Abstract
Introduced over 200 yr ago to the east coast of North America, Carcinus maenas now ranges from New York to Newfoundland. In the 1980s, a secondary invasion of European lineages, termed northern haplotypes, occurred in Nova Scotia. Young-of-the-year sampled in 2007 revealed that northern haplotypes were present in low frequencies at several northwestern Atlantic sites as far south as New York; a model predicted an increase in their range and frequency over time. We collected samples in 2013 and 2014 to determine the haplotypes of adult crabs from New York to Nova Scotia. Six haplotypes, encompassing previously identified northern and southern haplotypes, 1 novel southern haplotype, and 1 Scandinavian haplotype, were identified in 275 crabs sampled at 11 sites. Northern haplotypes were only found in Nova Scotia, Beals Island (Maine), and Mount Desert Island (Maine) at a frequency of 60, 8, and 24%, respectively; remaining sites were predominantly composed of a previously identified southern haplotype. Northern haplotypes are limited in adult crabs to Mount Desert Island and north, indicating that the southern haplotype is selectively favored at some point during their life history, recruitment of northern larvae is limited south of Mount Desert Island, or entire year-classes post-2007 were lost. Our results do not support the predictions of an increase in the range and frequency of northern haplotypes, at least among adults, and indicate that a more complete knowledge of factors affecting C. maenas life stages is necessary to understand the current distribution of haplotypes.
Keywords: Carcinus maenas, Population genetics, Mitochondrial DNA, Invasive species, Range
INTRODUCTION
Modification of marine ecosystems through the invasion of exotic species has been identified as a critical issue facing the world’s oceans (Carlton 1998). By successfully establishing themselves in foreign ecosystems, invaders can upset ecosystem processes such as primary production and nutrient cycling (Vitousek et al. 1996) as well as established feedback systems that maintain biodiversity and a stable community structure (Parker et al. 1999). One method for determining the establishment and spread of an invasive species is examining the genetics of their populations.
Population genetics predicts that the further apart 2 populations exist spatially, the more differentiated they should be at neutral genetic markers due to stochasticity. This correlation is often blurred by disruptions in patterns of gene flow caused by high migration rates and species invasions, such that invasive species often do not show classic phylogeographical distributions in non-native ranges (Novak & Mack 2005, Wares et al. 2005, Kolbe et al. 2007, Pringle et al. 2011, Gaither et al. 2013, Darling et al. 2014). The most important factor in the establishment of an invasive population may be propagule pressure, or the number of adult or larval individuals released into a non-native range (Allendorf & Lundquist 2003, Lockwood et al. 2005). Rapid population expansion following invasion events (Tepolt et al. 2009) and multiple invasions from different source populations may also aid in invasion success by mitigating diversity lost by genetic drift from the source population (Roman & Darling 2007, Darling et al. 2014). Despite empirical and theoretical work in the population genetics of invading species, it is still unclear which of these factors is most important in establishing, maintaining, and increasing populations of marine invaders.
The European green crab Carcinus maenas is a prime example of a successful marine invader, establishing along all continents with temperate coasts during the past 2 centuries (Compton et al. 2010). The crab was first discovered on the northeastern coast of the United States in 1817 in New York and southern Massachusetts, and it subsequently migrated north, reaching Casco Bay, Maine in the early 20th century (Audet et al. 2003, Carlton & Cohen 2003). By the 1950s, green crabs were ob served in Canada in Passamaquoddy Bay and the Bay of Fundy, from where they expanded over the next 50 yr to the Atlantic coast of Nova Scotia and the Gulf of St. Lawrence (Audet et al. 2003). Since that time, there has been an increase in Canadian populations, indicating expansion of populations introduced in the 19th century and recent invasions from northern European populations (Roman 2006, Blakeslee et al. 2010, Darling et al. 2014). The invasion events of the 1980s and 1990s potentially came in ballast water of European ships docking at the Strait of Canso (Nova Scotia) port, and have subsequently introduced novel northern haplotypes to the populations along the Canadian coast. In the most recent genetic study of C. maenas in the Gulf of Maine, from young-of-the-year samples collected in 2007, northern haplotypes were shown to have moved south, increasing in frequency by 25% between Louisbourg, Nova Scotia and Barnstable, MA (Cape Cod) between 1999 and 2007 (Pringle et al. 2011). The evolution of this genetic cline was attributed to an asymmetrical dispersal pattern, mediated by the prevailing southbound coastal currents (Pringle et al. 2011). Recent nuclear microsatellite data revealed that the cline shift may also be due to sex-biased reproductive dynamics and population size imbalances and not solely due to dispersal (Darling et al. 2014). Regardless of the cause, alleles and haplotypes representative of the northern cline were expected to increase in frequency in populations south of Nova Scotia by 10% by 2014 in young-of-the-year (Pringle et al. 2011).
