Abstract
Cancer cells utilize flexible metabolic programs to maintain viability and proliferation under stress conditions including nutrient deprivation. Here we report that phospholipase D1 (PLD1) participates in the regulation of metabolic plasticity in cancer cells. PLD1 activity is required for cancer cell survival during prolonged glucose deprivation. Blocking PLD1 sensitizes cancer cells to glycolysis inhibition by 2-deoxy-D-glucose (2-DG) and results in decreased autophagic flux, enlarged lysosomes, and increased lysosomal pH. Mechanistically, PLD1-regulated autophagy hydrolyzes bulk membrane phospholipids to supply fatty acids (FAs) for oxidation in mitochondria. In low glucose cultures, the blockade of fatty acid oxidation (FAO) by PLD1 inhibition suppresses adenosine triphosphate (ATP) production and increases reactive oxygen species (ROS), leading to cancer cell death. In summary, our findings reveal a novel role of PLD1 in sustaining cancer cell survival during metabolic stress, and suggest PLD1 as a potential target for anticancer metabolism therapy.
One hallmark of cancer cells is increased aerobic glycolysis, widely known as the Warburg effect, that maintains cell growth and proliferation through the generation of adenosine triphosphate (ATP) and precursors for macromolecular synthesis.1, 2 However, although the Warburg effect empowers cells to proliferate rapidly when glucose is abundant, a flexible and alternative metabolic program must also be available for cancer cells to respond readily to conditions leading to metabolic stresses, for example, rapid tumor growth and exposure to therapy.2, 3 This metabolic plasticity explains why, in theory, therapies aimed at inhibiting glucose utilization in ‘glucose-addicted' cancer cells should be efficacious at eliminating tumors, but in practice, cancer cells often resist to approaches that target glycolysis alone.2, 3 One major mechanism by which cancer cells adapt to nutrient scarcity is to shift their dependence on glycolysis to mitochondrial fatty acid oxidation (FAO) that serves to sustain ATP levels and counter oxidative stress.4, 5, 6 The biochemical basis for this metabolic reprogramming under metabolic stress is largely known.4, 5 However, how fatty acids (FAs) are generated, mobilized, and transferred into mitochondria for subsequent utilization remains unclear.
Autophagy is a tightly regulated process that maintains cellular homeostasis by lysosomal processing of damaged cellular organelles and long-lived proteins. It is generally accepted that autophagy suppresses tumor initiation at the early stages of cancer but promotes tumor growth by providing a source of nutrients during rapid tumor growth.7, 8 In fact, in many tumor cells, autophagy activation is required for proliferation and survival, as well as the development of resistance to the treatment.7, 8 Therefore, inhibiting autophagy may be beneficial for cancer therapy. One key mechanism by which autophagy promotes tumorigenesis is through the regulation of lipid metabolism.6, 9 However, despite the rapid progress in autophagy research, the underlying mechanisms by which autophagy promotes and maintains the growth of tumors remain undefined.
In this paper, we report that the activity of phospholipase D1 (PLD1) is required for metabolic reprogramming of cancer cells enduring prolonged glucose deprivation. PLD1 inhibition blocks autophagic flux and free FA production from bulk membrane phospholipids that in turn limits FAO in mitochondria. This results in a decrease in cytosolic ATP level and an increase in mitochondrial reactive oxygen species (ROS) production, promoting cancer cell death during glucose deprivation.
Results
PLD1 is required for cancer cell survival during prolonged glucose deprivation
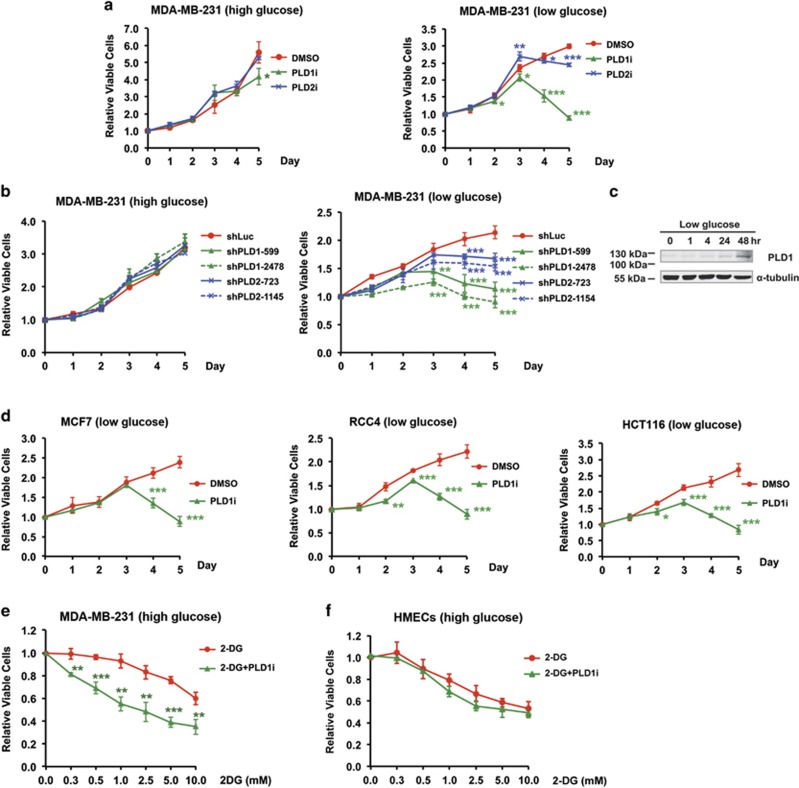
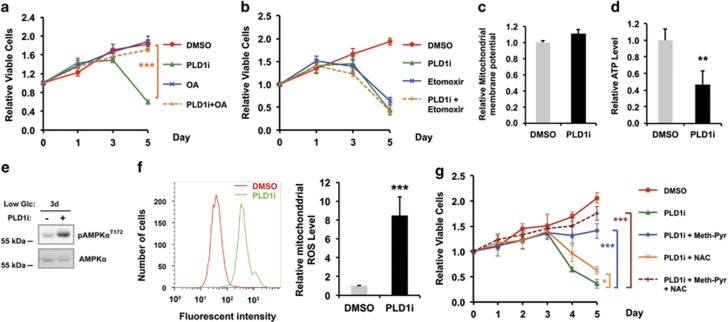
When treated with isoform-specific PLD inhibitors, the viability of MDA-MB-231 breast cancer cells grown in the regular culture medium that contained high glucose (25 mM) was not significantly altered by either the PLD1 inhibitor (PLD1i) or PLD2 inhibitor (PLD2i) (Figure 1a). However, both inhibitors, especially PLD1i, caused significant decreases in cell viability as early as day 3 in low glucose (1 mM) medium (Figure 1a). Furthermore, PLD1 and PLD2 knockdown by small hairpin RNAs (shRNAs)10, 11 (Supplementary Figure S1) also reduced the viability of MDA-MB-231 cells starting at day 3 in low, but not high, glucose medium (Figure 1b). As both PLD1 shRNAs and PLD1i showed greater effects on cell viability than those of PLD2 (Figures 1a and b), we focused on PLD1 in subsequent studies. Interestingly, the expression of PLD1 protein in MDA-MB-231 cells was increased 2 days after exposure to low glucose (Figure 1c), further supporting a critical role of PLD1 in protecting the cancer cells from glucose starvation.
