Abstract
Complexin (Cpx) is thought to be a major regulator of soluble N-ethylmaleimide-sensitive factor-attachment protein receptor (SNARE)-dependent membrane fusion. Although the inhibition of membrane fusion by Cpx has been frequently reported, its structural basis has been elusive and an anticipated disruption of the SNARE core has never been observed. In the present study, to mimic the natural environment, we assembled a single SNAREpin between two nanodisc membrane patches. Single-molecule FRET (smFRET) detects a large conformational change, specifically at the C-terminal half, whereas no conformational change is observed at the N-terminal half. Our results suggest that Cpx splits the C-terminal half of the SNARE core at least 10 Å (1 Å=0.1 nm), whereby inhibiting further progression of SNARE zippering and membrane fusion.
Keywords: complexin, fluorescence/Förster resonance energy transfer (FRET), nanodisc, single molecule, soluble N-ethylmaleimide-sensitive factor-attachment protein receptor (SNARE)
INTRODUCTION
In the neuron, soluble N-ethylmaleimide-sensitive factor-attachment protein receptors (SNAREs) mediate vesicle fusion that releases neurotransmitters to the synaptic cleft. Vesicle-associated SNARE (v-SNARE) associates with target membrane-SNARE (t-SNARE) to form a complex that facilitates fusion of two membranes [1,2]. If unregulated, however, vesicle fusion would be spontaneous and random. Complexin (Cpx) is a small soluble SNARE-binding protein [3,4] that is believed to suppress such spontaneous fusion [4–7]. When evoked, a major Ca2+ - sensor synaptotagmin 1 (Syt1) triggers fast fusion [8,9], perhaps by lifting the Cpx clamp [10–12].
Presumably, Cpx may achieve the inhibition of spontaneous membrane fusion by blocking SNARE complex formation. However, the structural basis for such an inhibitory function of Cpx is hotly debated [13–17]. An X-ray structure shows that Cpx binds to the surface groove of the SNARE core, which is a four-helix bundle [18–20], in a conformation that stabilizes the structure instead of disrupting it [21]. Another X-ray structure shows that Cpx cross-links two adjacent SNARE cores [22]. However, caveats of this study are that it was performed with a truncated v-SNARE in which a bulk of the C-terminal residues that have been proposed to compete with Cpx was removed and also the Cpx that was used was mutated to act as a super clamp. It has not yet been elucidated that the wild-type (WT) Cpx can form this cross-linking structure. Alternatively, Cpx is shown to be capable of displacing v-SNARE from the core structure but the results are purely computational and have not been verified experimentally [23].
Given the confusing results, one might wonder whether the recombinant soluble SNARE core is a good model system to investigate the function of Cpx. In the absence of two opposing membranes, the isolated SNARE core is likely to represent the post-fusion conformation [24]. Thus, previous structural studies of Cpx might be depicting the post-fusion conformation with the isolated SNARE core rather than its involvement in the fusion process.
There is compelling evidence that cognate SNAREs zipper, starting from membrane distal N-terminal region and proceed towards the membrane-proximal C-terminal region [25,26]. Consistent with this hypothesis, an intermediate in which the N-terminal half is zippered and the C-terminal half is frayed has been recently characterized [27–29]. One might wonder whether this half-zippered SNARE complex is the primary target of Cpx and other regulators.
In the present study, we investigated, using single-molecule FRET (smFRET), the effect of Cpx binding to a single trans-SNAREpin trapped in the nanodisc sandwich. Our results demonstrate that Cpx has the capacity to split t- and v-SNAREs at the C-terminal half while maintaining the core structure at the N-terminal, whereby Cpx inhibits SNARE complex formation and membrane fusion.
MATERIALS AND METHODS
Plasmid construct and site-directed mutagenesis
DNA sequences encoding syntaxin 1A (Syn1A, amino acids 1–288 with three cysteines replaced by alanines), SNAP-25 (amino acids 1–206 with four native cysteines replaced by alanines), VAMP2 (amino acids 1–116 with Cys103 replaced by alanine), soluble VAMP2 (amino acids 1–96), complexin 1 (Cpx I, amino acids 1–134), N-terminally truncated Cpx (Cpx 27–134, amino acids 27–134) and N-terminally mutated Cpx (M5E/K6E, amino acids 1–134, Met5 and Lys6 replaced with glutamic acid) were inserted into the pGEX-KG vector as N-terminal GST fusion proteins. A modified apolipoprotein A1 (apoA1, amino acids, 27–267) was inserted into pNFXeX vector as an N-terminal His-tagged protein.
