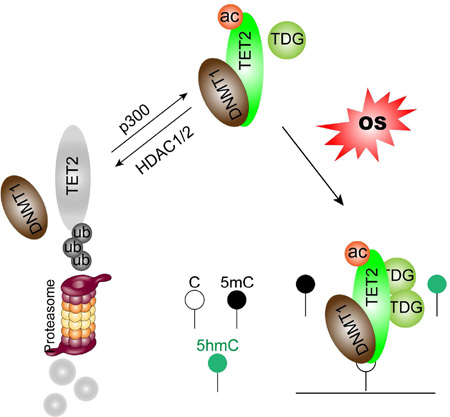
Summary
TET proteins, by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), are hypothesized, but not directly shown, to protect promoter CpG islands (CGIs) against abnormal DNA methylation (DNAm) in cancer. We define such a protective role linked to DNA damage from oxidative stress (OS) known to induce this abnormality. TET2 removes aberrant DNAm during OS through interacting with DNA methyltransferases (DNMTs) in a “Yin-Yang” complex targeted to chromatin and enhanced by p300 mediated TET2 acetylation. Abnormal gains of DNAm and 5hmC occur simultaneously in OS and knocking down TET2 dynamically alters this balance by enhancing 5mC and reducing 5hmC. TET2 reduction results in hypermethylation of promoter CGIs and enhancers in loci largely overlapping with those induced by OS. Thus, TET2 indeed may protect against abnormal, cancer DNAm in a manner linked to DNA damage.
Keywords: TET2, acetylation, DNA methylation, oxidative stress, 5hmC, DNA demethylation, cancer
Graphical abstract
Introduction
In the present study, we propose a mechanism for how inactivation of the ten-eleven translocation (TET) family of proteins may contribute to key DNA methylation (DNAm) abnormalities in cancer. Studies of these proteins have provided the most exciting recent development in DNAm research with the recognition that they initiate active DNA demethylation from the genome (Kohli and Zhang, 2013; Pastor et al., 2013). Through a base-flipping mechanism (Hashimoto et al., 2014; Hu et al., 2013), TET proteins oxidize 5mC to 5hmC (Ito et al., 2010; Tahiliani et al., 2009), with subsequent formation of 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011). The latter two marks can be excised by thymine-DNA glycosylase (TDG) and restored to unmodified cytosine through the base excision repair (BER) pathway (Wu and Zhang, 2014). Alternatively, conversion of 5mC to 5hmC can trigger passive, DNA replication dependent DNA demethylation by interfering with the DNAm maintenance process (Hashimoto et al., 2012; Wu and Zhang, 2014).
The discovery of TET proteins is also fundamental to fully understand DNAm abnormalities in cancer which involve abnormal widespread losses and more focal gains in promoter CpG islands (CGIs). This latter alteration affects hundreds of genes in individual tumors, including well characterized tumor suppressor genes that are transcriptionally silenced in association with this change (Baylin and Jones, 2011; Jones and Baylin, 2007; Shen and Laird, 2013). With regards to the above focal gains of promoter CGIs DNAm in cancer, loss of a normal protective role against unwanted DNAm has been proposed for TET proteins (Williams et al., 2012). Indeed, TET2 is frequently mutated in hematological malignancies (Delhommeau et al., 2009; Langemeijer et al., 2009), and patients carrying TET2 mutations often show significantly reduced global 5hmC levels (Ko et al., 2010). Interestingly, loss of 5hmC is also observed in many solid tumors (Haffner et al., 2011; Jin et al., 2011; Yang et al., 2013) where TET2 mutations are rarely detected, indicating other TET2 inactivation mechanisms may exist (Wu and Zhang, 2014). However, how TET2 inactivation and loss of 5hmC affect promoter CGIs DNAm has been controversial (Figueroa et al., 2010; Ko et al., 2010). Interestingly, recent studies suggest that loss of TET2 may lead to hypermethylation of enhancers (Hon et al., 2014; Lu et al., 2014; Rasmussen et al., 2015).
We now propose a new mechanism, tied to oxidative stress (OS), for the role of TET2 in protecting against abnormal DNAm in normally unmethylated promoter CGIs and enhancers. Chronic inflammation, and closely related OS, has long been accepted as an essential component of tumorigenesis and linked directly to cancer formation (Coussens and Werb, 2002; Federico et al., 2007). Recent studies revealed that epigenetic alterations, particularly DNA hypermethylation, likely play important roles in inflammation/OS associated carcinogenesis (Franco et al., 2008). We have recently defined DNMT1-containing repressive complexes that become tightly bound to chromatin during OS and induce abnormal gains of DNAm in basally low-expression gene promoters (O'Hagan et al., 2011), suggesting a key potential mechanism for inducing focal gains of cancer-specific aberrant DNAm. We now demonstrate TET2 forms “Yin-Yang” complexes with DNMTs and is targeted to chromatin during OS. TET2 actively removes abnormal DNAm induced by OS in promoter CGIs as well as enhancers by converting unwanted 5mC to 5hmC. Long-term reduction of TET2 caused even more DNA hypermethylation on these gene promoters and enhancers. In addition, we show TET2 is regulated by acetylation which increases its enzymatic activity, protein stability and partnering with DNMT1, thus enhancing its DNAm protection function. Deacetylation of TET2 by HDAC1/2 that are often overexpressed in cancers may represent another mechanism impairing TET2 functions and contributing to abnormal DNAm in cancer. Our data thus also suggest an additional translational implication for inhibiting HDACs in cancer.
Results
TET2 is acetylated by p300
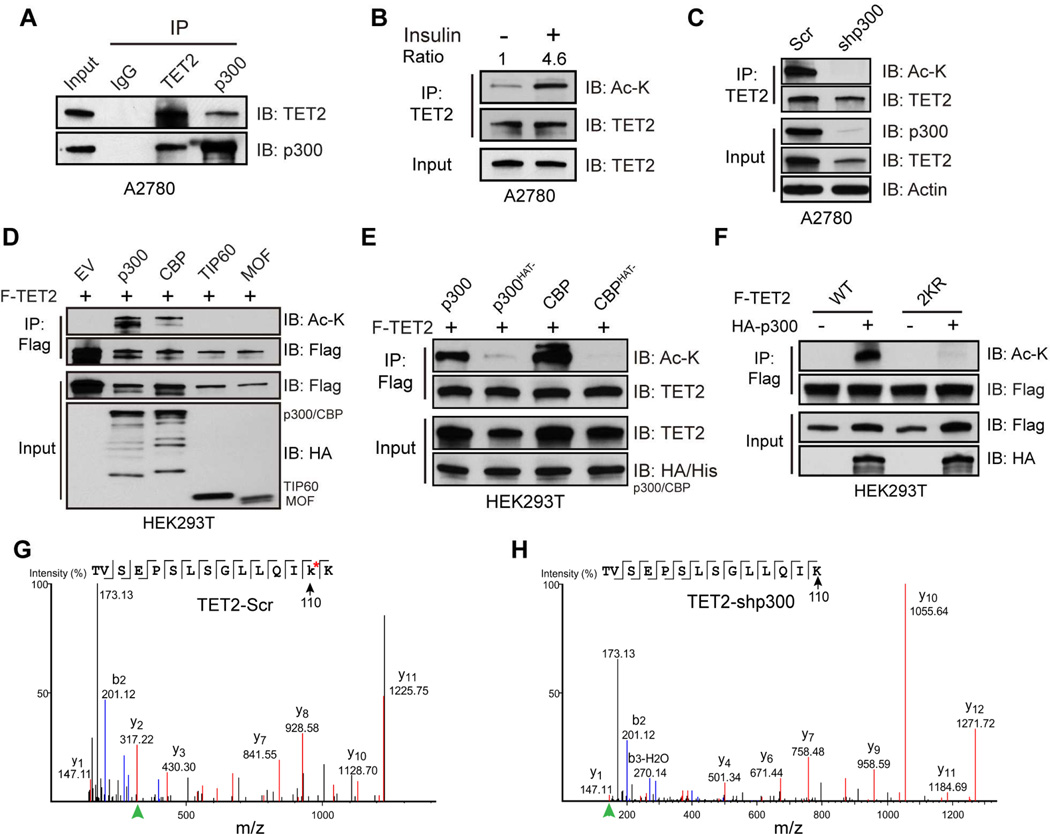
We have first explored the potential functional significance for the facts that 5hmC, as well as 5fC, is enriched significantly at binding sites for the acetyltransferase p300 throughout the genome (Song et al., 2013a; Yu et al., 2012) and that TET2 is an acetylated protein (Weinert et al., 2013). We have worked with ovarian cancer cell line A2780 and colorectal cancer cell line HCT116 that express endogenous TET2 within the range of levels in normal tissues (Figure S1A–C). The TET2 antibody employed detects endogenous TET2 proteins in most normal human tissues (Uhlen et al., 2015) and its specificity was further confirmed by shRNA-mediated TET2 knockdown and CRISPR-mediated TET2 knockout (Figure S1D and S1E). There is a robust interaction between endogenous TET2 and p300 (Figure 1A). Acetylation of endogenous TET2 is readily detected, and its level increases more than 4-fold upon insulin treatment (Figure 1B), which stimulates endogenous p300 acetyltransferase activity through the PI3K/Akt pathway (Inuzuka et al., 2012). Conversely, knocking down p300 completely abolishes TET2 acetylation (Figure 1C). Interestingly, acetylation of ectopic TET2 is enhanced by co-expressing p300 or CBP, but not TIP60 or MOF (Figure 1D), and is dependent on their acetyltransferase activities (Figure 1E). Moreover, all three TETs can be acetylated by p300 (Figure S1F), suggesting acetylation may represent a general regulatory mechanism for this family of proteins.