We investigated the genetic diversity of adult C. maenas populations spanning the Gulf of Maine from Nova Scotia to Cape Cod and south to Long Island Sound to evaluate whether the genetics of the adult populations have also shifted. Haplotypes of a 400 bp region of the mitochondrial cytochrome c oxidase I (COI) gene were examined as a neutral marker of genetic diversity and maternal gene flow among populations, the same marker that has been previously used to study green crab population genetics (Roman 2006, Darling et al. 2008, Darling 2011, Pringle et al. 2011). Adult crabs have significant negative effects on community structure (Leignel et al. 2014). Recent studies have suggested that crabs from Maine and Canada are better foragers and predators compared to southern populations (Rossong et al. 2006, Thompson 2007, League-Pike & Shulman 2009) and were causal in eelgrass loss in Maine during the summers of 2013 and 2014 (Neckles 2015). However, these studies did not determine the genetic background of the experimental animals, so differences in behavior or abundance cannot be linked to differences in genetics. One goal of our study was to document the adult genetic composition along the range. Of broader significance, knowledge of changes in the range at various life stages of C. maenas in nonnative habitats will allow for a better understanding of the complex dynamics underlying invasion in other marine species.
MATERIALS AND METHODS
Sample collection
Adult crabs Carcinus maenas (carapace width ≥30 mm) were collected from 11 sites along the northwestern Atlantic coast from Nova Scotia to Long Island Sound (Table 1; see Fig. 1). Specimens were either collected by hand from algae-covered rocks at low tide (sites: BI, PM, CA), trapped from shallow (<5 m) coastal waters (sites: MD, MB, WE, ES, DU, BA, and LI), or in one case, with a beach seine from a marsh (site: NS). To determine if there were differences in genetic composition across life stages at the same geographical location, young-of-the-year, determined by a carapace width <10 mm (Berrill 1982), were collected in 2014 by hand during low tide at 2 different sites in Maine: Mount Desert Island and Pemaquid (see Table 2). Prior to DNA extraction, demographic data were taken, including sex, carapace width, and coloration.
Table 1.
Carcinus maenas sampling locations, mean (±SD) carapace width, and collection date. See Fig. 1 for full US state names
| ID | Location | Lat. (°N), long. (°W) |
Carapace width (mm) |
Collection date |
|---|---|---|---|---|
| NS | Keji, Nova Scotia | 43.86, 64.82 | 49.7 ± 5.2 | Sep 30, 2013 |
| BI | Beals, ME | 44.48, 67.59 | 36.1 ± 6.1 | Sep 10, 2013 |
| MD | Mount Desert Island, ME | 44.45, 68.32 | 39.3 ± 5.5 | Sep 10, 2013 |
