Abstract
The lipid Phosphatidylinositol-3-phosphate [PtdIns3P or PI(3)P] plays many membrane trafficking roles and is primarily produced by the Class III PI3K, VPS34. Determining the level of cellular PI(3)P however can be complex. Extraction of cellular lipids by methanol/chloroform can struggle to separate and identify distinct phospholipid species. Alternately mass spectrometry may be utilised but this requires significant set up of specialised equipment and time to utilise.
Use of a PI(3)P-binding-specific recombinant protein domain is a quick method for ascertaining cellular PI(3)P levels and can also allow visualisation of sub-cellular localisation. The PX domain of p40phox (herein referred to as PX) is very specific for PI(3)P over other phospholipid species (Kanai et al., 2001). However, expressing PX directly in cells can be problematic, as it will act in a dominant negative manner to bind and sequester PI(3)P with greater affinity than endogenous proteins, thus disturbing cellular pathways and the normal balance of PI(3)P levels. Using fluorescently labelled PX following cell fixation is therefore more suitable, as it is able to highlight PI(3)P rich structures without risk of perturbing the system.
Materials and Reagents
Zeba™ spin desalting column (Thermo Fisher Scientific, catalog number: 89891)
22 × 22 mm cover glasses, Menzel Glaser (VWR, catalog number: 631-1336)
Millex-GS syringe filter unit, 0.22 µm (Merck Millipore, catalog number: SLGSV255F)
Slide-A-Lyzer dialysis cassettes, 10K MWCO 12 ml (Thermo Fisher Scientific, catalog number: 66810)
Microscope slides, 75 × 25 mm (superfrost) (VWR, catalog number: 48311-600)
BL-21 bacteria (New England BioLabs, catalog number: C2530H)
U2OS cells (ATCC, catalog number: HTB-96)
SOC medium (Thermo Fisher Scientific, catalog number: 15544034)
LB broth, Miller (Merck Millipore, catalog number: 71753)
LB agar, Miller (Merck Millipore, catalog number: 1102835000)
Ampicillin
IPTG (Sigma-Aldrich, catalog number: I6758)
Tris (VWR International, catalog number: 103157P)
Sodium chloride (NaCl) (VWR International, catalog number: 27810.364)
Triton X-100 (Fisher Scientific, catalog number: BP151-500)
β-mercaptoethanol (Sigma-Aldrich, catalog number: M6250)
PMSF (Sigma-Aldrich, catalog number: P7626)
Benzamidine (Sigma-Aldrich, catalog number: 12072)
Brij-35 (Merck Millipore Corporation, catalog number: 1.01894.1000)
EGTA (Sigma-Aldrich, catalog number: E4378)
Glutathione sepharose 4B (GE Healthcare, catalog number: 17-0756)
InstantBlue Coomassie stain (Expedeon, catalog number: ISB1L)
Alexa Fluor 488 TFP ester (Thermo Fisher Scientific, catalog number: A37570)
Alexa Fluor 594 NHS ester (Thermo Fisher Scientific, catalog number: A37572)
Paraformaldehyde (Sigma-Aldrich, catalog number: P6148)
HEPES (Formedium, catalog number: HEPES10)
Potassium chloride (KCl) (VWR International, catalog number: 26764.260)
Magnesium acetate (MgAc) (Sigma-Aldrich, catalog number: M5661)
Potassium glutamate (Sigma-Aldrich, catalog number: G1501)
DMEM (Thermo Fisher Scientific, Gibco®, catalog number: 11960-044
PBS pH 7.4 (Sigma-Aldrich, catalog number: P4417)
Bovine serum albumin fraction V (BSA) (Sigma-Aldrich, catalog number: 10735108001)
Liquid nitrogen
Sodium bicarbonate (Sigma-Aldrich, catalog number: S5761)
DMSO (Sigma-Aldrich, catalog number: D8418)
Glycerol, ultra pure (VWR, catalog number: IC800688)
Sucrose (Sigma-Aldrich, catalog number: S8501)
Sodium azide (Sigma-Aldrich, catalog number: S2002)
ProLong gold antifade mountant with DAPI (Thermo Fisher Scientific, catalog number: P36931)
Lysis buffer (see Recipes)
Equilibration buffer (see Recipes)