Figure 1. TET2 is acetylated by p300.
(A) Endogenous TET2 and p300 complexes were immunoprecipitated (IPed) from A2780 nuclear extract and analyzed by immunoblotting (IB). IgG was used as negative control. Whole cell extract (WCE) was used as input.
(B) Endogenous TET2 was IPed from A2780 cells treated with or without insulin (10 µg/ml, 1 hour) and analyzed by IB. Cells were serum starved for 24 hours, and pretreated with 2 µM TSA for 1 hour before insulin stimulation.
(C) Endogenous TET2 was IPed from A2780 cells infected with a scrambled (Scr) control shRNA or shp300 shRNA, and analyzed by IB.
(D and E) HEK293T cells were co-transfected with Flag-TET2 and various histone acetyltransferases (D), wildtype or catalytically inactive p300 or CBP. TET2 complexes were IPed and analyzed by IB.
(F) HEK293T cells were co-transfected with HA-p300 and Flag-TET2 wild type (WT) or 2KR mutant. TET2 complexes were IPed and analyzed by IB.
(G and H) Endogenous TET2 proteins from A2780 cells infected with scramble (G) or shp300 (H) virus were immunoprecipitated and analyzed by mass spectrometry. Residue 110 was identified as an acetylated lysine (highlighted with *) in control (G) but not p300 knockdown (H) cells. Peptide fragment ions used to calculate the mass of residue 110 (y2 in G and y1 in H) are highlighted by green arrowheads.
See also Figure S1.
Initial Mass spectrometry analysis of exogenous TET2 co-expressed with p300 reveals multiple potential acetylation sites, including two evolutionarily conserved (Figure S1J) lysine residues 110 and 111 (K110/111, or 2K, Figure S1G) in the N-terminus of TET2. Mutating these two residues completely abolishes p300 mediated TET2 acetylation (Figure 1F and S1H). Further mass spectrometry analysis reveals that lysine 110 is acetylated on endogenous TET2 proteins, and this acetylation is diminished with p300 knockdown (Figure 1G, 1F, and S1K). Interestingly, the full length of all three recombinant TET proteins possess higher enzymatic activities than their C-terminal catalytic domains (He et al., 2011; Hu et al., 2013), indicating the N-terminal domain may contain positive regulatory mechanisms. Two additional lysines (K53 and K1117) are also identified as bona fide acetylation sites (Figure S1G and S1H), but they do not affect full-length TET2 catalytic activity (Figure S1I) and are thus not studied further.
Deacetylation of TET2 by HDAC1/2
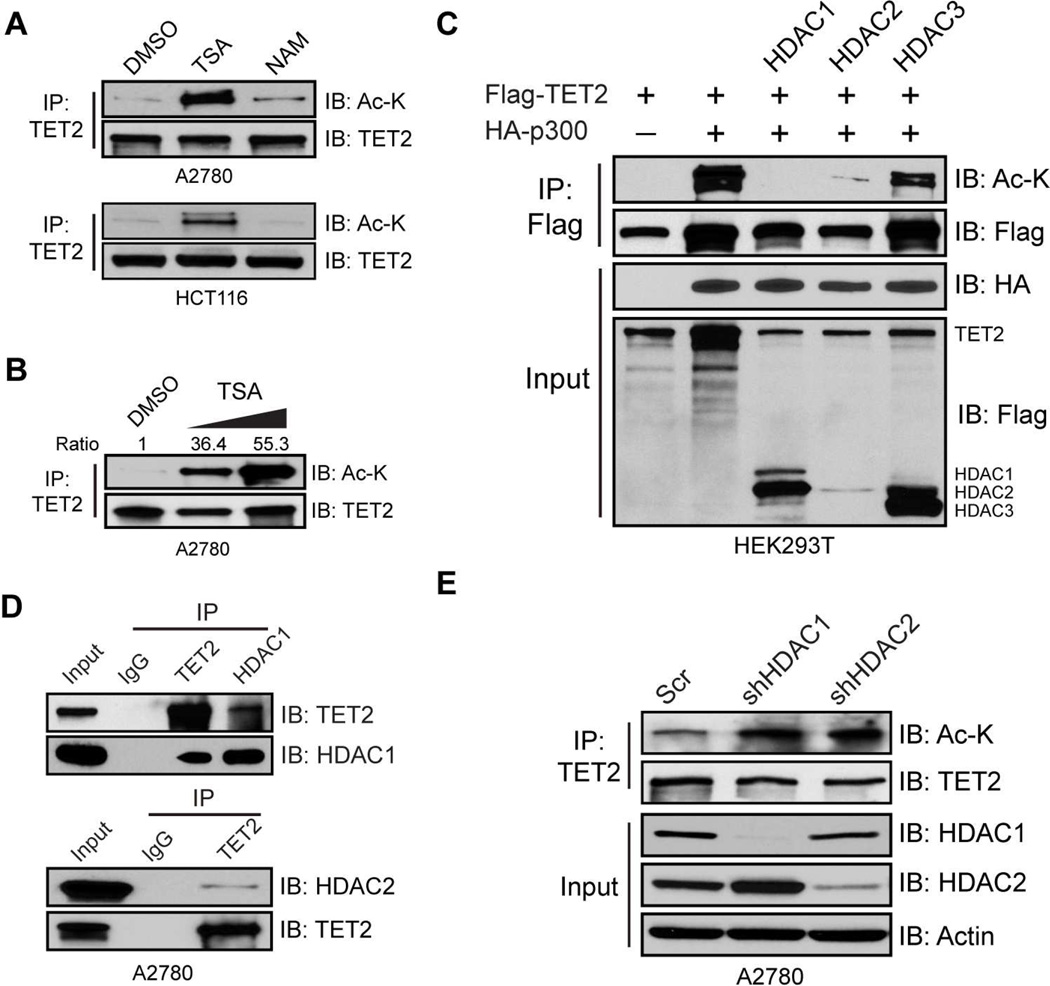
Treatment of A2780 cells with trichostatin A (TSA), or nicotinamide (NAM), class I, II, IV HDAC, and NAD+-dependent class III SIRT inhibitors, respectively, reveals the former, but not the latter, increases endogenous TET2 acetylation in a dose-dependent manner (Figure 2A and 2B). Induction of endogenous TET2 acetylation by TSA is also observed in HCT116 (Figure 2A), another cell line that expresses relatively high levels of TET2 (Figure S1A), suggesting TET2 acetylation may be a general phenomenon. Among the various HDACs, HDAC1/2/3 are the ones that predominantly localize in the nucleus and most sensitive to TSA. Co-expression of HDAC1 or HDAC2 with TET2/p300 completely abolishes TET2 acetylation while HDAC3 has much less influence (Figure 2C). Both HDAC1 and HDAC2 interact with TET2 (Figure 2D), and knocking down either increases endogenous TET2 acetylation (Figure 2E). Thus, HDAC1/2 are primary TET2 deacetylases.