Figure 1.
Requirement of PLD activity for cancer cell survival in low glucose medium. (a) PLD1 and PLD2 small-molecule inhibitors, PLD1i and PLD2i, reduced the viability of MDA-MB-231 cells when the culture medium contained low glucose (1 mM) but not when it had high glucose (25 mM); n=3. (b) shRNA knockdown of PLD1 and PLD2 in MDA-MB-231 cells reduced cell viability in low glucose but not high glucose medium. Two shRNA constructs were used for each PLD isoform; n=3. Statistical significances were labeled on days 3–5. (c) Glucose deprivation increased the expression of PLD1 protein after 24 h. The experiments were repeated twice with similar result. (d) PLD1i reduced the viability of MCF-7 (breast), RCC4 (renal), and HCT116 (colorectal) cancers cells grown in low glucose medium; n=3. (e) PLD1i sensitized MDA-MB-231 cells to cell death promoted by 2-DG. MDA-MB-231 cells grown in high glucose medium were treated with PLD1i (5 μM) and the indicated concentrations of 2-DG. Cell viability was measured on day 3; n=3. (f) PLD1i did not have an addition effect on the viability of 2-DG-treated HMECs. HMECs cells grown in high glucose medium were treated with PLD1i (5 μM) and the indicated concentrations of 2-DG. Cell viability was measured on day 3; n=3. *P<0.05; **P<0.01; ***P<0.001
We then measured the effect of PLD1 inhibition on the viability of three other cancer cell lines, including MCF-7 (breast), RCC4 (renal), and HCT116 (colorectal). Although PLD1i caused some moderate reduction in viable MCF-7 and RCC4 cells in high glucose (Supplementary Figure S2), it robustly inhibited the growth of all three cell types starting from day 3 in low glucose (Figure 1d). Therefore, the requirement for PLD1 activity to counter glucose deprivation is common among cancer cells.
As PLD1 inhibition decreased cancer growth in low glucose medium, we reasoned that this treatment might also sensitize cancer cells to glycolysis inhibition. Therefore, we treated MDA-MB-231 cells grown in the high glucose medium with different concentrations of 2-DG, a glycolytic inhibitor, in the absence and presence of PLD1i. Co-application of PLD1i significantly reduced the numbers of viable cells at all tested 2-DG concentrations (Figure 1e). In contrast, although 2-DG itself reduced the proliferation of nontumorigenic human mammary epithelial cells (HMECs), addition of PLD1i did not have additional effect (Figure 1f). These data support the potential use of PLD1i in cancer combination therapy.
PLD1 regulates autophagic flux independent of mTOR activity
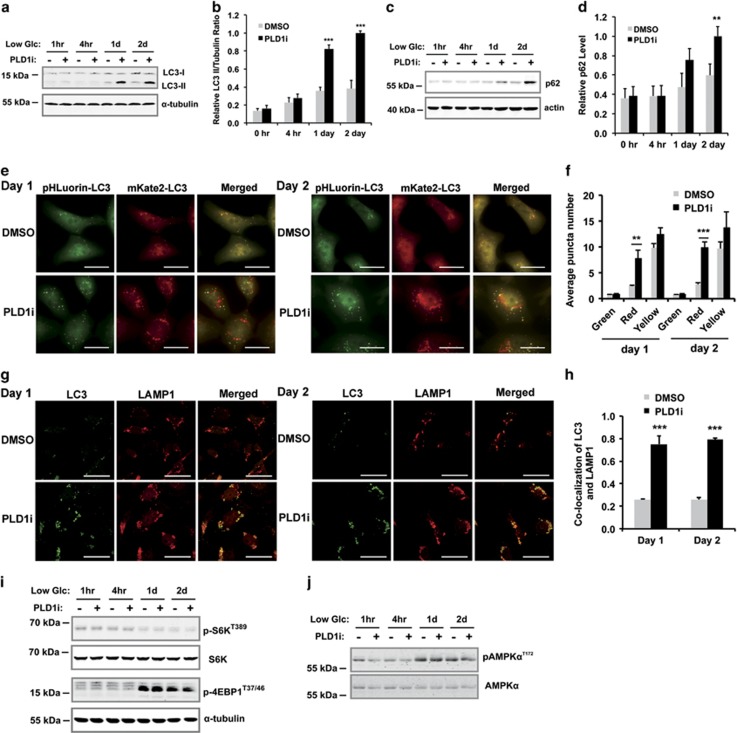
To understand the mechanisms through which PLD1 protects cells against metabolic stress, we examined the activity of autophagy, a common stress response pathway. The LC3-II level, a marker of autophagy, increased slightly after switching to low glucose by western blotting. In contrast, PLD1i treatment strongly and significantly increased LC3-II levels after 1 and 2 days (Figures 2a and b). The increased LC3-II levels could be a consequence of enhanced autophagy initiation or decreased autophagic flux.12, 13 To determine the cause for the increase in LC3-II, we measured the level of p62. Efficient flux of autophagy would result in a decrease in p62.12 PLD1i treatment resulted in an increase in p62 (Figures 2c and d), suggesting a blockage of autophagic flux that often signifies a failure of autolysosome function.12 We also measured the levels of LC3 and p62 in cells in which autophagy was blocked by shRNA knockdown of ATG5, an essential autophagy initiation protein.13, 14 ATG5 knockdown blocked the increased levels of LC3-II (Supplementary Figure S3), indicating the blockade of autophagy. ATG5 knockdown alone increased the levels of p62 as previously reported.12 However, combination of ATG5 knockdown and PLD1i did not further change the levels of p62 as compared with ATG5 knockdown alone (Supplementary Figure S3), suggesting that inhibition of autophagic degradation is the cause of p62 accumulation in PLD1i-treated cells, and is unlikely caused by the upregulation of transcription or inhibition of proteasomal degradation as reported in some stress conditions.12
Figure 2.