All cysteine mutants, including Syn1A I203C, Syn1A V241C, VAMP2 Q33C, VAMP2 A72C, soluble VAMP2 A72C and Cpx M5E/K6E were generated by the QuikChange site-directed mutagenesis (Stratagene). All DNA sequences were confirmed by the Iowa State University DNA Sequencing Facility.
Protein expression purification and fluorophore labelling
All recombinant proteins were expressed in Escherichia coli [BL21(DE3)] cells. Cells were first grown in LB medium at 37 °C with shaking at 200 rpm to an attenuance of 0.6–0.8 at 600 nm. IPTG (0.4 mM final concentration) was added to induce the protein production. For Syn1A I203C, Syn1A V241C, VAMP2 Q33C, VAMP2 A72C, cells were further grown at 16°C with shaking at 100 rpm for another 14–16 h. For soluble VAMP2 A72C, SNAP-25, apoA1 and Cpx, cells were grown at 20 °C with shaking at 100 rpm for another 14–16 h after induction.
For GST-tagged proteins, cell pellets were resuspended in 15 ml of PBST (PBS, pH 7.4, containing 0.2 Triton X-100) for membrane proteins and 15 ml of PBS (pH 7.4) for the soluble proteins with final concentration of 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF) and 4 mM DTT.
Cells were lysed by sonication in ice bath and centrifuged at 24000 g for 30 min at 4 °C. Except for apoA1, the supernatant was incubated with 2 ml of GST beads at 4 °C for 2 h. The proteins were then eluted by 0.02 unit/μl thrombin in cleavage buffer (PBS, pH 8.0, containing 2 mM DTT) with/without 0.8 n-octyl-D-glucopyranoside (OG) for membrane and soluble proteins respectively. ApoA1 was purified using the same protocol except for using the Ni-NTA column. Purified proteins were examined by SDS/15% (w/v) PAGE, and the purity was at least 85% for all proteins and the concentration was measured with RC DC kit (Bio–Rad Laboratories).
Single cysteine mutants of syntaxin 1A (I203C, V241C), soluble VAMP2 (A72C) and VAMP2 (Q33C, A72C) were desalted with a PD MiniTrap G-25 column (GE Healthcare) to eliminate free DTT and then incubated with a 10-fold molar excess of maleimide-derivative fluorophore indodicarbocyanine (Cy5) or indocarbocyanine (Cy3) respectively overnight at 4 °C. The labelled protein was purified using the PD MiniTrap G-25 column and free dye was further separated from the protein sample using centrifugal filters (Amicon). The labelling efficiency of each SNARE protein was measured spectrophotometrically (Beckman). Further details can be found in our recent paper [27].
Lipid reconstitution and purification of SNARE-incorporated nanodiscs
To generate homogeneously sized nanodiscs, POPC, DOPS, cholesterol, PIP2 and biotin–PEG–DSPE (Avanti Polar Lipids) lipid mixture with a molar ratio of 62.9:15:20:2:0.1 was used for t-SNARE nanodisc (t-nanodiscs) and cis-SNAREpin nanodiscs. The vesicle-SNARE nanodisc (v-nanodiscs) were consisted of POPC, DOPS and cholesterol with molar ratios of 75:5:20. The lipid mixture was initially dried with nitrogen gas and incubated under vacuum for 6–8 h and then resuspended with HEPES buffer (25 mM HEPES, pH 7.4, and 150 mM KCl) to a final concentration of 50 mM.
Then, 5 μl of 50 mM resuspended lipid mixture was first dissolved in sodium cholate such that the final concentration of sodium cholate was 50 mM after apoA1 and SNARE proteins were added. Then the t-SNARE binary complex (syntaxin 1A and SNAP-25 with a molar ratio of 1:2, pre-incubated at room temperature for 1 h), cis-ternary SNARE complex (syntaxin 1A, SNAP-25 and VAMP2 or soluble VAMP2 with a molar ratio of 1:2:1, pre-incubated at 4 °C overnight) or VAMP2 and apoA1 were added to the detergent-solubilized lipid mixture. The molar ratio of lipids, apoA1 and SNARE(s) was 300:5:1. After incubating for 20 min, the self-assembly of SNARE-incorporated nanodiscs was initiated via rapid removal of sodium cholate by adding 50% (w/v) SM-2 Bio-Beads (Bio–Rad Laboratories). The t-, v- or cis-nanodiscs were then purified through gel filtration using a Superdex™ 200 GL 10/300 column (GE Healthcare).