| PM | Pemaquid Point, ME | 43.83, 69.51 | 52.1 ± 4.3 | Jul 2, 2014 |
| MB | Montsweag Bay, ME | 43.92, 69.71 | 48.3 ± 2.9 | May 24, 2014 |
| CA | Casco Bay, ME | 43.82, 70.09 | 37.6 ± 16.5 | Sep 5, 2013 |
| WE | Wells, ME | 43.32, 70.56 | 39.2 ± 5.8 | Sep 13, 2013 |
| ES | Essex, MA | 42.66, 70.73 | 40.2 ± 2.1 | Sep 27, 2013 |
| DU | Duxbury, MA | 42.03, 70.65 | 44.2 ± 4.7 | Nov 18, 2013 |
| BA | Barnstable, MA | 41.70, 70.30 | 46.5 ± 3.6 | Oct 23, 2013 |
| LI | Long Island Sound, NY | 40.86, 72.45 | 52.3 ± 7.8 | Sep 27, 2013 |
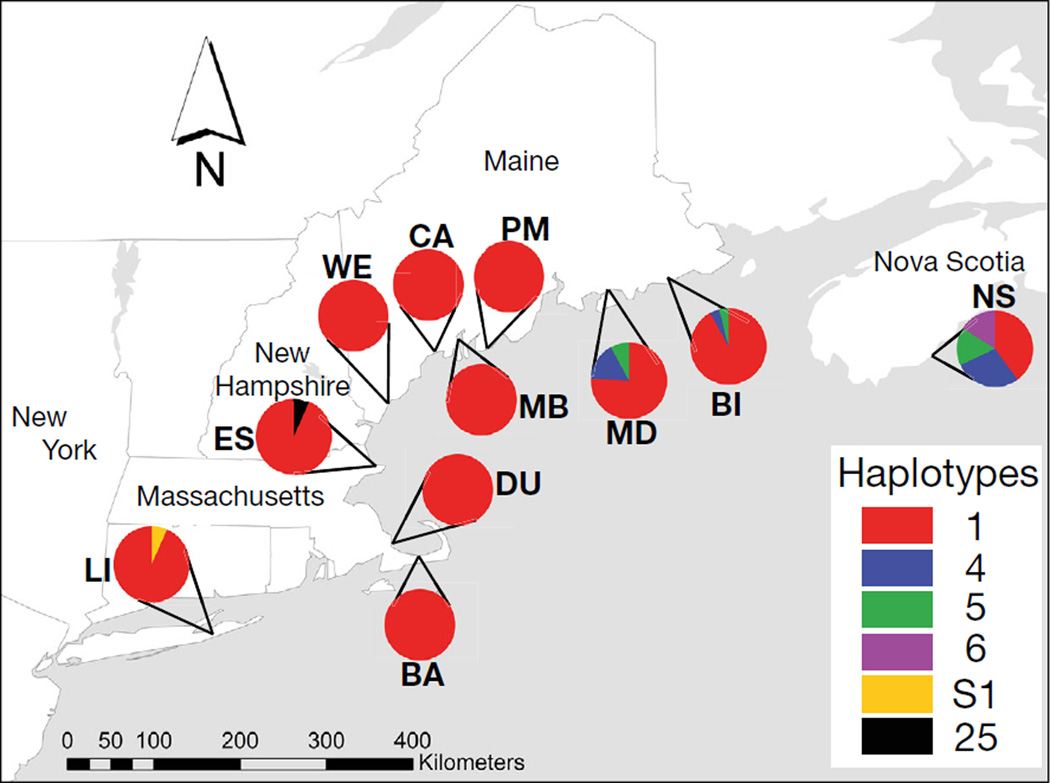
Fig. 1.
Adult Carcinus maenas haplotype frequencies of a 400 bp region of the cytochrome c oxidase I (COI) gene (n = 25 per site). Pie charts indicate proportion of haplotypes found from each population. See Table 1 for site abbreviations
Table 2.
Carcinus maenas young-of-the-year sampling locations, haplotype(s), average carapace width (±SD), and collection date. See Table 1 for site abbreviations
| ID | Haplotype name (% of individuals) |
Carapace width (mm) |
Collection date |
|---|---|---|---|
| MD | Haplotype 1 (75%) | 7.2 ± 0.3 | Jul 8, 2014 |
| Haplotype 4 (17%) | |||
| Haplotype 5 (8%) | |||
| PM | Haplotype 1 (96%) | 6.9 ± 0.2 | Jul 2, 2014 |
| Haplotype 4 (4%) |
DNA extraction and sequencing
DNA was extracted from pereopod muscle tissue using the Qiagen DNeasy Tissue kit and purified by ethanol precipitation from 25 individuals per sampling site. The DNA pellet was rinsed with EtOH (95%) before it was air-dried and re-suspended in molecular-grade water. A 400 bp region of COI was amplified by PCR (30 s at 94°C, then 35 cycles of 15 s at 95°C, 30 s at 56°C, 30 s at 68°C, then 68°C for 5 min) using OneTaq Hot Start DNA polymerase (New England Biolabs) and the following primers designed using GenBank accession no. KF369118 as a reference: forward, 5’-GCA TAG TAG GGA CTT CTT TGA G-3’; reverse, 5’-TTT CGG TCA GTT AGA AGT ATT G-3’. The amplified product was purified using the GeneClean kit (MP Biomedicals). The PCR products were sequenced directly using amplification primers in both directions.
Genetic analysis
Consensus sequences for each individual were produced from the forward and reverse sequences using ClustalW (Larkin et al. 2007); all populations were aligned using ClustalX 2.1 (Larkin et al. 2007). Genetic diversity was analyzed within and between populations by haplotype diversity (h), haplotype frequencies, pairwise distances between populations (FST), and number of migrants (Nm), calculated using DnaSP v.5.10.01 (Librado & Rozas 2009). Binomial distribution was used to determine the probability of observing particular patterns given published and predicted data.