PLD1 inhibition reduces autophagic flux without affecting initiation. (a) PLD1i treatment increased the levels of LC3-II and p62 in MDA-MB-231 cells cultured in low glucose medium (1 mM). Cells were treated with or without PLD1i for the indicated time period. Total cell lysates were analyzed by western blotting for the indicated proteins. (b) Quantification of the ratio of LC3-II and the loading control, α-tubulin, using ImageJ; n=3. (c) PLD1i treatment increased the levels of p62 in MDA-MB-231 cells cultured in low glucose medium. Cells were treated with or without PLD1i for the indicated time period. Total cell lysates were analyzed by western blotting for the indicated proteins. (d) Quantification of p62 levels using ImageJ, and normalized to the loading control, actin; n=3. (e) MDA-MB-231 cells stably expressing pHluorin-mKate2-LC3 were cultured in low glucose medium and treated with DMSO (control) or PLD1i (5 μM) for 1 (left) and 2 days (right) before images for pHluorin and mKate2 fluorescence were taken (scale bar: 20 μm). (f) Quantification of green (pHluorin) and red (mKate2) and yellow (both pHluorin and mKate2) puncta per cell in merged pictures. At least 20 cells were quantified in each experiment; n=3. (g) MDA-MB-231 cells cultured in low glucose medium were treated with DMSO or PLD1i for 1 (left) and 2 days (right) before immunostaining performed for LC3 (green) and LAMP1 (red) (scale bar: 20 μm). Cells were permeabilized with Triton X-100. (h) Quantification of colocalization between LC3 and LAMP1; n=3. (i) PLD1i treatment did not change mTOR activity as assessed by the phosphorylation of p70S6K and 4E-BP1. Note the low glucose culture increased phosphorylation of 4E-BP1 on day 1 and day 2, but PLD1i did not alter this effect. (j) PLD1i treatment did not change AMPK activity as assessed by the phosphorylation of AMPKα. **P<0.01; ***P<0.001
We next stably expressed a tagged LC3 that fused to a tandem pHluorin-mKate2,15 an improved tool of the monomeric red fluorescent protein (mRFP) –green fluorescent protein (GFP)–LC3 (tfLC3).16, 17 Because the pHluorin (green), but not the mKate2 fluorescent signal (far-red fluorescence, pseudocolored as red), is quenched by the acidic environment of lysosomal lumen, the coexistence of both pHluorin and mKate2 fluorescent signals (yellow) indicates LC3-containing vesicles before fusion with lysosomes, whereas those showing red represent autolysosomes.12, 17 We found that PLD1i significantly increased the red signal in pHluorin-mKate2-LC3-expressing cells (Figures 2e and f), indicating an accumulation of LC3 in autolysosomes. In support of this, PLD1i treatment also increased the colocalization of LC3 with LAMP1, a lysosome marker (Figures 2g and h). Taken together, PLD1i likely affects the terminal step of autophagic flux, autolysosomal degradation. On the other hand, treatment of PLD1i in the first 2 days did not exert a detectable effect on the activity of mTORC1 (measured by 4E-BP1 and p70S6K phosphorylation) (Figure 2i), AMPK (measured by AMPKα phosphorylation) (Figure 2j), and other signaling pathways, including p38, extracellular signal-regulated kinase (ERK), or AKT (Supplementary Figure S4). These results also support that PLD1 is involved in autophagic flux but not autophagy initiation.
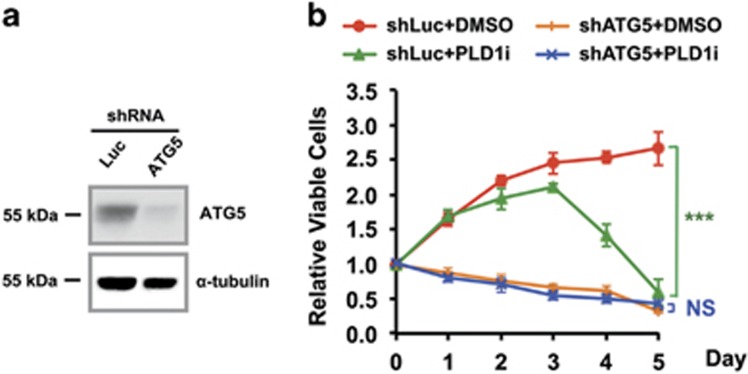
To investigate whether autophagy is required for the survival of MDA-MB-231 cells in low glucose, we measured viability in ATG5 knockdown cells (Figures 3a and b). The loss of ATG5 alone led to gradual decreases in cell viability beginning 1 day in the low glucose medium (Figure 3b). The addition of PLD1i did not further increase cell death (Figure 3b). These data suggest that cancer cells use autophagy to maintain growth under glucose-starved conditions and PLD1 activity is critically involved in this pathway through supporting autophagic flux.
Figure 3.