Preparation of trans- and cis-SNAREpin nanodiscs on the imaging surface for total internal reflection
The imaging surface was prepared by coating the quartz surface with a solution of methoxypoly(ethylene glycol) and biotin–PEG molecules (100:1). Flow chambers were assembled between the quartz slide and coverslip. Streptavidin (0.2 mg/ml) was introduced into and washed from the flow chamber for subsequent nanodisc immobilization via biotin–streptavidin conjugation.
For experiments with trans-SNAREpin nanodiscs, t-nanodiscs were immobilized on the surface and the unbound t-nanodiscs were washed out. Then v-nanodiscs were introduced into the flow chamber and incubated for 45 min to allow formation of a trans-SNAREpin between two nanodiscs (Figure 1). The unbound v-nanodiscs were then washed out.
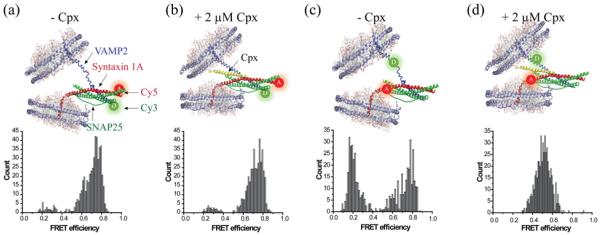
Figure 1. Cpx splits the C-terminal region of a single SNAREpin in the nanodisc sandwich.
(a) Nanodisc sandwich harbouring a single SNAREpin with the FRET pair at the N-terminal region (NN). VAMP2 Q33C–Cy3 and Syn1A I203C–Cy5 are used for NN. The majority of the population is distributed in the high FRET region. (b) Cpx at 2 μM does not affect the FRET distribution of NN. (c) Nanodisc sandwich harbouring a SNAREpin with the FRET pair at the C-terminal region (CC). VAMP2 A72C–Cy3 and Syn1A V241C–Cy5 were used to make CC. The distribution shows two distinct populations in the high and in the low FRET regions, but little in the mid FRET region. (d) Cpx at 2 μM pulls both the low and high FRET populations towards the mid FRET region. All experiments were independently conducted at least four times. Total of 499, 446, 502 and 524 traces were analysed for (a)–(c) and (d) respectively.
For experiments with WT and Cpx mutants, a 2 μM solution of the appropriate Cpx was injected into the flow chamber prior to the v-nanodisc injection and incubated for 10 min at room temperature (approximately 25 °C). Subsequent solutions were prepared such that the Cpx or Cpx mutant concentration was maintained throughout the experiments.
For experiments with cis-SNAREpins, a premixed solution of cis-SNAREpin nanodiscs and Cpx was prepared and injected into the flow chamber and unbound nanodiscs were washed out. A 2 μM concentration of Cpx was maintained throughout the experiments.
The imaging of the samples was performed at room temperature with the oxygen scavenger system [0.4% glucose (Sigma–Aldrich), 4 mM Trolox (Calbiochem), 1 mg/ml glucose oxidase (Sigma–Aldrich), 0.04 mg/ml catalase (Calbiochem)] in HEPES buffer with/without 2 μM Cpx. Further details can be found in our recent paper [27].
Data analysis of total internal reflection image recordings
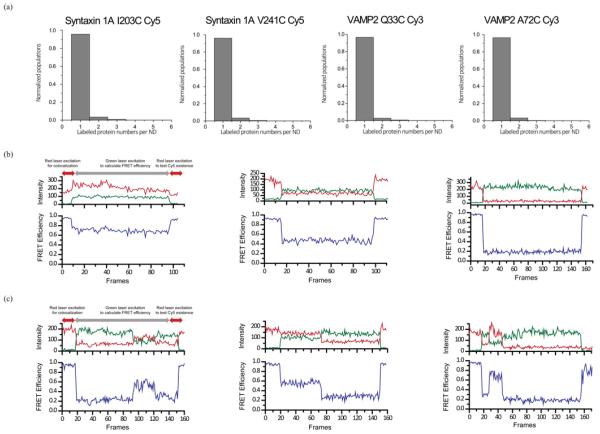
We analysed the traces from the total internal reflection (TIR) recordings using custom-designed software. The first 10–20 frames were recorded with excitation by a red laser (635 nm) in order to identify spots with t-nanodisc, with Cy5. Then, the light was switched to the green laser (532 nm) to select the trans-SNAREpin nanodisc sandwich or cis-SNAREpin nanodisc with both Cy3 (donor) and Cy5 (acceptor), which were characterized by the co-localized spots on both the acceptor and donor signal channel.