RESULTS
Six haplotypes were found in the 275 adult crabs Carcinus maenas sampled across 11 populations (Fig. 1), caused by polymorphisms in 5 loci. In the young-of-the-year, only 2 haplotypes, haplotype 1 (a southern haplotype) and haplotype 4 (a northern haplotype), were found (Table 2). Among all haplotypes, there were 2 possible nucleotide variants per locus. All polymorphisms were silent and in the 3rd position, except for the substitution creating haplotype S1, found in 1 individual from Long Island Sound, that resulted in a change of a non-polar methionine to a polar threonine at the 80th amino acid. Haplotype 1 (GenBank accession no. AY616437) was present in all populations, with adults from Pemaquid, Monts weag, Casco, Wells, Duxbury, and Barnstable exhibiting only this singular haplotype (Fig. 1). Of the 25 adults sequenced from each site, northern haplotype 4 (GenBank: DQ523684) was found in 7 individuals from Nova Scotia, 1 from Beals, and 4 from Mount Desert Island. Northern haplotype 5 (GenBank: AY616438) was found in 4 individuals from Nova Scotia and 2 from Mount Desert Island, while northern haplotype 6 (GenBank: AY616439) was found in 4 individuals from Nova Scotia and in no other populations. Haplotype 25 (GenBank: FJ159027), originating from Norway and Sweden (Darling et al. 2008), was found in 1 Essex adult. One novel southern haplotype, S1 (GenBank: KM114884), was found in 1 individual from Long Island Sound. In the young-of-the-year, similar patterns in adult haplotypes were found for Mount Desert Island individuals, with 75% of the individuals having haplotype 1, while 17% had haplotype 4, and 8% had haplotype 5. In Pemaquid, where all adults were haplotype 1, one young-of-the-year was haplotype 4; the remaining 24 individuals were haplotype 1.
The northern sites had the highest haplotype diversity (h) in the adults: Nova Scotia had the largest h (0.776), while Mount Desert Island had the second largest h (0.525). Beals, Essex, and Long Island Sound all had a low level of h (0.133), while all other populations had an h of zero.
Genetic differentiation between adult populations, measured by FST, was greatest amongst Nova Scotia versus all other populations (Table 3). Comparisons of Nova Scotia with other northern populations also show large population differentiation (Nova Scotia vs. Beals Island, FST: 0.445; Nova Scotia vs. Mount Desert Island, FST: 0.217). There are low levels of gene flow between Nova Scotia and the populations from Casco Bay south ward (Nm: 0.55 to 0.92). Nearly as segregated as Nova Scotia to the southern populations were the populations from Nova Scotia and Beals (Nm: 0.74). Although Mount Desert Island is farther south than Beals (by ~80 km), there was more gene flow between the Nova Scotia and Mount Desert Island populations (Nm: 1.92). There was a high level of gene flow between the 2 most northern Maine sites, Mount Desert Island and Beals (Nm: 4.51), due to the presence of northern haplotypes at both sites. There was a mid-level gene flow between Mount Desert Island and all other populations to the south as compared to other pairwise population comparisons (Nm: 2.2 to 3.0).
Table 3.