Autophagy is required for PLD1-regulated cell growth in low glucose medium. (a) Western blot of ATG5 knockdown by shRNA in MDA-MB-231 cells. (b) The knockdown of ATG5 by shRNA suppressed the growth of MDA-MB-231 cells right after culturing them in low glucose medium. PLD1i treatment did not aggravate the effect of autophagy inhibition by ATG5 shRNA. Luciferase shRNA (shLuc) was used as a negative control; n=3. ***P<0.001
PLD1 inhibition results in the perinuclear clustering, enlargement, and alkalinization of lysosomes
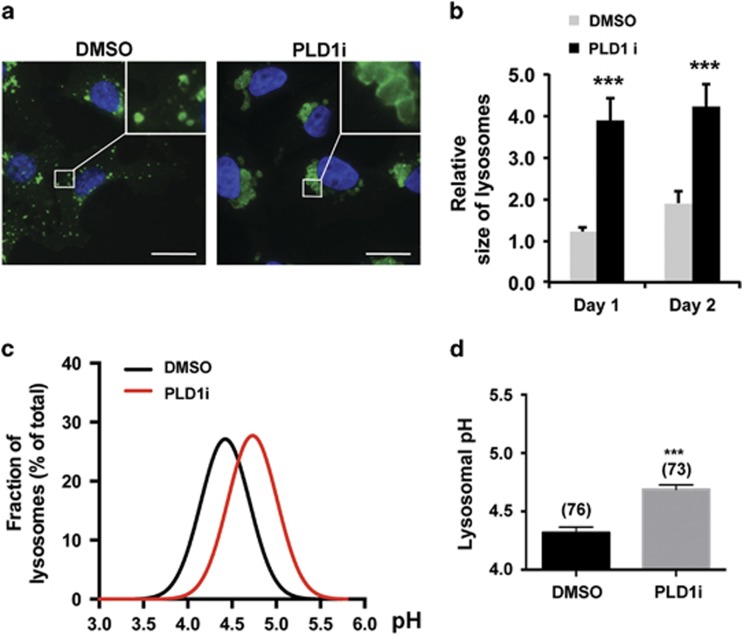
As reported in other cell types,18, 19 a significant amount of HA-tagged PLD1 was found in intracellular puncta that are labeled by the lysosomal marker, LAMP1 (Supplementary Figure S5). This lysosomal localization remained largely unchanged by glucose starvation and PLD1i treatment (Supplementary Figure S5). However, PLD1i treatment led to enlargement of lysosomes and their clustering around nuclei (Figures 4a and b). These changes were more prominent in cells permeabilized with a weak detergent, saponin (Figures 4a and b), than those treated with Triton X-100 (Figures 2e and f and Supplementary Figure S5).
Figure 4.
PLD1 inhibition impedes lysosomal functions. (a) PLD1 inhibition caused prominent lysosome enlargement. MDA-MB-231 cells were grown in low glucose medium and treated with DMSO or PLD1i for 2 days. Cells were fixed and then permeabilized with saponin that preserved good lysosomal morphology before staining for LAMP1 (green) and DAPI for nuclei (blue) (scale bar: 20 μm). (b) Quantification of lysosomal size in MDA-MB-231 cells as in (a). Lysosome size was determined by the areas of LAMP1-labeled vesicles using ImageJ; n=3. (c) PLD1 inhibition increased lysosomal pH. Distribution histograms of lysosomal pH values (fitted to Gaussian distributions) measured by ratiometric imaging from DMSO and PLD1i-treated MDA-MB-231 cells grown for 2 days in low glucose medium. (d) Mean lysosomal pH values of data points in (c); n=3. ***P<0.001
Macromolecule digestions in lysosomes are carried out by luminal hydrolytic enzymes that work optimally at acidic pH.20 We performed ratiometric lysosomal pH measurement in MDA-MB-231 cells loaded with pH-sensitive Oregon Green 488-dextran that exhibits pH-dependent changes in excitation and emission.21, 22 The PLD1i-treated cells displayed an elevated mean lysosomal pH value of 4.74 as compared with the control value of 4.30 obtained from untreated cells (Figures 4c and d). These observations suggest that inhibiting PLD1 disrupts lysosomal functions that may underlie the autophagy defect and reduced viability in MDA-MB-231 cells under glucose-deprived and PLD1-inhibited conditions.
PLD1-regulated FAO sustains cancer cell survival during glucose deprivation
As FA oxidation is critical to energy production and countering oxidative stress during metabolic stresses,4, 5, 6 the PLD1 inhibition-induced alteration of lysosomal functions might impair FA production needed for cancer cell survival during glucose deprivation. Indeed, supplementation of oleic acids, one of the abundant FAs in the cell, completely rescued cell death phenotype caused by PLD1i (Figure 5a). We next treated cells grown in low glucose with etomoxir, a FAO inhibitor. Similar to PLD1i, etomoxir caused a marked decrease in cell viability starting from day 3 in low glucose (Figure 5b). Co-application of PLD1i with etomoxir did not further reduce the viability (Figure 5b), indicating that the two inhibitors affect the same metabolic pathway. In addition, treatment of PLD1i had no effect on mitochondrial membrane potential, suggesting that PLD1 is not directly involved in mitochondrial function (Figure 5c).
Figure 5.
PLD1 regulates free FA generation to support mitochondrial FAO in MDA-MB-231 cells exposed to low glucose medium. (a) Addition of exogenous oleic acids fully rescued cell death induced by PLD1i; n=3. (b) Etomoxir, a FAO inhibitor, reduced viability of MDA-MB-231 cells similarly as PLD1i. The combined treatment with etomoxir and PLD1i had no additive effect; n=3. (c) PLD1i had no effect on mitochondrial membrane potential; n=3. (d) PLD1i reduced ATP production; n=3. (e) PLD1i treatment increased the phosphorylation of AMPKα on day 3 in low glucose medium. (f) PLD1i increased mitochondrial ROS level. The MDA-MB-231 cells stained with MitoSOX Red mitochondrial ROS indicator were analyzed by flow cytometry. Left, histograms of MitoSOX fluorescent intensity. Right, statistic results of the relative mitochondrial ROS level; n=3. (g) The cell death phenotype of PLD1i-treated cells was rescued by methyl-pyruvate (meth-Pyr) and N-acetylcysteine (NAC); n=3. *P<0.05; **P<0.01; ***P<0.001
We then measured intracellular ATP and ROS levels. PLD1i significantly decreased cytosolic ATP on day 3 of the low glucose culture when cell viability began to drop (Figure 5d). Consistent with ATP level, PLD1i treatment also significantly increased AMPK activity on day 3 of low glucose treatment (Figure 5e). PLD1i also caused a robust increase in mitochondrial ROS level (Figure 5f), whereas it had no effect on the cytosolic ROS level (Supplementary Figure S6), supporting the importance of PLD1 in facilitating mitochondrial FAO. Together with the unchanged mTOR and AMPK activities in PLD1i-treated cells on day 1 and 2 (Figures 2i and j), these data further support that PLD1 is not involved in the initiation of autophagy; however, PLD1-regulated autophagic degradation is responsible for fatty acid oxidation, ATP, and ROS production. Importantly, exogenous supplements of methyl-pyruvate, a membrane-permeable form of pyruvate that provides the substrate for ATP production from oxidative phosphorylation in mitochondria,23 greatly rescued the cells from PLD1i-induced death in low glucose (Figure 5g). N-acetylcysteine (NAC), an antioxidant, also had moderate rescue effect (Figure 5g). Co-application of both methyl-pyruvate and NAC with PLD1i further enhanced cell viability to the levels approaching the untreated cells (Figure 5g). Collectively, these findings suggest that PLD1 plays a critical role in FA production that supports FAO in mitochondria, therefore generating energy and antioxidative stress capacity needed for cancer cell survival under glucose-deprived conditions.