From the selected spots, the acceptor and donor time traces were analysed to obtain the FRET histograms. Because we seldom observed any significant transitions in the FRET value with our time resolution (200 ms per frame), we assigned a single FRET value (mean) for each trans- or cis-SNAREpin nanodisc (Figure 2b). We averaged FRET efficiency within the period in which both dyes were photon emitting. The rarely observed transitioning or fluctuating spots were not included in the histograms (Figure 2c).
Figure 2. smFRET for nanodisc sandwiches with SNAREs.
(a) Photobleaching experiment to confirm the number of labelled SNARE proteins in a single nanodisc. Distribution of Syn1A I203C–Cy5, Syn1A V241C–Cy5, VAMP2 Q33C–Cy3 and VAMP2 A72C–Cy3, from left to right. (b) Representative FRET traces from nanodisc sandwiches for high, mid and low FRET, from left to right. (c) Representative fluctuating FRET traces from nanodisc sandwiches for low–high–low, high–low and low–high–bleached FRET, from left to right. Such traces are rarely observed (<2%). The samples were initially excited with a red laser in order to find co-localized FRET pairs. The FRET efficiency of the specimen was calculated from the region depicted by grey arrows.
RESULTS
Cpx splits the C-terminal region of a single SNAREpin in the nanodisc sandwich
In order to understand the involvement of Cpx during the fusion process, we investigated single trans-SNAREpin in the chasm of two membranes using smFRET [27]. First, we prepared labelled proteins; the fluorescence donor Cy3 was attached site-specifically to an engineered cysteine on v-SNARE VAMP2 whereas the acceptor Cy5 was attached to t-SNARE Syn1A. We prepared two sets of dye pairs, one at the N-terminal residues (NN pair, Figures 1a and 1b) and the other at the C-terminal residues (CC pair, Figures 1c and 1d) to separately monitor the conformational changes in the N-terminal region from those in the C-terminal region.
The Cy3-labelled VAMP2 was incorporated into one population of the nanodisc (v-nanodisc). Meanwhile, the Cy5-labelled Syn1A was premixed with recombinant SNAP-25 and incorporated into another population of the nanodisc (t-nanodisc). The v- and t-nanodiscs were separately purified with FPLC. The protein concentration was adjusted in order to ensure that most nanodiscs had a single SNARE protein, which was later verified with photobleaching (Figure 2a).
The t-nanodiscs, doped with biotin–PEG–DSPE, were introduced into the flow cell and immobilized on to the PEGylated imaging surface. The v-nanodiscs were then injected into the flow cell to allow for the formation of the trans-SNAREpin in the middle of the nanodisc sandwich. The co-localized dots on the microscope image which have both the acceptor (Cy5) and donor signals (Cy3) were selected and the FRET efficiencies for the individual nanodisc pairs were analysed (Figures 1 and 2b).
For the NN pair, the population is distributed around the FRET efficiency E=0.75, consistent with short distance between the acceptor and the donor, which may reflect the robust helical structure at the N-terminal region. When 2 μM Cpx is added to this sample, no appreciable change in the population distribution of the FRET histogram is observed, indicating that Cpx has little effect on the SNARE conformation at the N-terminal region (Figures 1a and 1b).
For the CC pair, however, we observed two distinctly separated distributions, one peaked at E=0.2 and the other at E=0.8, with nearly equal counts (Figure 1c). The high FRET population reflects the fully zippered species whereas those at low FRET are from the half-zippered intermediate. It appears that these two species are energetically balanced to bear out near equal populations. We observe transitions between two states in a small number of time traces (<2%), indicating that it is likely to be a slow equilibrium (Figure 2c).