Wash buffer (see Recipes)
Elution buffer (see Recipes)
Dialysis buffer (see Recipes)
Formaldehyde (see Recipes)
Glutamate buffer (see Recipes)
DMEM + HEPES (see Recipes)
PBS + BSA (see Recipes)
Equipment
Water bath
Bacterial Shaker (e.g., Infors HT)
Benchtop Centrifuge (e.g., Beckman Coulter, model: Allegra X-12R)
Centrifuge (e.g., Beckman Coulter, model: Avanti J-26 XP)
Sonicator (e.g., Sonics Vibra Cell)
Tweezer
GSH-Sepharose beads
Magnetic stir bar (Fisher Scientific)
Gel electrophoresis equipment (e.g., Dual minislab, model: ATTO AE6500)
Fluorescence microscope (e.g., Nikon Eclipse Ti-E model equipped with appropriate filter set for Alexa-Fluor dye)
Procedure
-
Purification of GST-PX probe
The PX domain utilised is M1-R148 of the p40phox protein and has been previously characterised to selectively detect PI(3)P (Kanai et al., 2001). M1-R148 p40phox was cloned into a bacterial (pGEX) expression vector with N-terminal GST tag and linker region (SDLEVLFQGPLGS).
Transform BL21 E.coli cells with 2 ng of plasmid by heat-shock at 42 °C for 45 sec.
Add 900 μl SOC media and incubate in a water bath at 37 °C for 1 h to allow cell recovery.
Spread 100 μl onto LB plates with appropriate selection marker (Ampicillin in the case of pGEX vectors).
Incubate at 37 °C overnight. Plates may be stored at 4 °C before proceeding to the next step.
Inoculate 25 ml of LB media (containing selection marker) with a single colony and incubate for 16 h at 37 °C and agitation at 180 rpm in an Infors HT bacterial shaker.
Inoculate 500 ml of LB media (containing selection marker) with 10 ml of overnight culture and grow until OD600 = 0.4-0.6 at 37 °C with agitation at 180 rpm in an Infors HT bacterial shaker. Once OD600 is achieved, reduce temperature to 26 °C and induce expression with 10-400 μM IPTG for 16 h (50 μM IPTG gave best results in our hands).
Pellet cells by centrifugation at 4,000 × g for 30 min at 4 °C. The pellet may be stored at - 20 °C if required prior to continuing the process.
Add 35 ml of lysis buffer to bacterial pellet and resuspend.
Sonicate at high power on ice, pulse for 15 sec and rest for 15 sec, repeat 3-5 times.
Spin at 15,000 x g for 30 min at 4 °C to clarify lysate and separate the lysate for pelleted cell debris.
Add 10 ml of Equilibration buffer to 3.5 ml (~50% slurry) of GSH-Sepharose, vortex briefly and spin at 1,000 x g for 2 min. Aspirate off supernatant to waste and repeat once more.
Add the washed GSH-Sepharose beads to the clarified lysate and incubate on a roller at 4 °C for 45 min.
Spin down beads at 1,000 × g for 2 min, remove supernatant and add wash buffer up to 40 ml total, vortex briefly and repeat three times.
Add 5 ml of elution buffer to the beads and incubate on ice for 10 min. Spin at 1,000 × g for 2 min and then recover the supernatant. Add 2.5 ml back to the beads and incubate a further 10 min on ice. Spin once more at 1,000 × g for 2 min and recover the supernatant.
Combine the elutions and pass through a 0.22 μm filter to remove any residual resin.
Determine protein concentration of eluted protein (e.g., by Bradford Assay) and check the purity by running on a polyacrylamide gel and staining with Coomassie blue.
Dialyse the eluted material at 4 °C for 16 h by loading sample into a Slide-A-Lyzer cassette with 10 kDa molecular weight cut off and floating in dialysis buffer with a rotating magnetic stir bar. Dialysis is important to remove glutathione and is better for long-term storage of the protein at -80 °C. Alternately if planning to label the PX protein immediately then it can be dialysed directly into PBS + DTT as required in step 2a.