PLD1 regulates free FA production from membrane phospholipids during prolonged glucose starvation
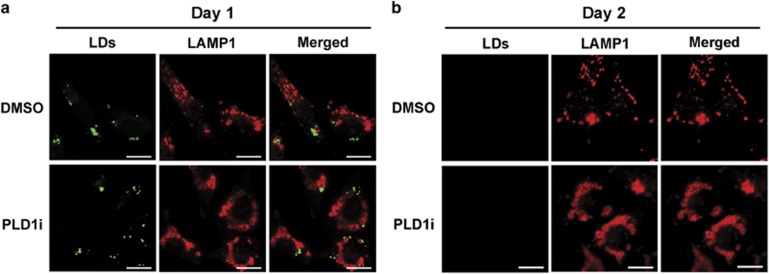
During nutrient starvation, free FAs can be released either from lipid droplets by lipase-mediated lipolysis or from autophagic digestion of membrane-bound organelles (e.g., the endoplasmic reticulum) or lipid droplets (by a form of autophagy called lipophagy).24, 25 We first examined whether PLD1 inhibition caused accumulation of lipid droplets. Lipid droplets were abundant in both control and PLD1i-treated cells on day 1 after exposure to low glucose (Figure 6a), consistent with the previous observations in short-term starvation.26, 27, 28 By day 2, the lipid droplets were barely detectable (Figure 6b), presumably because of depletion of stored lipids under the low glucose conditions. This result also suggests that lipids previously stored in lipid droplets contribute minimally to cell survival under long-term glucose starvation (beyond 3 days in our condition). PLD1 inhibition had no effect on the depletion of the lipid droplets (Figures 6a and b), suggesting that PLD1 is not involved in the utilization of lipid droplets during glucose starvation. Furthermore, PLD1 did not affect the colocalization between lipid droplets and LAMP1 (Figures 6a and b).
Figure 6.
PLD1 is not involved in the homeostasis of lipid droplets. MDA-MB-231 cells grown in low glucose medium for 1 day (a) or 2 days (b) were stained with BODIPY493/503 for lipid droplets and LAMP1 for lysosomes (scale bar: 20 μm). Cells were permeabilized with saponin
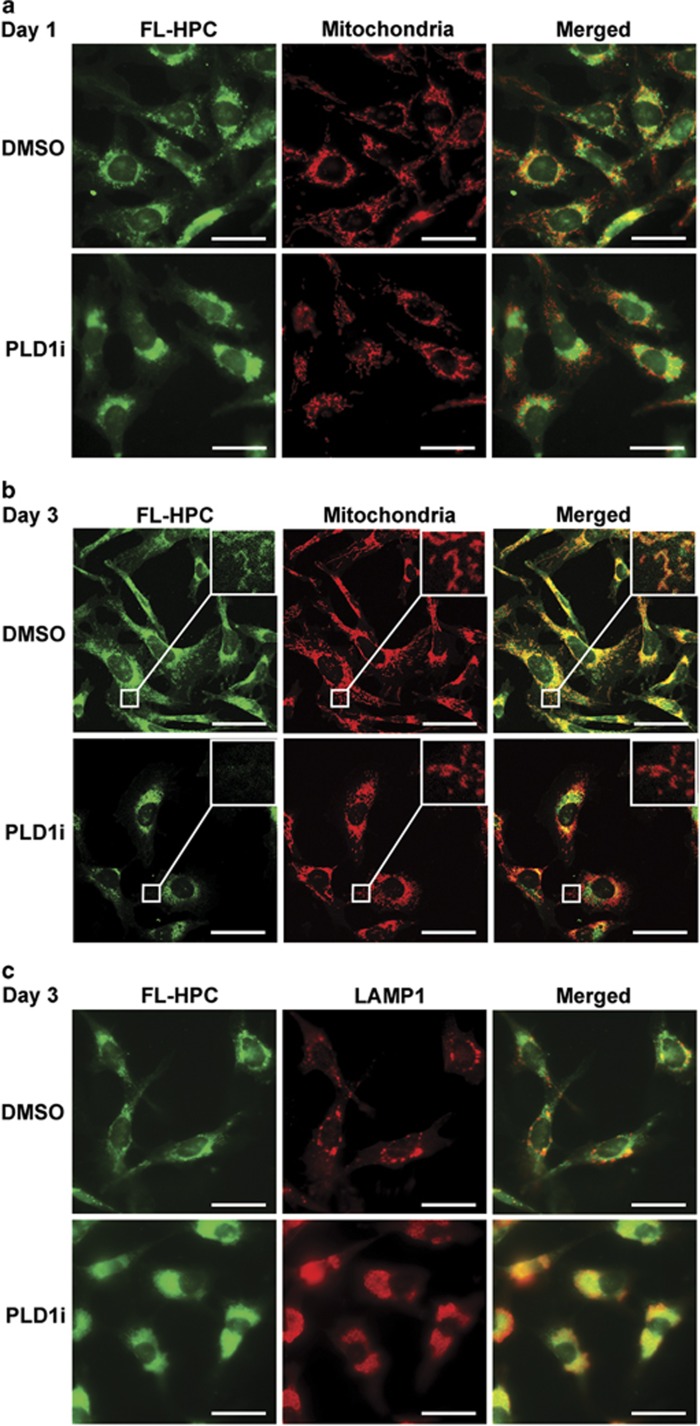
To determine the source of free FAs, we labeled membranes with a phosphatidylcholine species that contained a fluorescence tag at its FA tail (FL-HPC). Similar to fibroblasts,28 the green fluorescence of FL-HPC labeled various cellular membranes and only partially colocalized with mitochondria in MDA-MB-231 cells 1 day after labeling (Figure 7a), and remained largely unchanged on day 2. By day 3 when cells started to die in the low glucose medium (Figure 1), all mitochondria were strongly labeled with the green fluorescence, presumably by channeling free fatty acids hydrolyzed from FL-HPC into mitochondria as demonstrated before28 (Figure 7b). Although PLD1 inhibition did not seem to change the FL-HPC labeling pattern on day 1 (Figure 7a), it greatly reduced green fluorescence in the mitochondria (especially those in cell peripherals) on day 3 (Figure 7b), concurrent with an accumulation of the green fluorescence in autolysosomes (Figure 7c). Taken together, these results suggest that PLD1-regulated autophagy hydrolyzes membrane phospholipids rather than triglycerides in lipid droplets to supply FAs for oxidation in mitochondria.