Surprisingly, when Cpx is added into the sample with the CC pair, a single distribution peaked at E=0.5 emerges. The population at low FRET and those at high FRET are both pulled into the middle (Figure 1d). Average distance changes from high FRET to mid FRET and from low FRET to mid FRET are estimated to be approximately 10–15 Å (1 Å=0.1 nm). Such rearrangements of the distribution reflect a significant conformational change for both fully zippered and half-zippered conformations. In particular, the disappearance of the high FRET distribution indicates that t- and v-SNAREs in the C-terminal region split at least 10 Å on average, most likely due to the insertion of Cpx into the SNARE core.
The cis-SNARE complex shows no splitting by Cpx
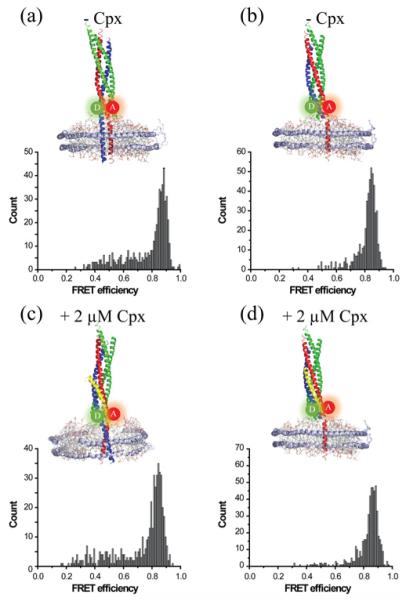
It has been previously shown that Cpx binds to the surface groove of the isolated SNARE core without causing structural disruption [21]. As controls, we investigated the effect of Cpx binding to cis-SNARE complexes. We prepared two types of cis-complexes; one sample with VAMP2, Syn1A and SNAP-25 anchored to the nanodisc with the transmembrane domains of VAMP2 and Syn1A (Figure 3a). In another sample, we prepared a nanodisc with soluble VAMP2, such that the cis-SNAREpin is anchored to the nanodisc with one transmembrane helix (Figure 3b). In both cases, in the absence of Cpx, we observe a single high FRET population with some minor populations in the low FRET region, consistent with fully zippered SNARE complex. Even when Cpx is added to samples we did not observe any appreciable change in the FRET distribution, indicating that no splitting between t- and v-SNAREs occurs in the presence of Cpx. Thus, the results show that, for the cis-SNAREpin, Cpx is incapable of inserting into the SNARE core and that trans-binding of v- and t-SNAREs to opposite membranes is necessary for Cpx insertion into the SNARE core. We observe a slight increase in the low FRET population when both transmembrane helices are present (Figures 3a and 3c) compared with just one, reflective of relative dynamic movement of the two transmembrane helices in the nanodisc membrane. Also, the high FRET peak of the cis-complex is shifted to a higher FRET value (Figure 3a), compared with the trans-complex (Figure 1c), which is indicative of a more compact structure.
Figure 3. The cis-SNARE complex shows no splitting by Cpx.
The cis-SNAREpin with the CC FRET pair with two (a and c) or one (b and d) transmembrane domains without (a and b) or with (c and d) 2 μM Cpx. Cpx does not affect the cis-SNARE complex. All experiments were independently conducted at least four times. Total of 516, 499, 489 and 517 traces were analysed for (a)–(c) and (d) respectively.
The N-terminal region of Cpx plays a role in restructuring the trans-SNAREpin
It was previously shown that Cpx mutants M5E/K6E, in which two N-terminal positions 5 and 6 are altered, and Cpx-(27–134), in which N-terminal 26 residues were deleted, both suppress spontaneous fusion significantly [30,31]. We investigate these mutants with our SNARE zippering assay to look for the structural basis for the enhanced fusion-suppressing activity of the Cpx mutants.
When the Cpx double mutant M5E/K6E was added to the nanodisc sandwich, harbouring the SNAREpin with the CC pair, we observed the disappearance of the high FRET population and the appearance of the mid FRET population as it was observed for WT Cpx. However, the low FRET population did not shift towards the mid FRET region in contrast with what was observed for WT Cpx (Figure 4). Thus, the results show that, although the mutant still maintains the ability to insert into and split the SNARE core, it loses some capacity to induce a conformational change that causes the v-SNARE to come closer to the t-SNAREs. We found that the effect of Cpx-(27–134) is very similar to that of Cpx M5E/K6E (Figures 4a and 4b). Thus, the results show that the N-terminal sequence of Cpx plays a role in inducing a conformational change in the v-SNARE VAMP2 to keep v- and t-SNAREs closer than that in the half-zippered intermediate.