Figure 2. HDAC1/2 deacetylate TET2.
(A and B) A2780 or HCT116 cells were treated with 2 uM TSA or 5 mM NAM (A) or increasing amounts of TSA (2uM and 5 uM) (B) for 6 hours. Endogenous TET2 was IPed and analyzed by IB. DMSO was used as a vehicle control.
(C) HEK293T cells were co-transfected with Flag-TET2/HA-p300, and HDAC1/2/3 constructs. TET2 proteins were IPed and analyzed by IB.
(D) TET2 and HDAC1/2 complexes were IPed from A2780 nuclear extract and analyzed by IB. IgG was used as a negative control. WCE was used as input.
(E) Endogenous TET2 proteins were IPed from A2780 cells infected with Scr, or HDAC1/2 shRNAs and analyzed by IB.
See also Figure S2
Acetylation enhances TET2 enzymatic activity
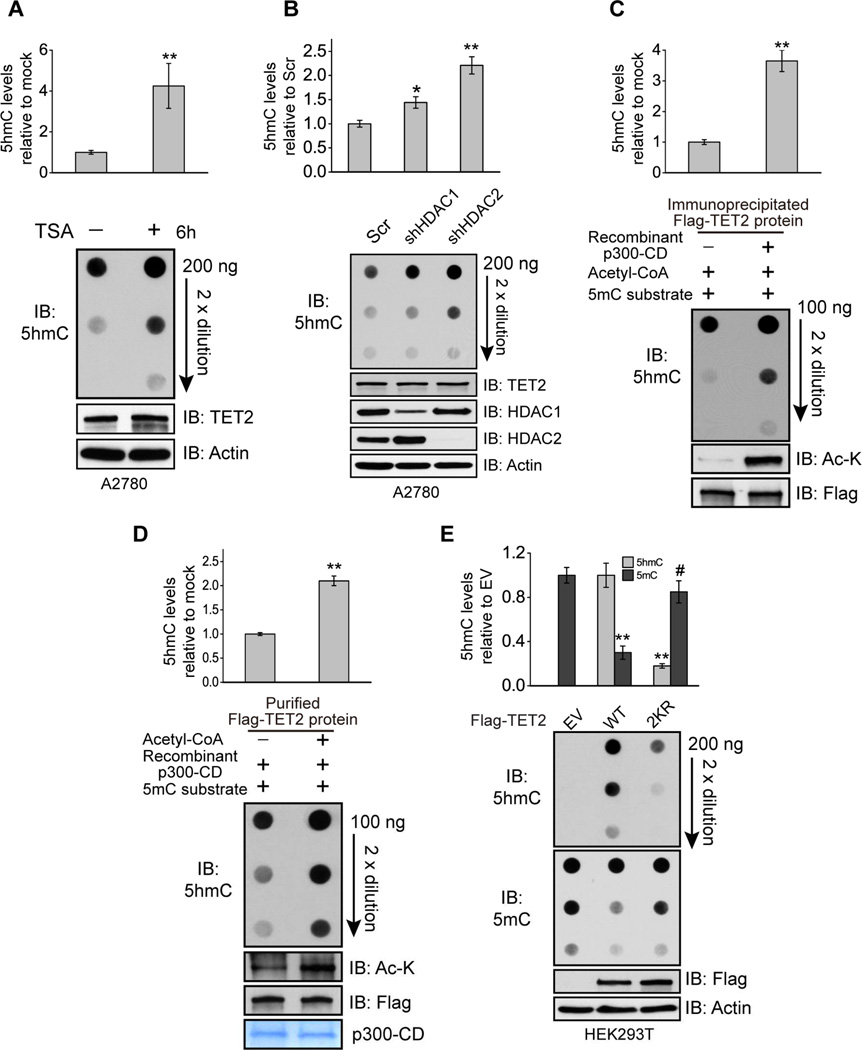
The above transient treatment (6 hours) with TSA, in addition to inducing endogenous TET2 acetylation (Figure 2A and 2B), significantly increases global 5hmC levels without affecting TET2 protein levels at this time point (Figure 3A). Similarly, knocking down either HDAC1 or HDAC2 also increases 5hmC levels (Figure 3B). When immunoprecipitated TET2 protein is pre-incubated with purified p300 catalytic domain, both TET2 acetylation and catalytic activity are enhanced (Figure 3C). Similarly, incubation of purified recombinant TET2 with p300 catalytic domain in the presence of Acetyl-CoA increases TET2 acetylation and the production of 5hmC (Figure 3D and S3A). Co-expression of wild type, but not a catalytically inactive mutant, p300 with TET2 increases global 5hmC levels by 3-fold along with increased TET2 acetylation (Figure S3B). While overexpression of wild type TET2 dramatically increases global 5hmC levels, and triggers DNA demethylation, the 2KR mutant, which blocks TET2 acetylation (Figure 1F), generates markedly less 5hmC/DNA demethylation and can no longer be activated by p300 (Figure 3E and Figure S3C). Taken together, these data demonstrate that acetylation of TET2 enhances its enzymatic activity.
Figure 3. Acetylation enhances TET2 enzymatic activity.
(A and B) Global 5hmC levels from genomic DNA extracted from A2780 cells treated with or without TSA for 6 hours (A), or infected with Scr or HDAC1/2 shRNAs (B) were quantified by 5hmC ELISA (upper) and 5hmC dot blot (bottom). Values represent mean ± SEM (n=3). *: P < 0.05, **: P < 0.01 (student’s t-test).
(C) Immunoprecipitated TET2 proteins bound on beads were divided equally and incubated with or without recombinant p300 catalytic domain. After wash, beads were incubated with 5mC substrates. The level of generated 5hmC was quantified by 5hmC ELISA (upper) and dot blot (bottom). Values represent mean ± SEM (n=3). **: P < 0.01 (student’s t-test).
(D) Purified recombinant TET2 protein (bound on beads) were divided equally and incubated with recombinant p300 catalytic domain in the presence or absence of acetyl-CoA. After wash, beads were incubated with 5mC substrates and the level of generated 5hmC was quantified by 5hmC ELISA (upper) and dot blot (bottom). Values represent mean ± SEM (n=2). **: P < 0.01 (student’s t-test).
(E) Global 5hmC and 5mC levels from genomic DNA extracted from HEK293T cells transfected with empty vector (EV), TET2 WT, or 2KR mutant were quantified by 5hmC ELISA (upper) and dot blot (bottom). Values represent mean ± SEM (n=3). **: P < 0.01; #: not significant (student’s t-test).
See also Figure S3.