Figure 7.
PLD1 regulates the mobilization of phospholipids from cellular membranes to mitochondria during glucose starvation. (a and b) MDA-MB-231 cells were labeled with FL-HPC overnight, and chased in low glucose medium for 1 day (a) or 3 days (b) in the presence or absence of PLD1i (scale bar: 20 μm). Cells were permeabilized with saponin. Mitochondria were labeled by TOM22. (c) Inhibition of PLD1 led to the accumulation of FL-HPC in LAMP1-labeled autolysosomes on day 3 (scale bar: 20 μm)
Discussion
PLD1 regulation of autophagy and lysosome functions
PLD1 activity has been implicated in autophagy in several previous studies.29, 30, 31, 32 However, the mechanism and functional implication of PLD1 regulation of autophagy remain unclear. We found that PLD1 inhibition has no effect on mTOR activity, but rather it increases LC3 and p62 accumulation in autolysosomes, lysosomal size, and lysosomal pH (Figures 2 and 4), supporting that PLD1 functions in the late stages of autophagy, most likely autolysosomal degradation, during prolonged glucose starvation. Because an optimal acidic pH is critical to the activity of lysosomal enzymes,20 the increased pH may compromise digestion and be responsible for the detected accumulation of LC3, p62, and phospholipids. The failure of phospholipid hydrolysis reduces the production of free FAs that in turn limits FAO for ATP production and ROS reduction under glucose deprivation conditions. Therefore, our current study reveals a mechanism and the functional consequence of PLD1-regulated autophagy in long-term glucose starvation. It is worth noting that the involvement of PLD1 in lysosomal pH regulation does not necessarily rule out the previously proposed role for PLD1 in autophagosome–lysosome fusion29, 31 that can be compromised by an elevation in lysosomal pH.33
Similar to Vps34,34 PLD1 appears to regulate autophagy in a context-dependent manner. Depending on cell types and states, and experimental conditions, PLD1 may regulate both initiation and completion of autophagy, and function as both positive and negative regulator of autophagic flux.30, 31, 32 Jang et al.30 reported that PLD1 inhibition increases LC3-II levels, whereas it reduces p62 levels and promotes autophagic flux. Dall'Armi et al.29 reported that PLD1 inhibition reduces the LC3-II levels but increases p62 levels. Bae et al.31 and our current study showed that PLD1 inhibition increases the levels of both LC3-II and p62 as a result of inhibition of autophagic flux. It is unclear how PLD1 functions at the different steps of autophagy. One possibility is that different autophagy-induction conditions, for example, complete depletion of all nutrients29, 30 versus low glucose condition in our study, differently modulate the activities of PLD1 or other autophagy regulators at various steps in autophagy. It is also possible that the uses of different sources or concentrations of PLD1 inhibitors cause unexpected off-target effects.
A remaining question is how PLD1 regulates lysosomal pH. Lysosome pH is controlled primarily by the vacuolar-type H+ ATPase (V-ATPase) that translocates protons into lysosomal lumen and facilitated by transporters and channels that either promote the influx of cytosolic anions into the lysosomes or the efflux of luminal cations to cytosol to compensate for change in the transmembrane electrical potential generated by luminal proton accumulation.20 PLD1 generates the signaling lipid phosphatidic acid (PA). It has been known that phospholipids are important regulators of many channels.35 It is likely that PLD1-generated PA is involved in the regulation of the transportation of protons and/or counterions in lysosomes. How PA regulates the activities of lysosomal V-ATPase, transporters, and channels would be an important area to explore in the future.
Generation of FAs from bulk membrane phospholipids during nutrient deprivation
Recent studies suggest that pleiotropic role of autophagy in lipid metabolism may be tissue or condition specific.25, 28, 36 Although lipolysis of lipid droplets is the major source of intracellular FAs in fibroblasts during acute starvation in Hanks' balanced salt solution (HBSS) that lacks glucose, amino acids, and growth factors,28 lipophagy plays a more critical role in conditions of serum depletion in the presence of amino acids and glucose.25, 28, 36 In the current study, lipolysis by cytosolic lipases and lipophagy of lipid droplets occurred within 2 days after exposure of the cancer cells to low glucose; therefore, they do not seem to play a significant role in PLD1-regulated cell viability that did not show an effect until 3 days after glucose starvation (Figures 1 and 6). In contrast, we demonstrated that PLD1-regulated autophagic flux supplies FAs to mitochondria from bulk membrane phospholipids in response to prolonged glucose starvation (Figure 7). Noticeably, the aforementioned studies25, 28, 36 focused on relatively short-term starvation (hours to 1 day), and/or performed experiments in an extreme condition, for example, HBSS. It remains to be addressed how different sources of FAs, that is, lipolysis, lipophagy, and autophagy of bulk membranes, contribute to different cellular functions, such as proliferation and viability, during early and late phases of starvation conditions lacking different forms of nutrients, for example, amino acids and glucose. One possibility is that different nutrient deprivation stresses activate overlapping yet distinct signaling pathways that determine the use of different mechanisms to obtain intracellular FAs. Another possibility is that regardless of the nutrient stress type, membrane phospholipids are the major substrates of FA production in the late phase after exhaustion of other sources. Although it has been speculated for some time that membrane phospholipids may supply free FAs to cells via autophagy,8, 37 no experimental evidence has appeared until the current study. More studies need to be done to further understand how autophagic degradation of membrane phospholipids contributes to cellular functions, for example, energy production, ROS reduction, and resynthesis of lipids.
Targeting PLD1-regulated metabolic reprogramming in cancer therapy
A major obstacle of targeting cancer metabolism is the metabolic plasticity that allows cancer cells to adapt to harsh conditions for proliferation and/or survival, and developing resistance to metabolic cancer therapeutics.2, 3, 6 Our data show that during prolonged glucose deprivation, cancer cells rely on FAs recycled from membrane phospholipids for survival. Although FAs may be produced by lipolysis and lipophagy from preexisting lipid droplets, they do not seem to affect cell survival during prolonged glucose starvation. It has been previously shown that autophagy provides FAs to maintain growth and survival of a dormant population of tumor cells.9, 38 Our data demonstrate that during prolonged glucose starvation, membrane phospholipids are the source of autophagy-derived FAs that support ATP synthesis and suppress excess ROS production in mitochondria; and this process is regulated by PLD1 (Figures 5 and 7). Our data underscore the importance of recycling membrane phospholipids via autophagy to cancer cell survival during nutrient scarcity and establish PLD1 as a critical player in this important metabolic adaption pathway.
