a class III molar bite (otherwise known as an anterior bite).