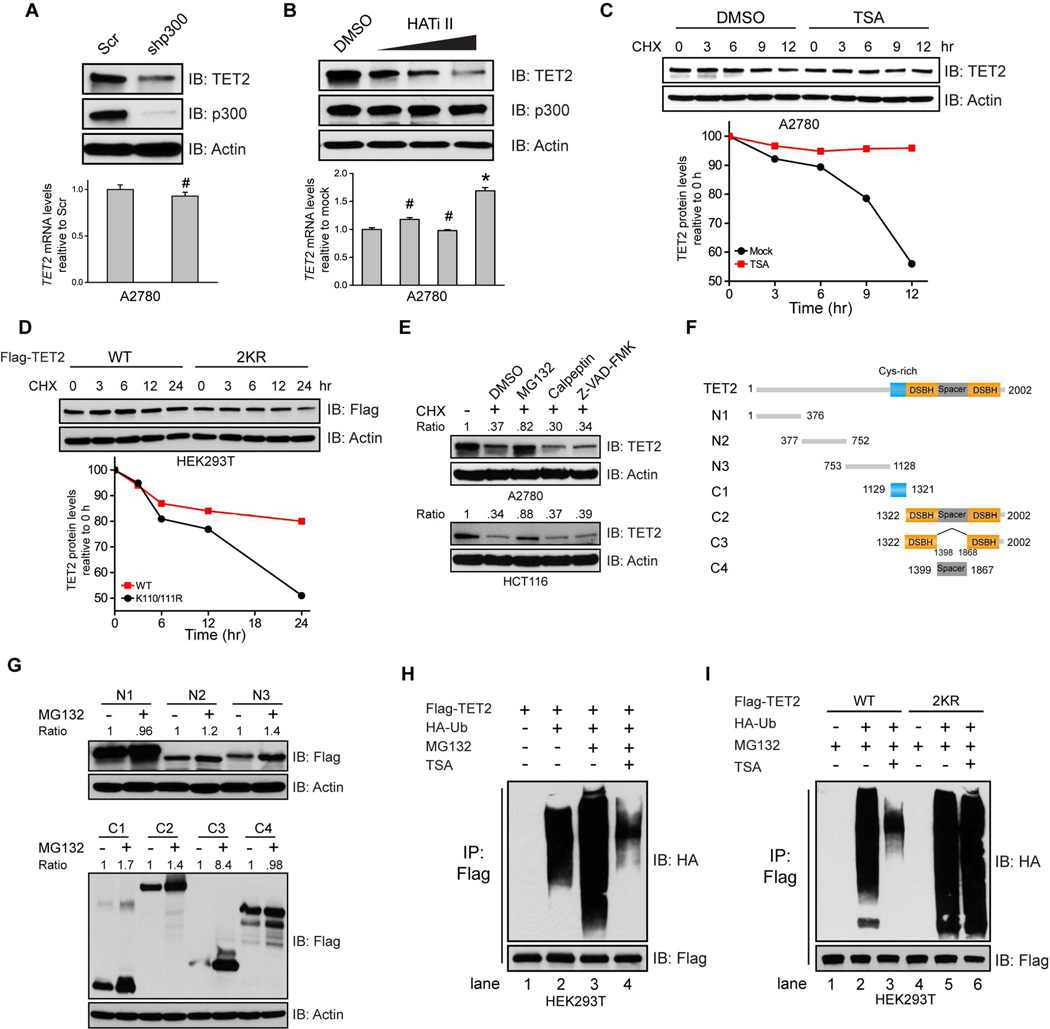
Acetylation stabilizes TET2 protein by inhibiting ubiquitination
Knocking down p300 not only abolishes TET2 acetylation (Figure 1C), but also dramatically reduces steady state TET2 protein levels without affecting its transcription (Figure 4A). Similarly, blocking p300 acetyltransferase activity by a p300 selective inhibitor which keeps p300 protein intact also decreases TET2 protein levels in a dose-dependent manner (Figure 4B). Moreover, TSA treatment which rapidly increases endogenous TET2 acetylation within 6 hours significantly extends TET2 half-life (Figure 4C, 9 hours and after). In contrast, the 2KR mutant is starkly less stable than the wild type protein (Figure 4D), demonstrating that acetylation stabilizes TET2
Figure 4. Acetylation stabilizes TET2 by inhibiting ubiquitination.
(A and B) A2780 cells were infected with either Scr or p300 shRNAs (A), or treated with DMSO, or increasing amount of p300 inhibitor (5, 10, and 20 uM) (B). Cell lysates were analyzed by IB (upper). TET2 mRNA was quantified by qRT-PCR (lower). Values represent mean ± SEM (n=3). #: not significant, *: P < 0.05 (student’s t-test).
(C and D) A2780 cells were treated with DMSO or 2 uM TSA (C), or HEK293T cells were transfected with WT or 2KR mutant for 24 hours (D), incubated with Cycloheximide (CHX, 100 ug/ml) and collected at indicated time points. Cell lysates were analyzed by IB. Band intensities were quantified by ImageJ and normalized to Actin.
(E) A2780 (upper) or HCT116 (lower) cells were treated with CHX (100 ug/ml) and various proteolysis inhibitors for 24 hours. TET2 protein levels were analyzed by IB. DMSO was used as a vehicle control. Cells without any treatment were used as control. MG132, proteasome inhibitor; Calpeptin, calpain inhibitor; Z-VAD-FMK: caspase inhibitor.
(F) Schematic presentation of TET2 truncations used in (G).
(G) HEK293T cells were transfected with various TET2 truncation constructs showed in (F) and treated with DMSO, or MG132 for 12 hours. Cell lysates were analyzed by IB.
(H and I) HEK293T cells were transfected with Flag-TET2 (H), TET2 WT or 2KR mutant (I), with or without HAUb for 24 hours, treated with DMSO or MG132 overnight. TSA were added 6 hours before harvest. TET2 proteins were IPed and analyzed by IB.
See also Figure S4.
It has been previously reported that the stability of TET proteins may be regulated by degradation pathways mediated by caspase and calpains (Ko et al., 2013; Wang and Zhang, 2014). Surprisingly, we find that in the cells under study, TET2 is stabilized by proteasome inhibitor MG132, but not calpains or caspase inhibitors (Figure 4E), and the conserved double-strand beta helix (DSBH) domain in the catalytic domain exhibits the most dramatic protein accumulation after MG132 treatment (Figure 4F and 4G). These findings indicate that TET2 protein may be primarily regulated through the ubiquitin-proteasome pathway and that the ubiquitination site(s) likely reside in the C-terminal DSBH domain. Indeed, we readily detected TET2 ubiquitination which is enhanced upon MG132 treatment (Figure 4H, lanes 1, 2, and 3). TSA treatment, while increasing TET2 acetylation (Figure 2A and 2B), significantly reduces TET2 ubiquitination (Figure 4H, lanes 3 and 4). Conversely, the acetylation deficient 2KR mutant exhibits stronger ubiquitination than does the wild type (Figure 4I, lanes 2 and 5) and resists reducing ubiquitination after TSA treatment (Figure 4I, lanes 5 and 6, compared to lanes 2 and 3). Since the ubiquitination site(s) reside in the C-terminal DSBH domain (Figure 4G), and the regulatory acetylation site K110 is in the N-terminus, our data suggest that acetylation may stabilize TET2 through recruiting other proteins which in turn protect the distant lysines from ubiquitination site(s).