Figure 4. The N-terminal region of Cpx plays a role in restructuring the trans-SNAREpin.
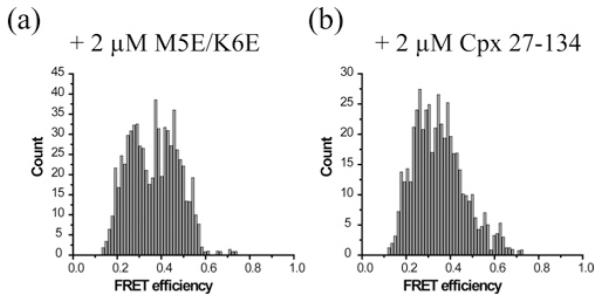
Both N-terminal Cpx mutants (a) M5E/K6E and (b) Cpx-(27–134) can pull the high FRET population down to mid FRET for CC. However, they are incapable of pushing the low FRET population to the mid FRET region. All experiments were independently conducted four times. Total of 704 and 494 traces were analysed for (a) and (b) respectively.
DISCUSSION
In the present study, we demonstrate that Cpx has the capacity to insert into and split the SNARE core, specifically in the C-terminal region, which may serve as a mechanism to inhibit SNARE zippering and membrane fusion. Such splitting is observed only when the SNAREpin is attached in trans to two opposed membrane patches but it is not seen when SNAREpin is attached to one nanodisc, either by one or two transmembrane domains. Thus, our results show that the repulsive force between two membranes is necessary for the proper Cpx function and two opposed membranes are an integral part of the regulatory machinery for synaptic vesicle fusion.
A previous EPR study revealed that the low FRET species corresponds to the half-zippered SNARE intermediate in which the C-terminal half of the VAMP2 SNARE motif is completely unstructured with an average distance of 65 Å [27]. Our results show that Cpx has the ability to pull the extreme ends of v-and t-SNAREs to be approximately 50 Å. We speculate that it involves a conformational change in C-terminal half of VAMP2, most likely from a random coil to an α-helix. A hypothetical model that represents such conformational changes is depicted in Figure 1(d).
Although our data suggest that Cpx might insert into a single SNARE core it is possible that Cpx might be able bring together the two opposing lipid membranes to a point where the half-zippered trans-SNAREpin exhibits mid FRET (CC pair) but the steric hindrance of Cpx could cause the high FRET population to be shifted lower, converging into a single mid FRET population. We also note that Rothman and co-workers proposed the possibility of Cpx binding to one SNAREpin and inserting, in trans, to a neighbouring SNAREpin [22]. We do not observe such in trans insertion in our single-molecule SNARE zippering assay. But our experiments were conducted under very low concentrations (100 pM range). The fusion site between synaptic vesicles and the plasma membrane bears a clouded environment. Therefore, we cannot rule out the possibility of such cross-binding occurring in synaptic vesicle fusion.
Care must be taken when relating Cpx-induced conformational changes in the SNARE core to the evoked release. In evoked release, the three-way interactions among Cpx, Syt1 and SNAREs would determine the efficiency and the time scales of vesicle fusion. Therefore, more work is definitely necessary to fully understand the Cpx phenotypes in evoked fusion. Whether Cpx inhibits spontaneous fusion or whether clamping of vesicle fusion by Cpx is necessary for synchronization or not are hotly debated issues [13–15]. Certainly, further work is needed to elucidate the Cpx function in synaptic vesicle fusion. Studies of the interaction between Cpx and SNAREpin will serve as a good starting point towards understanding the exquisite regulatory mechanism of synaptic vesicle fusion.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant number R01 GM051290 (to Y.-K.S.)].