Our current study shows that inhibition autophagy by ATG5 knockout causes significant death of MDA-MB-231 cells in low glucose condition, and PLD1 inhibition is unable to further increase cell death in this condition (Figure 3). This result suggests that PLD1 regulates cell survival mainly through autophagy, and metabolic cancer therapeutics attacking the Warburg effect would be more effective when PLD1 or PLD1-regulated signaling pathways are inhibited. Our data also support a previous finding that MDA-MB-231 cells are very sensitive to autophagy inhibition.39 In contrast, it was proposed by Jang et al.30 that the combination of PLD1 and autophagy inhibition might be of benefit to cancer patients, based on the result that PLD1 inhibition reduced cell viability in ATG7 knockdown MDA-MB-231 cells. This result suggests that PLD1 regulates cell death mainly through an autophagy-independent role. It remains unclear whether this apparent discrepancy is caused by different glucose concentrations (1 mM in current study and 0 mM and in Jang et al.30, respectively), different degrees of autophagy inhibition by ATG5 (our study) and ATG7 knockdown (Jang et al.30), or different concentrations and sources of PLD1 inhibitors (5 μM from Avanti Polar Lipids in current study and 10 μM from Cayman Chemical in Jang et al.30, respectively). Further clarifying the mechanism through which PLD1 inhibition results in cancer cell death would provide a better cancer therapy.
Materials and Methods
General reagents and antibodies
PLD1i (VU0359595) and PLD2i (VU0285655-1) were from Avanti Polar Lipids (Alabaster, AL, USA). Antibodies for phosphorylated and/or total AKT (1 : 1000), p70S6K (1 : 1000), 4E-BP1 (1 : 1000), p38 (1 : 1000), ATG5 (1 : 1000), and LC3 (1 : 1000) were from Cell Signaling Technology (Danvers, MA, USA). Mouse monoclonal p62 antibody was from BD Biosciences (San Jose, CA, USA). FIPI, antibodies for α-tubulin (1 : 5000), phospho-ERK1/2 (1 : 40 000), and total ERK1/2 (1 : 40 000) were from Sigma-Aldrich (St. Louis, MO, USA). LAMP1 was from the Developmental Studies Hybridoma Bank (Iowa City, IA, USA). Rabbit monoclonal PLD1 antibody (1 : 1000) was from Abcam (Cambridge, MA, USA). Rabbit polyclonal PLD2 antibody (1 : 500) has been described before.40, 41 Goat anti-mouse and anti-rabbit IgGs conjugated with Alexa Fluor 488 or 594 were from Life Technologies (Grand Island, NY, USA). Goat anti-mouse and anti-rabbit IgGs conjugated with IRDye 680CW or IRDye 800CW (1 : 5000) were from Rockland Immunochemicals (Gilbertsville, PA, USA).
Cell culture and viability assays
All cancer cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % fetal bovine serum (Life Technologies). The immortalized nontumorigenic HMECs were cultured in 1/2 MEBM Mammary Epithelial Cell Growth Medium from Lonza (Allendale, NJ, USA) and 1/2 DMEM/F12 supplemented with insulin (5 μg/ml), EGF (10 ng/ml), and hydrocortisone (0.5 μg/ml) as described before.42 For cell viability measurement, cells were seeded in 96-well plates at a density of 3000 cells per well in 100 μl medium containing either 25 or 1 mM glucose. When needed, PLD1i or PLD2i was added to the medium at the final concentration of 5 μM. In some experiments, cells were also treated with different concentrations of 2-DG (0.25, 0.5, 1, 2.5, and 5 mM), 20 μM oleic acid (Sigma-Aldrich), 200 μM etomoxir (Sigma-Aldrich), 1 μM methyl-pyruvate (Santa Cruz Biotechnology, Dallas, TX, USA), or 0.1 mM NAC (Sigma-Aldrich). The numbers of viable cells were measured at indicated time points by CyQuant Cell Proliferation Assay Kit (Life Technologies).
Lentivirus production and transduction
The shRNAs for PLD1 and PLD2 have been described in previous studies.11, 41, 43 Lentiviruses were generated by co-transfection of the lentiviral vector (luciferase, PLD1, PLD2, and ATG5 shRNAs in pLKO vector, or pHluorin-mKate2-LC3), pMDLg/pRRE, pRSV-Rev, and pMD2.G in TLA-293T cells (Thermo Fisher, Waltham, MA, USA) using Lipofectamine and Plus reagent (Life Technologies). After infection with lentiviruses, MDA-MB-231 cells expression shRNAs were selected with puromycin before experiments.
Western blotting
Whole-cell lysates were generated in radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitor cocktails on ice. After brief sonication, protein samples were load on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes that was blocked in 0.1% casein and probed with the indicated primary antibodies. Fluorescently labeled secondary antibodies were used for western blotting and detected by the Li-COR Odyssey infrared imaging system from Li-COR Biotechnology (Lincoln, NE, USA).
Confocal and fluorescence microscopy
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, then permeabilized with 0.1% Triton X-100 or 0.1% Saponin in PBS for 10 min. After blocking with 5% goat serum in PBS, cells were incubated with the desired primary antibody for 1 h at room temperature. Subsequently, cells were washed in PBS and incubated with Alexa 488 or Alexa 594-labeled secondary antibody, or BODIPY493/503 (0.5 μg/ml) (Life Technologies) for 1 h at room temperature. After washing with PBS, the coverslips were mounted on glass slides with 4% n-propyl gallate. Cells were visualized with a Zeiss Axiophot epifluorescence microscope (Carl Zeiss, Thornwood, NY, USA) or a Nikon A1 confocal microscope (Nikon Instruments Inc., Melville, NY, USA). The colocalization and measurement were quantified using ImageJ software (Bethesda, MD, USA).