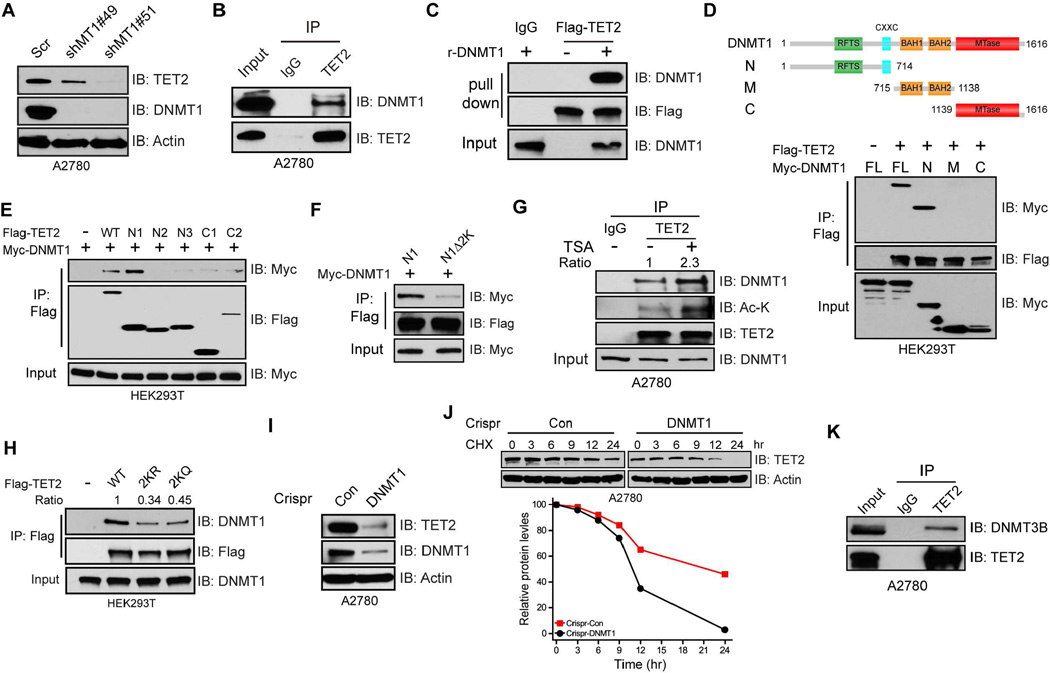
TET2 acetylation enhances DNMT1 binding to promote protein stability
In searching for the consequences of the above modifications of TET2, we wondered whether the acetylation defined above influences interactions of TET2 with other key DNAm regulating proteins, even those with opposing activities, such as DNMTs. We observed a reduction, or even loss, of TET2 protein upon acute DNMT1 knockdown (Figure 5A, and Figure S5A, S5B). Moreover, endogenous TET2 and DNMT1 interact (Figure 5B) in a TET2 enzymatic activity independent manner (Figure S5C). A direct binding between these two proteins, demonstrated using immunoprecipitated TET2 and purified recombinant DNMT1, is also readily detected (Figure 5C). We further determined that the N-terminal regulatory domain of DNMT1, known to interact with multiple proteins, is solely responsible for interacting with TET2 (Figure 5D), and that the N1 region of TET2 containing the K110/111 residues possesses the strongest association with DNMT1 (Figure 5E). A deletion mutant of TET2 N1 region lacking K110/111 shows severely reduced association with DNMT1 (Figure 5F), suggesting these two lysines are important for mediating DNMT1 interaction. Interestingly, treatment of cells with TSA, which increases TET2 acetylation (Figure 2A and 2B) and protein stability (Figure 4C), enhances interactions between endogenous TET2 and DNMT1 (Figure 5G). In contrast, the acetylation deficient 2KR mutant reduces this interaction by ~70% (Figure 5H and Figure S5D). Surprisingly, a TET2 2KQ mutant, which mimics constitutively acetylated status by neutralizing the positive charge but avoids acetylation, also decreases DNMT1 binding (Figure 5H). This finding suggests that it is perhaps the presence of acetylation, per se, that fosters interactions between DNMT1 and TET2. Lastly, DNMT1 binding seems essential for TET2 stability as the half-life of TET2 protein decreases significantly (Figure 5J) in a pool of cells that have been infected with DNMT1 CRISPR virus (Figure 5I). Taken together, our data demonstrate that acetylation of TET2 enhances DNMT1 binding and promotes its stability.
Figure 5. TET2 acetylation enhances DNMT1 binding to promote protein stability.
(A) A2780 cells were infected with Scr or DNMT1 shRNAs. Cell lysates were analyzed by IB.
(B) Endogenous TET2 complexes were IPed from A2780 nuclear extract and analyzed by IB.
(C) Immunoprecipitated TET2 bound on beads was incubated with or without recombinant DNMT1 (500 ng) overnight. Bound proteins were analyzed by IB. Rabbit IgG was used as a negative control.
(D and E) HEK293T cells were transfected with full length Flag-TET2 and various DNMT1 truncations (D) or full length Myc-DNMT1 and various TET2 fragments (E). TET2 complexes were IPed and analyzed by IB.
(F) HEK293T cells were transfected with full length DNMT1 and TET2-N1 or TET2-N1 deletion lacking K110/111. TET2 complexes were IPed and analyzed by IB.
(G) Endogenous TET2 complexes were IPed from A2780 cells treated with or without TSA and analyzed by IB. IgG was used as a negative control.
(H) TET2 complexes were IPed from HEK293T cells transfected with TET2 WT, 2KR, or 2KQ mutants and analyzed by IB. Non-transfected (Mock) cells were used as a negative control.
(I and J) A2780 cells were infected with non-targeting (Con) or DNMT1 LentiCRISPR viruses and selected by puromycin. Lysates from pooled cells were analyzed by IB (I). Control and DNMT1 knockdown cells treated with cycloheximide (CHX, 100 ug/ml) were collected at indicated time points and TET2 protein levels were analyzed by IB.
(K) TET2 complexes were IPed from A2780 nuclear extract and analyzed by IB. Rabbit IgG was used as a negative control.
See also Figure S5.
Finally, we show that all three TETs interact with the major maintenance methyltransferase, DNMT1 (Figure S5E), and that TET2 associates with not only DNMT1, but also the de novo methyltransferase, DNMT3B (Figure 5K and Figure S5F). These findings indicate a general relationship between TETs and DNMTs, and potential functional relevance of these TETs/DNMTs complexes.
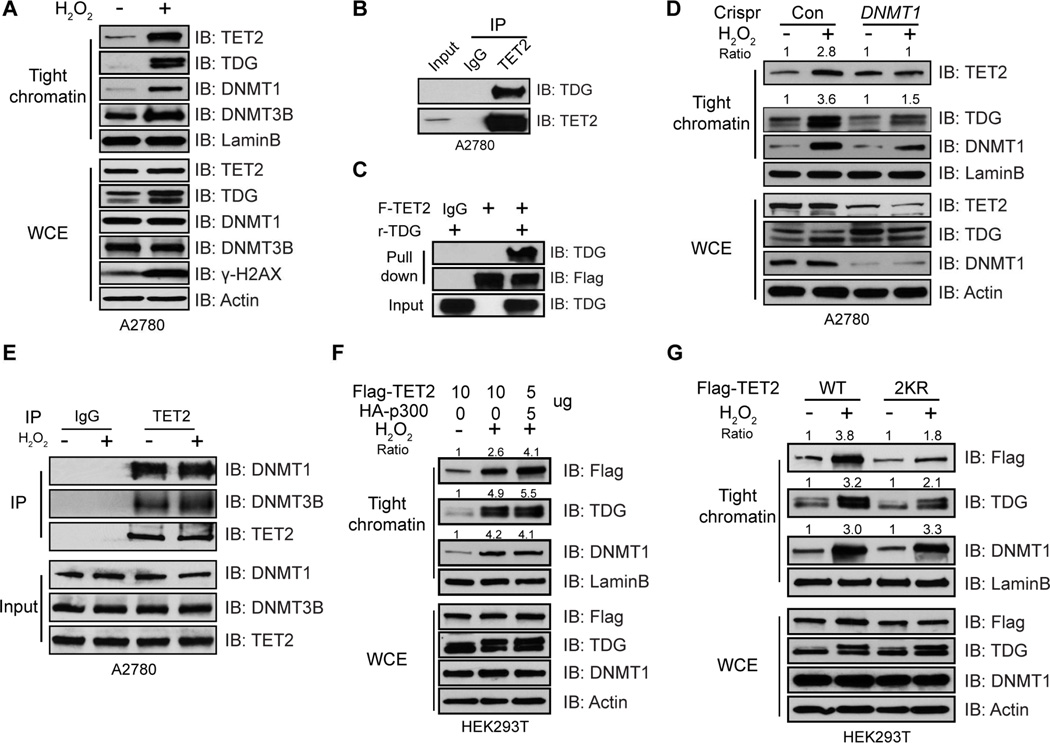
Targeting of TET2 to chromatin during OS to protect against abnormal DNAm
We have previously demonstrated that abnormal methylation may occur during repair of double strand breaks (DSB) and especially after exposure to reactive oxygen species (ROS) (O'Hagan et al., 2008; O'Hagan et al., 2011). During this process, we found rapid movement to CGIs of protein complexes, initiated by DNA repair proteins, that contains the counterpart activity to TET2, DNMTs (Ding et al., 2015; O'Hagan et al., 2011). Interestingly, recent reports also show that 5hmC is present at DNA damage sites and loss of TETs impair DNA damage repair (An et al., 2015; Kafer et al., 2016). We now tie together these key processes and show that TET2, as well as TDG, a protein that is required to complete the final demethylation steps initiated by TET2, participate in these DNMT complexes in a manner influenced by TET2 acetylation. Moreover, TET2 functionally protect against cancer-specific abnormal DNAm on normally unmethylated promoter CGIs and enhancers during OS.