Abbreviations
- apoA1
apolipoprotein A1
- Cpx
complexin
- Cy3
indocarbocyanine
- Cy5
indodicarbocyanine
- DOPS
1,2-dioleoyl-sn-glycero-3-phospho-L-serine
- DSPE
1,2-distearoyl-sn-glycero-3-phosphoethanolamine
- Ni-NTA
Ni2+ -nitrilotriacetate
- PIP2
1,2-dioleoyl-sn-glycero-3-phospho-(1′-myoinositol-4′,5′-bisphosphate)
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- smFRET
single-molecule FRET
- SNAP-25
25 kDa synaptosome-associated protein
- SNARE
soluble N-ethylmaleimide-sensitive factor-attachment protein receptor
- Syn1A
syntaxin 1A
- Syt1
synaptotagmin 1
- TIR
total internal reflection
- t-nanodisc
t-SNARE nanodisc
- t-SNARE
target membrane-SNARE
- VAMP-2
vesicle-associated membrane protein 2
- v-nanodisc
vesicle-SNARE nanodisc
- v-SNARE
vesicle-associated SNARE
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Yeon-Kyun Shin and Linxiang Yin designed experiments. Linxiang Yin performed all experiments. Jaewook Kim wrote the computer program for the data analysis. Yeon-Kyun Shin, Linxiang Yin and Jaewook Kim wrote the paper.
REFERENCES
- 1.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner W, Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishizuka T, Saisu H, Odani S, Abe T. Synaphin: a protein associated with the docking/fusion complex in presynaptic terminals. Biochem. Biophys. Res. Commun. 1995;213:1107–1114. doi: 10.1006/bbrc.1995.2241. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 4.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 5.Maximov A, Tang J, Yang X, Pang ZP, Südhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobson RJ, Liu Q, Watanabe S, Jorgensen EM. Complexin maintains vesicles in the primed state in C. elegans. Curr. Biol. 2011;21:106–113. doi: 10.1016/j.cub.2010.12.015. CrossRef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous and synaptic growth. Nat. Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Chacón R, Königstorfer A, Gerber SH, García J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Südhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 9.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 10.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 12.Schaub JR, Lu X, Doneske B, Shin Y, Mcnew JA. Hemifusion arrest by complexin is relieved by Ca2+ –synaptotagmin I. Nat. Struct. Mol. Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 13.Xue M, Stradomska A, Chen H, Brose N, Zhang W, Rosenmund C, Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7875–7880. doi: 10.1073/pnas.0803012105. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Cao P, Südhof TC. Deconstructing complexin function in activating and clamping Ca2+ -triggered exocytosis by comparing knockout and knockdown phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20777–20782. doi: 10.1073/pnas.1321367110. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trimbuch T, Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat. Rev. Neurosci. 2016;17:118–125. doi: 10.1038/nrn.2015.16. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 16.Trimbuch T, Xu J, Flaherty D, Tomchick DR, Rizo J, Rosenmund C. Re-examining how complexin inhibits neurotransmitter release: SNARE complex insertion or electrostatic hindrance? eLife. 2014;3:e02391. doi: 10.7554/eLife.02391. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnakumar SS, Li F, Coleman J, Schauder CM, Pincet F, Rothman JE, Reinisch KM. Re-visiting the trans insertion model for complexin clamping. eLife. 2015;4:e04463. doi: 10.7554/eLife.04463. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 1998;5:765–769. doi: 10.1038/1799. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 19.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic resolution exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Su TC. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 22.Kümmel D, Krishnakumar SS, Radoff DT, Li F, Giraudo CG, Pincet F, Rothman JE, Reinisch KM. Complexin cross-links prefusion SNAREs into a zigzag array. Nat. Struct. Mol. Biol. 2011;18:927–933. doi: 10.1038/nsmb.2101. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraudo CG, Garcia-Diaz A, Eng WS, Chen Y, Hendrickson WA, Melia TJ, Rothman JE. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lou X, Shin Y. SNARE zippering. Biosci. Rep. 2016;36:e00327. doi: 10.1042/BSR20160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sørensen JB, Wiederhold K, Müller EM, Milosevic I, Nagy G, de Groot BL, Grubmüller H, Fasshauer D. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 1999;6:117–123. doi: 10.1038/5803. PubMed. [DOI] [PubMed] [Google Scholar]
- 27.Shin J, Lou X, Kweon D, Shin Y, Korea S. Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre-fusion states. Biochem. J. 2014;459:95–102. doi: 10.1042/BJ20131668. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1344. doi: 10.1126/science.1224492. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Min D, Kim K, Hyeon C, Cho YH, Shin Y, Yoon T. Mechanical unzipping and rezipping of a single. Nat. Commun. 2013;4:1705–1710. doi: 10.1038/ncomms2692. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue M, Reim K, Chen X, Chao H-T, Deng H, Rizo J, Brose N, Rosenmund C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat. Struct. Mol. Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue M, Craig TK, Xu J, Chao H-T, Rizo J, Rosenmund C. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat. Struct. Mol. Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]