| NS | BI | MD | PM | MB | CA | WE | ES | DU | BA | LI | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NS | – | ||||||||||
| BI | 0.445 | – | |||||||||
| MD | 0.217 | 0.094 | – | ||||||||
| PM | 0.480 | 0 | 0.182 | – | |||||||
| MB | 0.480 | 0 | 0.182 | 0 | – | ||||||
| CA | 0.480 | 0 | 0.182 | 0 | 0 | – | |||||
| WE | 0.480 | 0 | 0.182 | 0 | 0 | 0 | – | ||||
| ES | 0.367 | 0 | 0.050 | 0 | 0 | 0 | 0 | – | |||
| DU | 0.480 | 0 | 0.182 | 0 | 0 | 0 | 0 | 0 | – | ||
| BA | 0.480 | 0 | 0.182 | 0 | 0 | 0 | 0 | 0 | 0 | – | |
| LI | 0.462 | 0 | 0.160 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | – |
DISCUSSION
Our data reveal that the southern limit of novel northern haplotypes (4, 5, and 6) in the adult stage of the invasive green crab Carcinus maenas is Mount Desert Island, Maine. Adult haplotype data from Nova Scotia and northern Maine (Fig. 1) is concordant with young-of-the-year data collected in 2007 (Pringle et al. 2011, Darling et al. 2014) that show extensive population admixture between northern and southern haplotypes. The lack of novel northern haplotypes and dominance of southern haplotype 1 in the adult stage south of Mount Desert Island (Fig. 1) is, however, surprising, given that novel northern juveniles composed 8.8% of individuals sampled along a similar geographical distribution in 2007. Using an average lifespan estimate of 4 to 7 yr (Klassen & Locke 2007), populations of adult green crabs analyzed in our study reflect year-class recruits as far back as the last genetic study in 2007 (Pringle et al. 2011) and up to 2009. With our sample size of 250 individuals for sites south of Mount Desert Island, and an expected probability of at least 0.088 if northern young-of-the-year had survived (Pringle et al. 2011), we would have expected to sample >8 adult individuals with a probability of 0.999 (binomial probability: x = 8, n = 250, p = 0.088). If the expected probability of having northern haplotypes was as low as 0.05, we would have expected to sample >8 individuals with a probability of 0.930 (binomial probability: x = 8, n = 250, p = 0.05). With statistical certainty, we can conclude that there were no northern adult haplotypes (4, 5, and 6) present in our 8 sampling sites ranging from mid-coast Maine to New York. We did, however, find 1 adult in Essex, MA, of a novel European H25 lineage (Darling et al. 2008) and 1 of a novel haplotype (S1) that has never been sequenced. While H25 has never been sequenced in North America, its presence in one Massachusetts site may have been mediated through the secondary invasion of the 1980s and 1990s and subsequent transport via currents (Roman 2006). Haplotype S1, found at Long Island Sound, may also be a cryptic European haplotype. Given the overwhelming lack of adults with novel northern haplotypes along much of the northeastern US coastal cline, selection against those northern haplotypes may be occurring at one or more life stages.
Temperature may be one of the factors contributing to the current range of the northern haplotype, since it is hypothesized to be a major regulatory mechanism of green crab distribution (Yamada et al. 2005, Compton et al. 2010), but evidence for its role in the data presented here is inconclusive. The lack of novel northern adults south of Mount Desert Island may be due to temperature-driven and spatially variable selection during the larval, juvenile, or adult stage as a result of warmer waters in the southern compared to the northern end of the range. Between 2009 and 2013, Massachusetts Bay average monthly maximum sea surface temperatures (SST) (at 1 m depth) were 21.27 ± 0.33°C in the summer (June to September) compared to 17.57 ± 0.49°C in Halifax Harbor (Northeastern Regional Association of Coastal and Ocean Observing Systems, http://neracoos.org/). Larval (Dawirs 1985, Nagaraj 1993, deRivera et al. 2007) and adult (Tepolt & Somero 2014) temperature tolerance is known in the green crab, whereas in the lab, larvae can survive up to 25°C (Dawirs 1985) and adults up to 37.3°C (Tepolt & Somero 2014); thus, as the animal matures, its ability to survive broader temperature ranges increases. Temperature also affects developmental time and larval transport, where increased environmental temperatures re duce developmental time and transport, and de creased environmental temperatures increase developmental time and transport (Berrill 1982, Reitzel et al. 2004, Byers & Pringle 2006). Differences in SST between the northern and southern end of the range may have resulted in mortality or changes in connectivity between the populations. What is still un clear is whether novel northern haplotypes, at any stage, have a narrower temperature tolerance compared to their southern counterpart. Tepolt & Somero (2014) showed that there was no difference in adult heat tolerance between crabs collected (in 2011) from Newfoundland to New Jersey. Because southern haplotype young-of-the-year had already arrived at their sample site of North Harbor, Newfoundland in 2007, and were found to be at a frequency of 59%(Blakeslee et al. 2010), there is a 99.99%chance that that at least 11 individuals (binomial probability: x = 11, n = 32, p = 0.59) sampled in 2011 (Tepolt & Somero 2014) were southern haplotypes. Our young-of-the-year data supports the environmental temperature hypothesis, whereby south of Mount Desert Island, there were northern young-of-the-year but no northern adults; at Mount Desert Island, there were both northern young-of-the-year and northern adults. However, the lack of data that connects thermal tolerance to haplotype leaves open the question of whether there actually is a difference in temperature tolerance between northern and southern haplotypes that may account for the lack of northern adults in the typically warmer waters south of Mount Desert Island (Shearman & Lentz 2010).