First, with respect to DNA damage, exposing A2780 cells for 30 min to hydrogen peroxide (H2O2) increases total γ-H2AX protein levels and induces formation of γ-H2AX foci (Figure S6A), as well as tighter binding of DNMT1 and DNMT3B to chromatin (Figure 6A and Figure S6B). Intriguingly, both TET2 and TDG are also tightened to chromatin (Figure 6A). The involvement of TDG in the complexes is validated by a direct association between TDG and TET2 (Figure 6B and 6C) which is further confirmed by identifying regions of TET2 that interact with TDG (Figure S6C). Such interaction is independent of TET2 acetylation (Figure S6D), but enhanced by OS (Figure S6E), and is conserved among all three TET proteins (Figure S6F). Moreover, recruitment of TET2/TDG to chromatin during OS is dependent on DNMT1, as knocking down DNMT1 abolishes this tightening (Figure 6D). It appears, in this regard, that OS mainly induces recruitment of TET2/TDG to chromatin through DNMT1, but does not affect interactions between TET2 and DNMT1/DNMT3B (Figure 6E). Although DNMT3B interacts with TET2 (Figure 5K and Figure 6E) and tightens to chromatin during OS (Figure 6A and Figure S6B), it is not required for targeting of TET2 to chromatin (Figure S6G). The importance of TET2 acetylation in these above chromatin targeting events, through interacting with DNMT1, is also evident. While H2O2 treatment alone dramatically increases binding of DNMT1/TET2/TDG to chromatin, the amount of chromatin bound TET2 and TDG are further increased by co-expressing p300 to promote TET2 acetylation (Figure 6F). The transfection conditions are optimized so that equal amounts of TET2 proteins are produced in all samples (Figure S6H). Conversely, although the wild type TET2 and TDG become more tightly bound to chromatin after treatment, the 2KR mutant significantly reduces the enrichments of both proteins (Figure 6G). Tightening of DNMT1, however, is not affected in either case, indicating the increased/decreased targeting of TET2/TDG, as a function of TET2 acetylation, is attributed to enhanced/reduced association between TET2 and DNMT1, as demonstrated above (Figure 5G and 5H). Thus, by enhancing association with DNMT1, TET2 acetylation positively regulates its tightening to chromatin during OS.
Figure 6. Acetylation enhances targeting of TET2 to chromatin during OS.
(A) A2780 cells treated with or without H2O2 for 30 min were extracted sequentially with various buffers. Proteins resistant to 0.45M NaCl buffer are considered as tight chromatin fraction and analyzed by IB. Tight chromatin DNMT3B is referred to DNMT3B that is resistant to 2 M NaCl buffer. WCE was used as input.
(B) Endogenous TET2 complexes were IPed from A2780 nuclear extract and analyzed by IB.
(C) Immunoprecipitated TET2 bound on beads was incubated with or without 500 ng recombinant TDG overnight. Bound proteins were analyzed by IB. IgG was used as negative control.
(D) A2780 Con and DNMT1 knockdown cells were treated with or without H2O2 for 30 min. Tight chromatin and WCE were analyzed by IB. Band intensities were quantified by ImageJ and normalized to LaminB loading control.
(E) Endogenous TET2 complexes were IPed from A2780 cells treated with or without H2O2 for 30 min and analyzed by IB. Spermine and Spermidine were added to extract proteins bound to chromatin.
(F and G) HEK293T cells were transfected with Flag-TET2, in the presence or absence of HA-p300 (F), or TET2 WT, or 2KR mutant (G) for 2 days and treated with or without H2O2 for 30 min. Tight chromatin proteins and WCE were analyzed by IB. Band intensities were quantified by ImageJ and normalized to LaminB loading control. A pre-determined ratio between TET2 and p300 constructs was used in (F) to achieve equal levels of total TET2 proteins.
See also Figure S6.
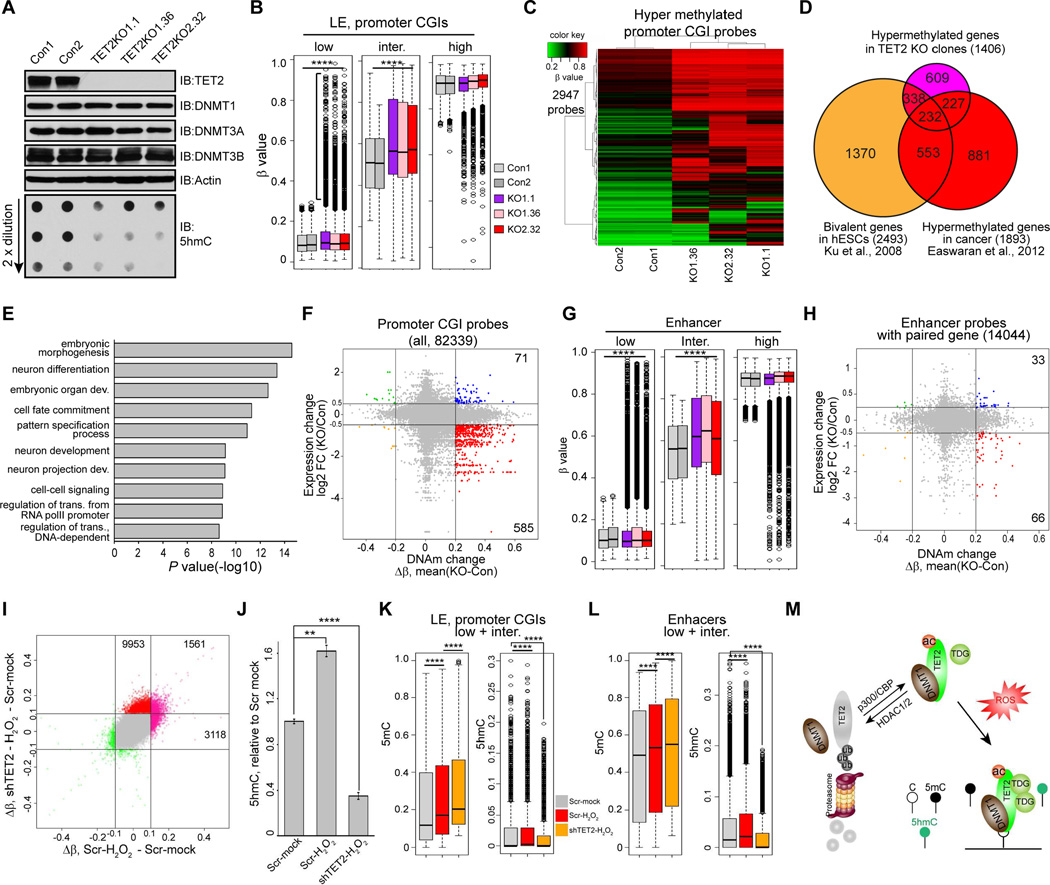
Importantly, for all of the above interactions, we find TET2 has a distinct influence on levels of DNAm. First, at a genome-wide level as assayed by the Infinium 450 methylation array, complete disruption of this protein in A2780 cells by CRISPR/Cas9 technology (Shalem et al., 2014), yields single selected clones with markedly reduced global 5hmC levels, and unchanged levels of all three DNMTs (Figure 7A). Second, and most importantly, in all three TET2 KO clones, there is not only a predominant gain of global DNAm (Figure S7A) but some of the gains are specifically targeted to a subset of probes for normally unmethylated promoter CGIs (basal β < 0.25) of low basal expression genes (CGI LE) (Figure 7B and Figure S7B). Gains can also be seen for LE promoter CGIs probes which have intermediate basal methylation levels (0.25 < β < 0.75) (Figure 7B). These genes, frequently marked by bivalent chromatin in ESC and adult stem cells, are most likely to gain de novo, promoter CGI hypermethylation in cancer (Easwaran et al., 2012; O'Hagan et al., 2011). Indeed, among those probes with gains of DNAm in TET2 KO clones (Figure 7C, mean (Δβ) > 0.2), a striking 40.4% (568/1406) of the target genes are marked by bivalent chromatin in ESCs (Figure 7D). A total of 459 genes, 232 (50.5%) of which are bivalent genes, that gained methylation in TET2 KO clones, also frequently become hypermethylated in various cancer types (Figure 7D and Table S1) (Easwaran et al., 2012). These genes are also significantly enriched in the biological functions operative for those with bivalent chromatin in ESCs such as development, differentiation, and transcriptional regulation (Figure 7E). Importantly, hypermethylation of these promoter CGIs is associated with down regulation of the involved genes (Figure 7F), further signifying the biological significance of the abnormal DNAm gains induced by TET2 KO.