Measurement of intracellular ATP and mitochondrial ROS levels and mitochondrial membrane potential
The intracellular ATP levels were measured using the ATPlite Luminescence ATP Detection Assay System (Perkin Elmer, Waltham, MA, USA). The ATP levels of the treated cells were normalized to the mean value obtained from untreated cells. Cytosolic and mitochondrial ROS were detected by flow cytometry after loading the cells with 5 μM DCFH-DA [2-(2,7-dichloro-3,6-diacetyloxy-9H-xanthen-9-yl)-benzoic acid] (Cayman Chemical, Ann Arbor, MI, USA) and 2 μM MitoSOX Red mitochondrial superoxide indicator (Thermo Fisher), respectively. Mitochondrial membrane potential was measured by the fluorescent intensity of TMRE (Tetramethylrhodamine, ethyl ester) (Thermos Fisher) using flow cytometry after staining the cells with 50 nM TMRE for 15 min at 37 °C.
Determination of lysosomal pH
Cells seeded on 35-mm glass-bottom dishes (MatTek, Ashland, MA, USA) were incubated with 5 mg/ml Oregon Green 488 dextran 10 000 MW (Life Technologies) overnight, followed by 2 h chase in dextran-free medium. At the end of the chase period, the cells were rinsed with and maintained in the imaging buffer (140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM HEPES, 10 mM Glucose, pH 7.4). Fluorescence images were acquired using an inverted Nikon ECLIPSE TE200 microscope at excitation wavelengths of 440 nm and 490 nm and emission wavelength of 545±50 nm, with a CCD camera controlled by InCytIM-2 software (Intracellular Imaging Inc., Cincinnati, OH, USA). The 490/440 ratio was calculated for each region of interest representing individual lysosomes or lysosomal clusters and converted into a pH value using the pH calibration curve generated from the same coverslip after the initial ratio measurement. To generate the lysosomal pH calibration curve, potassium isotonic solution (10 mM HEPES, 10 mM MES, 140 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM glucose) of pH 3.5, 4, 4.5, 5, 5.5, 6, 6.5, and 7.0 containing 10 μg/ml nigericin were sequentially added to cells and each was equilibrated for 10 min before ratio images were taken. Lysosomal pH values were obtained by fitting the obtained intensity ratios to the standard curves.
Statistical analysis
All statistical analyses were evaluated using unpaired two-sample Student's t-test. Data are shown as mean±S.D. *P<0.05; **P<0.01; ***P<0.001.
Acknowledgments
We thank Dr Ilya Levental for oleic acids. This study was supported by grants from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP130425 and RP160775 to GD) and a grant from National Institutes of Health (NIH) (NS092377 to MZ).
Author contributions
MC, GW, MZ and GD designed the experiments; MC and GD analyzed data; MC, JH, JX, LWRT, ZW, CR, JW, YX and FL performed experiments; YB generated PLD2 antibody; MC, MZ and GD wrote the manuscript.
Glossary
- 2-DG
2-deoxy-D-glucose
- ATP
adenosine triphosphate
- ERK
extracellular signal-regulated kinase
- FA
fatty acid
- FAO
fatty acid oxidation
- NAC
N-acetylcysteine
- PA
phosphatidic acid
- PBS
phosphate-buffered saline
- PLD
phospholipase D
- PLD1
phospholipase D1
- PLD1i
PLD1 inhibitor
- PLD2i
PLD2 inhibitor
- shRNA
small hairpin RNA
- ROS
reactive oxygen species
- tfLC3
tandem monomeric red fluorescent protein (mRFP)
- GFP
green fluorescent protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by S Lavandero
Supplementary Material
References
- Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science 2014; 345: 1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015; 17: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 2013; 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, Farese RVJr. Cellular fatty acid metabolism and cancer. Cell Metab 2013; 18: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol 2014; 24: 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and cancer. Clin Cancer Res 2015; 21: 5037–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes H, Rubio N, Garg AD, Agostinis P. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med 2013; 19: 428–446. [DOI] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 2013; 27: 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Huang P, Liang BT, Frohman MA. Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol Biol Cell 2004; 15: 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Chardin P, Yamazaki M, Kanaho Y, Du G. RhoA-mediated phospholipase D1 signaling is not required for the formation of stress fibers and focal adhesions. Cell Signal 2006; 18: 469–478. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 2015; 25: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5: 886–897. [DOI] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Uchiyama Y. A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS One 2014; 9: e110600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3: 452–460. [DOI] [PubMed] [Google Scholar]
- Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT et al. Phospholipase D1 localises to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr Biol 1998; 8: 835–838. [DOI] [PubMed] [Google Scholar]
- Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ et al. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J Cell Biol 2003; 162: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol 2012; 74: 69–86. [DOI] [PubMed] [Google Scholar]
- Canton J, Grinstein S. Measuring lysosomal pH by fluorescence microscopy. Methods Cell Biol 2015; 126: 85–99. [DOI] [PubMed] [Google Scholar]
- Steinberg BE, Huynh KK, Brodovitch A, Jabs S, Stauber T, Jentsch TJ et al. A cation counterflux supports lysosomal acidification. J Cell Biol 2010; 189: 1171–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene 2005; 24: 4165–4173. [DOI] [PubMed] [Google Scholar]
- Zechner R. FAT FLUX: enzymes, regulators, and pathophysiology of intracellular lipolysis. EMBO Mol Med 2015; 7: 359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M et al. Autophagy regulates lipid metabolism. Nature 2009; 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep 2014; 9: 349–365. [DOI] [PubMed] [Google Scholar]
- Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A et al. HIF2alpha-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov 2015; 5: 652–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 2015; 32: 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun 2010; 1: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YH, Choi KY, Min DS. Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ 2014; 21: 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae EJ, Lee HJ, Jang YH, Michael S, Masliah E, Min DS et al. Phospholipase D1 regulates autophagic flux and clearance of alpha-synuclein aggregates. Cell Death Differ 2014; 21: 1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruntz RC, Taylor HE, Lindsley CW, Brown HA. Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J Biol Chem 2014; 289: 600–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 2007; 3: 154–157. [DOI] [PubMed] [Google Scholar]
- Yoon MS. Vps34 and PLD1 take center stage in nutrient signaling: their dual roles in regulating autophagy. Cell Commun Signal 2015; 13: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis DE, Petrou VI, Zhang M, Mahajan R, Meng XY, Adney SK et al. Phosphoinositide control of membrane protein function: a frontier led by studies on ion channels. Annu Rev Physiol 2015; 77: 81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA et al. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 2013; 203: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab 2011; 13: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014; 514: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res 2014; 74: 2579–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y et al. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem Biophys Res Comm 2000; 278: 140–143. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y et al. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol Cell Biol 2014; 34: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Frohman MA. A lipid-signaled myosin phosphatase surge disperses cortical contractile force early in cell spreading. Mol Biol Cell 2009; 20: 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.