The lack of northern haplotype adults could be due to a decrease or deficiency of recruitment of northern haplotypes after 2007 in the southern region of the cline. The frequency of northern haplotype young-of-the-year from Maine to New York was small in 2007, averaging 8.8% (Pringle et al. 2011). Compared to 2000, however, the frequency of the northern haplotypes in 2007 young-of-the-year increased by 25% and allele and haplotype frequency was expected to increase further in populations south of Nova Scotia by approximately 10% by 2014 (Pringle et al. 2011). It was proposed that the northern haplotypes arrived and increased in frequency in the southern region of the cline between 2000 and 2007 due to current-mediated dispersal (Pringle et al. 2011). Thus, small changes in local recruitment of novel haplotypes or a loss of northern haplotype larval input from upstream sources (Byers & Pringle 2006, Banas et al. 2009) could have led to a decrease and eventual loss of the novel haplotypes in future young-of-the-year and would be reflected in the adult population. While the expansion of the northern haplotypes was occurring between 2000 and 2007 in the southward direction, so too was the expansion of southern haplotypes in the northward direction (Pringle et al. 2011, Darling et al. 2014). Since allelic diversity in the southern region of the range is mainly set by that of the northern, retentive, upstream edge in northern Maine and Canada (Pringle & Wares 2007, Wares & Pringle 2008, Pringle et al. 2011), it is possible that the transport of larvae after 2007 in the downstream direction was mainly composed of southern haplotypes. While this scenario is highly unlikely, the frequency of southern haplotype larvae each year (post-2007) may have increased to a point where northern haplotypes were lost from the larval pool and the resultant adults that we sampled in 2013 and 2014 were reflective of that change. Furthermore, if only a small founder population of northern haplotype(s) existed in each of the downstream sink populations (sampling locations from mid-coast Maine to New York), they would be subject to drift; without rapid population expansion (Tepolt et al. 2009) and additional settlement, these novel haplotypes would be lost quickly and would not have been sampled as adults in our study. While we did not collect young-of-the-year from each sampling site, young-of-the-year data from Pemaquid and Mount Desert Island also offer some support for our larval recruitment theory. In Mount Desert Island, where one quarter of the adults are of a northern haplotype, young-of-the-year follow a similar genetic pattern (Table 2). In Pemaquid, though, where 100% of the adults are southern haplotype 1, only 4% of the young-of-the-year are a northern haplotype (Table 2). Thus, unless a steady and sufficient influx of northern larvae from a local or upstream source is maintained over time, the adult population is composed of the most dominant haplotype.
While temperature and/or recruitment may be the contributing factor(s) to the lack of adult northern haplotypes from New York to northern Maine, another potential reason for the lack of adult novel haplotypes may be that year-classes of both southern and northern haplotypes could have been completely lost post-2007 in one or more seasons. While this is merely speculative, perhaps disease, predation, and/ or temperature caused the loss. Since there is no way to accurately age green crabs (though a new method for aging crustaceans appears promising; Kilada et al. 2012), we are unable to determine the age of our crabs; we can say that our crabs were ≥3 and ≤7 yr old (Berrill 1982). Thus, the loss of these year-classes may have occurred from 2007 up through 2009 or 2010. Sampling in future field seasons may be able to resolve whether loss of year-classes is the cause of our observed lack of northern haplotypes south of Mount Desert Island, Maine.
The reason for the lack of adult northern haplotypes in much of the northeast US coast is not known and has not been tested, but it is likely because one or more biotic and/or abiotic factors are regulating the dispersal, recruitment, survival, and sustainability of these haplotypes across this cline. It will be important to collect data such as larval abundance and recruitment as well as physiological tolerance across life stages in light of genetic background in order to be able to better predict and measure future genetic cline changes of the green crab in marine systems.
Acknowledgments
Thank you to Brian Beal, John Brawley, Jane Disney, Dan Harrington, Chris McCarthy, Hilary Neckles, Alyssa Novak, Bradley Peterson, Pete Thayer, and Kristin Wilson for their help with sample collection. Thank you to Arnaud Germain and Ludovic Giloteaux of Maureen Hanson’s lab, Cornell University, for their instruction in sequence alignment to C.L.N. Conversations between W.G.A. and A. I. Wanamaker as well as between W.G.A., L.M.W., and P. Petraitis were very helpful in developing the discussion. This project was supported by grants from the National Center for Research Resources (5P20RR016463) and the National Institute of General Medical Sciences (8 P20 GM103423) from the National Institutes of Health, by the National Oceanic and Atmospheric Administration (NOAA) through funds from Maine Sea Grant, as well as the Bates College Student Research Fund.