Figure 7. TET2 protects against abnormal DNAm.
(A) A2780 cells were infected with Con or TET2 LentiCRISPR viruses and single clones were selected. Cell lysates of single clones were analyzed by IB, and global 5hmC levels were measured by dot blot.
(B) Box plot showing increase of DNAm of unmethylated (β < 0.25) and intermediately methylated (0.25 < β < 0.75) promoter CGI probes of low expression genes (LE) in TET2 KO clones. ****: P < 0.0001 (student’s t-test).
(C) Heatmap showing LE promoter CGI probes hypermethylated in TET2 KO clones (Δβ (KO - WT) > 0.2).
(D) Venn diagram showing overlaps between genes hypermethylated in TET2 KO clones (C), bivalent genes in hESCs, and cancer specific hypermethylated genes.
(E) Gene ontology enrichment analysis for genes hypermethylated in TET2 KO clones.
(F) Scatter plot showing correlation between DNAm and gene expression changes in TET2 KO clones. Hypermethylated promoter CGIs probes (Δβ (KO - WT) > 0.2) associated with down-regulation of gene expression (log2 (KO/Con)>0.5) are highlighted in red.
(G and H) (G) Box plot showing gains of DNAm in enhancer probes with low and intermediate basal DNAm in TET2 KO clones. (H) Scatter plot showing correlation between DNAm and gene expression changes on potential enhancer-target gene pairs. Hypermethylated enhancer probes (Δβ (KO - WT) > 0.2) associated with down-regulation of gene expression (log2 (KO/Con)>0.5) are highlighted in red.
(I) Scatter plot showing correlation between probes which gain DNAm during OS alone (x-axis, Δβ (Scr-H2O2 –Scr-mock) and during OS with TET2 knockdown (double treatment) (y-axis, Δβ (shTET2-H2O2 – Scr-mock). |Δβ|>0.1 was used as cut-off.
(J) Global 5hmC levels in mock (Scr-mock), OS (Scr-H2O2), and double treated (shTET2-H2O2) cells were measured by 5hmC ELISA. Values represent mean ± SEM (n=3). **: P < 0.01, ***: P <0.001 (student’s t-test)
(K and L) Box plot showing dynamic 5mC and 5hmC changes for LE CGI (K) and enhancer (L) probes that gain DNAm during OS (compare red bar to gray bar). Further increase of 5mC levels coincide with decrease of 5hmC levels on these probes in double treated cells (compare orange bar to red bar). ****: P <0.0001 (student’s t-test).
(M) Model of how TET2 actively protects against abnormal DNA methylation during OS. OS induces recruitment of TET2/DNMTs complex(es) to chromatin. DNA methylation installed by DNMTs, which would be abnormal if retained, is actively removed by TET2 through converting 5mC to 5hmC. p300-mediated TET2 acetylation enhances TET2/DNMT1 interaction and positively regulates this process. Deacetylation of TET2 reduces its enzymatic activity, weakens its interaction with DNMT1 and triggers protein degradation.
Another important aspect of the above protection against DNAm is changes we observe in enhancer elements. These are also of particular interest especially since p300, which acetylates TET2 as we now show, marks active enhancers (Creyghton et al., 2010). Studies in mouse systems also show that enhancers are prone to DNA hypermethylation upon loss of TETs (Hon et al., 2014; Lu et al., 2014; Rasmussen et al., 2015). Similar to promoter probes, with the present TET2 KO, gains of DNAm are seen for a subgroup of enhancer probes that have low and intermediate basal methylation levels (Figure 7G, and Figure S7C), and a group of hypermethylated enhancers functionally repress the expression of their putative target genes (Figure 7H). To confirm the potential biological meaning of these enhancer results, we subjected the 2107 hypermethylated enhancer regions identified by Rasmussen et al. (Rasmussen et al., 2015) to similar subgroups based on their basal methylation levels and find a very similar pattern for gains of DNAm in their TET2 KO scenario (Figure S7D).
How mechanistically does TET2 protect against abnormal DNAm? Using the OS system discussed above as a way to introduce aberrant DNAm, we show that targeting of TET2 to chromatin, through interaction with DNMT1, actively removes unwanted DNAm in the process. In TET2 wild type cells treated with H2O2 for 30 min and examined for DNAm at this time point (30 min) and 2.5 hours after (3 h) (Figure S7E), there are overall gains of DNAm at the later time point (Figure S7F). These findings expand, to a genome-wide level, the concept that tightening of DNMTs to chromatin induced by OS causes increase of DNAm, as demonstrated previously in selected candidate genes (O'Hagan et al., 2011). Again, the abnormal gains are well seen for the promoter CGIs of low expression genes, as well as the low and intermediate methylated enhancer probes (Figure S7G–I). Moreover, the majority of the promoter CGIs and enhancer probes that gained DNAm during OS in TET2 wild type cells are also hypermethylated in TET2 KO clones (Figure S7J and S7K), suggesting TET2 may function through similar pathways to protect against abnormal DNAm in KO and OS induced scenarios.
Finally, we further define the role for TET2 in protecting against abnormal DNAm in the above OS model by examining levels of 5hmC in parallel with assessing 5mC levels. First, increase of global 5hmC levels coincides with the gains of abnormal DNAm at the 3-hour time point after H2O2 treatment (Figure S7L), indicating TET2 is actively converting 5mC to 5hmC during this process. Second, acute depletion of TET2 by shRNA, which by itself imposes minimal impact on global DNAm during this short period of time, dramatically reduces global 5hmC levels (Figure S7M and S7N), and, most importantly, induces greater gains of DNAm when combined with H2O2 treatment (double treatment) (Figure 7I, 7J, and Figure S7O, S7P). Third, adapting a prior treatment procedure which allows simultaneous assessment of 5mC and 5hmC by the Infinium 450K assay (Stewart et al., 2015), we confirm that the group of low expression promoter CGIs probes that become aberrantly hypermethylated during OS indeed show further increases of 5mC and simultaneously decreases of 5hmC in the presence of double treatment (Figure 7K, compare Scr-H2O2 to shTET2-H2O2). Similar trends are also evident in a group of vulnerable enhancers (Figure 7L). Finally, the promoter CGI and enhancer probes that become hypermethylated in TET2 KO clones (Figure 7C and Figure S7C) also tend to gain abnormal DNAm during OS, and gain further DNAm during induction of OS when TET2 is depleted (Figure S7Q and S7R). All of the above findings then further indicate the essential role of TET2 in protecting against abnormal DNAm in the above OS induced scenario.
Discussion
Our present data suggest a paradigm for the participation of TET2, and possibly other TETs, in potentially preventing aberrant DNAm in cancer. We demonstrate that through a balance of acetylation and deacetylation at key lysine, the catalytic activity, stability, and protein partnering of TET2 may participate in a dynamic protective process against abnormal DNAm in cancer during OS. While a building body of work now is outlining the biochemistry by which TET proteins mediate active DNA demethylation, and the importance of TETs in development, reprograming, and tumorigenesis, much less has been studied about how these proteins are dynamically regulated and their mechanistic roles in regulating DNAm in cancer. This highlights the importance of our uncovering of a balance of acetylation and deacetylation as a key post-translational process regulating TET2 functions. The immediate effect of increasing TET2 enzymatic activity by acetylation as well as the long-term impact on protein stability well illustrates the functional relevance. Such importance is further emphasized by the enhanced interaction with DNMT1 and subsequent function in preventing aberrant DNAm during OS. TET1/3 are also found to be acetylated (Figure S1E), likely at different lysines, as K110 is not conserved in TET1/3 (data not shown). In line with a recent report (Bauer et al., 2015), we also identify multiple potential phosphorylation sites all in the N-terminus (Figure S1E, lower panel). The biological significance of this phosphorylation remains to be determined, but together with the N-terminal acetylation we now define, the importance of post translational modifications for TET2 functions is reinforced.