-
Labelling the GST-PX domain with AlexaFluor-594
The N-terminal GST tag allows the probe to be stained using a GST recognising antibody and appropriate secondary conjugated to a fluorophore. Alternately the GST-PX domain can be labelled directly with a reactive Alexa Fluor dye to reduce the time to stain permeabilised cells. The GST-PX domain was labelled utilising a reactive Alexa Fluor (AF) dye, such as AF-594 (A37572) or AF-488 (A37570). Labelling was carried out as recommended in manufacturer’s instructions, briefly:- 0.5 ml of 1 mg/ml purified GST-PX protein was first dialysed with a 10 kDa molecular weight cut off membrane into PBS pH 7.4 + 1 mM DTT for 3 h at 4 °C, the buffer was then changed and dialysed overnight for 16 h at 4 °C. It is important to dialyse the GST-PX protein efficiently as earlier buffer components may interfere with the subsequent labelling efficiency.
- 1 M sodium bicarbonate was prepared fresh in ddH2O and 50 µl added to the GST-PX protein solution.
- A 100 µg vial of reactive dye was prepared fresh in 10 µl of DMSO and added immediately to the protein and shaken in a thermomixer at 1,100 rpm for 1 h at room temperature. As the dye is light sensitive, cover with aluminium foil to protect the dye as much as possible during the protocol.
- Unconjugated dye was separated from protein by adding sample to a Zeba™ desalting column and centrifugation according to manufacturer’s instructions. After removing unconjugated dye, the protein solution should still retain a colour change indicating that dye has conjugated to the protein. Determining the degree of labelling is not essential but can be calculated by following manufacturer’s instructions available with the Alexa Fluor dye.
- Cleared sample was added to glycerol to create a 50% (v/v) solution for long-term storage. The labelled protein should be separated into small aliquots and stored long-term at -80 °C, a working aliquot can be retained at -20 °C.
The Alexa Fluor™-PX probe is now complete and ready for use in cellular staining.
-
Preparation of formaldehyde fixative
Formaldehyde was prepared in advance from paraformaldehyde powder. Ensure all work is carried out in a fume hood as heating formaldehyde causes the release of toxic gases. For the preparation of 250 ml:- Weigh out 9.25 g of paraformaldehyde and add to 200 ml ddH2O.
- Mix with a magnetic stir bar and heat to 60 °C.
- Add NaOH dropwise until the solution turns clear.
- Remove from heat and add 50 ml of 1 M HEPES (pH 7.4).
- Allow to cool before removing from fume hood and store at 4 °C.
-
Freeze-thaw permeabilisation
To maintain cellular PI(3)P levels, a detergent-free method of fixing and permeabilizing cells is needed.
Cells to be stained were seeded onto glass coverslips (22 × 22 mm) in 6-well dishes a minimum of 16 h prior to use to allow cells to attach and settle. U2OS cells attach well to glass and settle very flat for clear imaging. Other cell types may require pre-coating of coverslips with gelatin or other reagents.
After treating cells as required for stimuli being investigated, move the 6-well dish onto ice.
Aspirate media and wash coverslips 2 × 2 ml with ice-cold PBS.
Aspirate PBS and add 2 ml of ice-cold glutamate buffer.
Pick up a single coverslip with tweezers and dab the edge against some paper towel to remove excess buffer. Submerse the coverslip directly into liquid nitrogen until it stops bubbling (~2-3 sec). Remove and place cell side up on paper towel to allow the coverslip to thaw back to room temperature (approximately 1 min or less). During this time it is possible to repeat the freezing procedure with any other coverslips.
-
Once the coverslip has thawed, move back into the 6-well dish on ice.
Note: Take care with coverslips, after the freeze-thaw coverslips become more brittle and more prone to cracking.
Wash coverslips 2 × 2 ml with ice-cold glutamate buffer, cells can now be moved off ice and onto bench at room temperature.
Aspirate glutamate buffer and add 2 ml, 3.7% formaldehyde to coverslips for 10 min. After 10 min, remove and replace 2 ml, 3.7% formaldehyde and incubate a further 20 min.
Wash coverslips 2 × 2 ml with DMEM/HEPES followed by 10 min incubation in 2 ml DMEM/HEPES to quench formaldehyde.
Wash each coverslip 2 × 2 ml and then incubate for 15 min in 2 ml PBS/1% BSA to block.