LITERATURE CITED
- Allendorf FW, Lundquist LL. Introduction: population biology, evolution, and control of invasive species. Conserv Biol. 2003;17:24–30. [Google Scholar]
- Audet D, Davis DS, Miron G, Moriyasu M, Benhalima K, Campbell R. Geographical expansion of a nonindigenous crab, Carcinus maenas (L.), along the Nova Scotian shore into the southeastern Gulf of St. Lawrence, Canada. J Shellfish Res. 2003;22:255–263. [Google Scholar]
- Banas N, McDonald PS, Armstrong D. Green crab larval retention in Willapa Bay, Washington: an intensive Lagrangian modeling approach. Estuar Coasts. 2009;32:893–905. [Google Scholar]
- Berrill M. The life cycle of the green crab Carcinus maenas at the northern end of its range. J Crustac Biol. 1982;2:31–39. [Google Scholar]
- Blakeslee AMH, McKenzie CH, Darling JA, Byers JE, Pringle JM, Roman J. A hitchhiker’s guide to the Maritimes: anthropogenic transport facilitates long-distance dispersal of an invasive marine crab to Newfoundland. Divers Distrib. 2010;16:879–891. [Google Scholar]
- Byers JE, Pringle JM. Going against the flow: retention, range limits and invasions in advective environments. Mar Ecol Prog Ser. 2006;313:27–41. [Google Scholar]
- Carlton JT. Apostrophe to the ocean. Conserv Biol. 1998;12:1165–1167. [Google Scholar]
- Carlton JT, Cohen AN. Episodic global dispersal in shallow water marine organisms: the case history of the European shore crabs Carcinus maenas and C. aestuarii. J Biogeogr. 2003;30:1809–1820. [Google Scholar]
- Compton TJ, Leathwick JR, Inglis GJ. Thermogeography predicts the potential global range of the invasive European green crab (Carcinus maenas) Divers Distrib. 2010;16:243–255. [Google Scholar]
- Darling JA. Interspecific hybridization and mitochondrial introgression in invasive Carcinus shore crabs. PLoS ONE. 2011;6:e17828. doi: 10.1371/journal.pone.0017828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling JA, Bagley MJ, Roman J, Tepolt CK, Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus . Mol Ecol. 2008;17:4992–5007. doi: 10.1111/j.1365-294X.2008.03978.x. [DOI] [PubMed] [Google Scholar]
- Darling JA, Tsai Y-HE, Blakeslee AMH, Roman J. Are genes faster than crabs? Mitochondrial introgression exceeds larval dispersal during population expansion of the invasive crab Carcinus maenas. R Soc Open Sci. 2014;1:140202. doi: 10.1098/rsos.140202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawirs RR. Temperature and larval development of Carcinus maenas (Decapoda) in the laboratory; predictions of larval dynamics in the sea. Mar Ecol Prog Ser. 1985;24:297–302. [Google Scholar]
- deRivera C, Hitchcock N, Teck S, Steves B, Hines A, Ruiz G. Larval development rate predicts range expansion of an introduced crab. Mar Biol. 2007;150:1275–1288. [Google Scholar]
- Gaither MR, Bowen BW, Toonen RJ. Population structure in the native range predicts the spread of introduced marine species. Proc R Soc B. 2013;280:20130409. doi: 10.1098/rspb.2013.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilada R, Sainte-Marie B, Rochette R, Davis N, Vanier C, Campana S. Direct determination of age in shrimps, crabs, and lobsters. Can J Fish Aquat Sci. 2012;69:1728–1733. [Google Scholar]
- Klassen GJ, Locke A. Can Manuscr Rep Fish Aquat Sci 2818. Ottawa: Fisheries and Oceans Canada; 2007. A biological synopsis of the European green crab, Carcinus maenas. [Google Scholar]
- Kolbe JJ, Glor RE, Schettino LR, Lara AC, Larson A, Losos JB. Multiple sources, admixture, and genetic variation in introduced Anolis lizard populations. Conserv Biol. 2007;21:1612–1625. doi: 10.1111/j.1523-1739.2007.00826.x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- League-Pike PE, Shulman MJ. Intraguild predators: behavioral changes and mortality of the green crab (Carcinus maenas) during interactions with the American lobster (Homarus americanus) and Jonah crab (Cancer borealis) . J Crustac Biol. 2009;29:350–355. [Google Scholar]