Several pathways regulating TETs protein stability have been delineated, such as the caspase (Ko et al., 2013), and calpains (Wang and Zhang, 2014) pathways. We now present compelling evidence showing that TET2 stability could also be regulated by the ubiquitin-proteasome pathway. These seemingly discrepant observations may result from different origins and/or states of cells used. Differing from our findings that polyubiquitination of the conserved DSBH domain destabilizes TET2, Nakagawa recently identified mono-ubiquitination at lysine 1299 which promotes binding of TET2 to DNA (Nakagawa et al., 2015). The mono- and poly-ubiquitination patterns observed in these two studies are likely due to the use of the proteasome inhibitor, MG132 or not (Figure S4).
We further revealed the biological significance of acetylation in regulating TET2 function through modulating its partnering with DNMT1 which in turn stabilizes TET2 protein. We link these dynamics to how TET2 may participate in dynamically protecting against abnormal DNAm during OS in a process enhanced by TET2 acetylation through enhanced binding to DNMT1. Similar to tightening of DNMT1 to GC rich regions during OS which induces abnormal gain of DNAm (O'Hagan et al., 2011), we now show TET2 is also targeted to chromatin through interacting with DNMT1 and prevents accumulation of abnormal DNAm by converting 5mC to 5hmC (Figure 7M, proposed model). Although de novo methyltransferase DNMT3B also interacts with TET2 and tightens to chromatin during OS, and likely catalyzes the abnormal gains of DNAm occurred within 3 hours in our oxidative stress model, it is not required for targeting of TET2 to chromatin. This model is further supported by the observation that transient knocking down of TET2 reduces 5hmC and further enhances gains of 5mC when combined with H2O2 treatment. Since acute depletion of TET2 by itself does not induce gains of 5mC in the time frame tested (4 days), the sharp reduction of 5hmC resulted from TET2 knockdown likely plays a causal role in the further gains of 5mC during double treatment. Many of such CG rich areas, such as promoter CGIs, are normally protected from DNAm but frequently hypermethylated in cancer (Baylin and Jones, 2011; Jones and Baylin, 2007; Shen and Laird, 2013). We now define a TET2/DNMTs complex(es) as a key regulator of DNAm which responds to OS and prevents accumulation of abnormal DNAm, and propose that loss of TETs function may contribute to aberrant DNA hypermethylation phenotype often observed in cancer.
Loss of 5hmC has been observed in various cancers (Haffner et al., 2011; Jin et al., 2011; Yang et al., 2013), yet TET2 mutations are only frequently detected in leukemia (Scourzic et al., 2015). IDH1/2 mutations resulting in production of oncometabolite D-2-HG which inhibits TETs and many other epigenetic enzymes, are also highly restricted to gliomas and leukemia (Dang et al., 2009; Yan et al., 2009). In addition to down regulation of TETs by microRNAs (Song et al., 2013b), our data now suggest altered post-translational modification as another mechanism for deregulating TET2 functions in cancers. Deacetylation of TET2 by HDAC1/2 which are often over expressed in cancers (Glozak and Seto, 2007) may down-regulate the “failsafe” function of TET2 for preventing unwanted DNAm in focal regions such as normally unmethylated, promoter CGIs. In supporting this hypothesis, we found that mRNA levels of TETs are comparable between normal and tumor samples in the majority of cancers analyzed, while HDAC1/2 are almost always up-regulated (Figure S2). Such deacetylation of TET2 triggered by elevated HDAC1/2 could be extremely harmful when cells, during tumor initiation and progression, are continuously challenged by chronic inflammation and/or ROS which have been linked directly to tumorigenesis (Coussens and Werb, 2002). Decreased TET2 activities and reduced interaction with DNMT1 during OS might, thus, worsen the situation and ultimately result in focal gains of abnormal DNAm often observed in cancer (Baylin and Jones, 2011; Jones and Baylin, 2007; Shen and Laird, 2013). From a translational standpoint, our findings provide additional implications for why the use of HDAC inhibitors in cancer could be beneficial.
Experimental Procedures
Detailed materials and methods can be found in Supplemental Experimental Procedures.
Cell culture, treatments, and tight chromatin preparation
A2780 and HCT116 cells were maintained in RPMI-1640 and McCoy’s 5A with 10% FBS. For H2O2 exposure, 30% H2O2 (Sigma) was diluted in fresh medium immediately before use. After 30 min, cells were collected and tight chromatin fractions were prepared as indicated in the Supplemental Experimental Procedures.
Immunoprecipitation and pull down
Nuclear extracts were incubated with primary antibodies overnight at 4 °C, and additional 3 hours with magnetic beads. For pull down, immunoprecipitated TET2 was incubated with purified recombinant DNMT1 or TDG overnight at 4 °C. After wash, beads were boile d in sample buffer and subjected to SDS-PAGE analysis. Band densitometry was quantified using ImageJ software.
shRNA knockdown and CRISPR knockout
For shRNA knockdown, cells were infected with virus for 2 days and selected with puromycin for 5 days. For CRISPR knockout, cells were infected for 2 days, and selected for 10 days before isolating single cell clones.
Dot blot and ELISA
Dot blot assays were performed as described previously (Ko et al., 2010). For ELISA, genomic DNA was denatured and coated onto 96-well microplates. Global 5hmC levels were measured using 5hmC specific antibody.
Global DNA methylation and 5hmC analysis
For regular DNA methylation analysis, 500 ng DNA was bisulfite converted (Zymo EZ DNA Methylation Kit) and hybridized to Illumina 450K arrays according to manufacturer’s instructions. For 5hmC analysis, DNA samples were split evenly into two aliquots and processed through either bisulfite (BS), or oxidative bisulfite (oxBS) workflows (Cambridge Epigenetix), and hybridized to Illumina 450K arrays. Probes with poor signals (P > 0.01) were removed from further analysis. 5hmC levels were obtained by subtracting the oxBS β value from the BS β value in each sample and negative 5hmC values were considered to be 0.
Supplementary Material
Acknowledgments
We thank Drs. Jianyuan Luo, Xiaochun Yu, Alan Friedman, Ed Seto, and A-Lien Lu for DNA constructs, Dr. Wayne Wei Yu at the SKCCC Microarray Core Facility for Infinium 450K assays. We thank members of the Baylin laboratory for discussion and Kathy Bender for manuscript preparation. This work was supported by NIH grants R01ES011858, R01CA170550, R01CA043318, U01HL099775, and Hodson Trust, all to SBB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
All genome-wide DNA methylation and gene expression data used in the study has been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE81428.
Author Contributions
Y.W.Z and S.B.B conceived and designed the experiments. Y.W.Z performed the experiments and analyzed the data. Z.W. purified proteins. W.X, J.L., and H.E. helped with bioinformatics. L.X. helped with tight chromatin experiments. R.C.Y. helped with cell culture. Y.L. helped with suspension cell culture. Y.W.Z. and S.B.B wrote the manuscript.
References
- An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, Rao A. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071. doi: 10.1038/ncomms10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Gobel K, Nagaraj N, Colantuoni C, Wang M, Muller U, Kremmer E, Rottach A, Leonhardt H. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT) J. Biol. Chem. 2015;290:4801–4812. doi: 10.1074/jbc.M114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Ding N, Bonham EM, Hannon BE, Amick TR, Baylin SB, O'Hagan HM. Mismatch repair proteins recruit DNA methyltransferase 1 to sites of oxidative DNA damage. J. Mol. Cell. Biol. 2015 doi: 10.1093/jmcb/mjv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int. J. Cancer. 2007;121:2381–2386. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N, Correa IR, Jr, Zheng Y, Cheng X. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer GR, Li X, Horii T, Suetake I, Tajima S, Hatada I, Carlton PM. 5-Hydroxymethylcytosine Marks Sites of DNA Damage and Promotes Genome Stability. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.01.035. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D'Alessio AC, Nakayama K, Fan HY, Chen X, Xiong Y. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol. Cell. 2015;57:247–260. doi: 10.1016/j.molcel.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013a;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013b;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SK, Morris TJ, Guilhamon P, Bulstrode H, Bachman M, Balasubramanian S, Beck S. oxBS-450K: a method for analysing hydroxymethylation using 450K BeadChips. Methods. 2015;72:9–15. doi: 10.1016/j.ymeth.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6:278–284. doi: 10.1016/j.celrep.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.