Aspirate off buffer and add 100 μl PX-594 probe to each coverslip at 1:200-1:500 (dilution may vary between cell types) in PBS/1% BSA and cover samples with foil from this point on to protect from light.
Incubate for 1 h at RT.
Wash coverslips 3 × 2 ml with PBS/1% BSA.
Pipette ~10 μl of mounting solution (ProLong Gold Antifade Mountant with DAPI) directly onto a glass slide in a “T” shape.
Wash each coverslip briefly by dipping into a beaker containing ddH2O, remove excess water by touching the edge against paper towel. Slowly lower the coverslip (cell side down toward the glass slide). Lower gently so that the coverslip comes in contact with the slide from the top of the “T” and downwards.
Ensure all coverslips are covered by foil to protect from light and leave overnight to dry at RT.
Cells can be analysed using a fluorescent microscope with appropriate filters to excite the Alexa-Fluor dye.
Representative data
Notes
The selectivity of staining for PI(3)P may also be confirmed by generating an additional fluorescently labelled PX domain probe mutated in the Zinc Finger domain (Such as an R57Q mutation) to prevent lipid binding (Kanai et al., 2001).
The Alexa Fluor dye is light sensitive, so it is important to protect the labelled PX domain probe from light as much as possible by storing in light-resistant tubes and using aluminium foil to protect samples during and after the staining procedure to minimise loss of signal.
Recipes
-
Lysis buffer
50 mM Tris (pH 7.5)
250 mM NaCl
1% (v/v) Triton X-100
0.1% β-mercaptoethanol
0.2 mM PMSF
1 mM benzamidine
Store at 4 °C
Add β-Me, PMSF and benzamidine fresh before use
-
Wash buffer
50 mM Tris (pH 7.5)
250 mM NaCl
0.03% Brij-35
0.1 mM EGTA
0.1% β-mercaptoethanol
mM PMSF
1 mM benzamidine
Store at RT
Add β-Me, PMSF and benzamidine fresh before use
-
Equilibration buffer
50 mM Tris (pH 7.5)
250 mM NaCl
0.1% β-mercaptoethanol
0.2 mM PMSF
1 mM Benzamidine
Store at RT
Add β-Me, PMSF and benzamidine fresh before use
-
Elution buffer (Make fresh)
Wash buffer
20 mM glutathione
Adjust pH to 7.5
-
Dialysis buffer (Make fresh)
50 mM Tris (pH 7.5)
150 mM NaCl
0.1 mM EGTA
270 mM sucrose
0.03% Brij-35
0.07% β-mercaptoethanol
1 mM benzamidine
0.1 mM PMSF
-
Glutamate buffer
25 mM HEPES (pH 7.4)
25 mM KCl,
2.5 mM MgAc
5 mM EGTA
150 mM potassium glutamate
Store at 4 °C
-
Formaldehyde
200 mM HEPES (pH 7.4)
(see protocol for preparation, step 3)
3.7% (w/v) formaldehyde
Store at 4 °C
-
DMEM + HEPES
DMEM
10 mM HEPES (pH 7.4)
Store at 4 °C
-
PBS + BSA
PBS
1% (w/v) BSA
0.02% sodium azide
Store at 4 °C
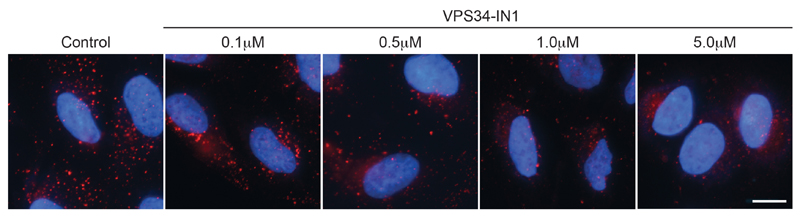
Figure 1. Example staining with PX-594 in U2OS cells.
U2OS cells were treated with VPS34-IN1 inhibitor as indicated for 1 h prior to fixation and staining as detailed here. Bar = 10 μm
Acknowledgments
This work was supported by the Medical Research Council (MRC) and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KGaA, Janssen Pharmaceutica and Pfizer).
References
- 1.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3(7):675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]