- Leignel V, Stillman JH, Baringou S, Thabet R, Metais I. Overview on the European green crab Carcinus spp. (Portunidae, Decapoda), one of the most famous marine invaders and ecotoxicological models. Environ Sci Pollut Res Int. 2014;21:9129–9144. doi: 10.1007/s11356-014-2979-4. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lockwood JL, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasions. Trends Ecol Evol. 2005;20:223–228. doi: 10.1016/j.tree.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Nagaraj M. Combined effects of temperature and salinity on the zoeal development of the green crab, Carcinus maenas (Linnaeus, 1758) (Decapoda: Portunidae) Sci Mar. 1993;57:1–8. [Google Scholar]
- Neckles H. Loss of eelgrass in Casco Bay, Maine, linked to green crab disturbance. Northeast Nat. 2015 (in press) [Google Scholar]
- Novak S, Mack R. Genetic bottlenecks in alien plant species: influence of mating systems and introduction dynamics. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- Parker IM, Simberloff D, Lonsdale WM, Goodell K, et al. Impact: toward a framework for understanding the ecological effects of invaders. Biol Invas. 1999;1:3–19. [Google Scholar]
- Pringle J, Wares J. Going against the flow: maintenance of alongshore variation in allele frequency in a coastal ocean. Mar Ecol Prog Ser. 2007;335:69–84. [Google Scholar]
- Pringle JM, Blakeslee AMH, Byers JE, Roman J. Asymmetric dispersal allows an upstream region to control population structure throughout a species’ range. Proc Natl Acad Sci USA. 2011;108:15288–15293. doi: 10.1073/pnas.1100473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Miner BG, McEdward LR. Relationships between spawning date and larval development time for benthic marine invertebrates: a modeling approach. Mar Ecol Prog Ser. 2004;280:13–23. [Google Scholar]
- Roman J. Diluting the founder effect: cryptic invasions expand a marine invader’s range. Proc R Soc B. 2006;273:2453–2459. doi: 10.1098/rspb.2006.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J, Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Rossong MA, Williams PJ, Comeau M, Mitchell SC, Apaloo J. Agonistic interactions between the invasive green crab, Carcinus maenas (Linnaeus) and juvenile American lobster, Homarus americanus (Milne Edwards) J Exp Mar Biol Ecol. 2006;329:281–288. [Google Scholar]
- Shearman RK, Lentz SJ. Long-term sea surface temperature variability along the US East Coast. J Phys Oceanogr. 2010;40:1004–1017. [Google Scholar]
- Tepolt CK, Somero GN. Master of all trades: thermal acclimation and adaptation of cardiac function in a broadly distributed marine invasive species, the European green crab, Carcinus maenas. J Exp Biol. 2014;217:1129–1138. doi: 10.1242/jeb.093849. [DOI] [PubMed] [Google Scholar]
- Tepolt CK, Darling JA, Bagley MJ, Geller JB, Blum MJ, Grosholz ED. European green crabs (Carcinus maenas) in the northeastern Pacific: genetic evidence for high population connectivity and current-mediated expansion from a single introduced source population. Divers Distrib. 2009;15:997–1009. [Google Scholar]
- Thompson W. Population-level effects of the European green crab (Carcinus maenas L.) in an eelgrass community of the southern Gulf of St. Lawrence. MSc thesis. Fredericton: University of New Brunswick; 2007. [Google Scholar]
- Vitousek PM, D’Antonio CM, Loope LL, Westbrooks R. Biological invasions as global environmental change. Am Sci. 1996;84:218–228. [Google Scholar]
- Wares J, Pringle J. Drift by drift: effective population size is limited by advection. BMC Evol Biol. 2008;8:235. doi: 10.1186/1471-2148-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wares J, Hughes A, Grosberg R. Mechanisms that drive evolutionary change: insights from species introductions and invasions. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- Yamada S, Dumbauld B, Kalin A, Hunt C, Figlar-Barnes R, Randall A. Growth and persistence of a recent invader Carcinus maenas in estuaries of the northeastern Pacific. Biol Invas. 2005;7:309–321. [Google Scholar]