Abstract
Eight different types of ubiquitin (Ub) linkages are present in eukaryotic cells that regulate diverse biological processes. Proteins that mediate specific assembly and disassembly of atypical Lys6, Lys27, Lys29 and Lys33 linkages are largely unknown. We here reveal how the human Ovarian Tumor (OTU) domain deubiquitinase (DUB) TRABID specifically hydrolyzes both Lys29- and Lys33-linked diubiquitin (diUb). A crystal structure of the extended catalytic domain reveals an unpredicted Ankyrin repeat (Ank) domain that precedes an A20-like catalytic core. NMR analysis identifies the Ank domain as a new Ub binding fold termed AnkUBD, and DUB assays in vitro and in vivo show that this domain is crucial for TRABID efficiency and linkage-specificity. Our data are consistent with a role of the AnkUBD as an enzymatic S1' Ub binding site, which orients a Ub chain such that Lys29 and Lys33 linkages are cleaved preferentially.
Protein ubiquitination is a versatile posttranslational modification that regulates an increasingly large number of cellular processes. This versatility originates from the ability of ubiquitin (Ub) to form eight structurally and functionally distinct polymers, in which Ub molecules are linked via one of seven lysine (Lys) residues or through the N-terminus 1,2. It is clear from proteomic analysis that all Ub linkages exist in eukaryotic cells 3, however the functions of the so-called atypical Ub chain types linked via Lys6, Lys11, Lys27, Lys29, or Lys33 remain obscure. Recent proteomics data show that Lys29-linkages may be the third most common chain type in mammalian cells, accounting for ~10% of all linkages 4. Several HECT E3 ligases were suggested to mediate Lys29-linkage assembly in vivo 5–7, and amongst cellular substrates of Lys29-ubiquitination are signaling molecules including microtubule affinity regulating kinases (MARKs) 8, Deltex 7, and β-catenin 6. In yeast, the Ub fusion degradation (Ufd) pathway involves Lys29-linkages 9,10.
Atypical Ub chains as in vitro reagents have only become available recently through chemical biology methods. Ub Lys residues are exchanged genetically or by peptide synthesis for Lys derivatives that can be chemically ubiquitinated, either by steps of protection, specific deprotection and activated ligation (GOPAL method) 11, or when a thio-Lys derivative is incorporated, by direct ubiquitination using native-chemical ligation protocols 12–14. Both strategies allow generation of isopeptide-linked diUb molecules with atypical linkages, and enable comprehensive analysis of linkage specificity in DUBs.
Human cells comprise ~90 DUBs that can be sub-classified into five families 15,16. Amongst these, ubiquitin-specific proteases (USPs) show little linkage specificity 11,17, while the JAMM metalloproteases comprise Lys63-specific members 18,19. The OTU family comprises 14 enzymes in humans with distinct specificity profiles, including Lys11- and Lys48-specific members 20,21. OTU family DUBs contain a papain-like catalytic core of ~180 amino acids (aa) 15. The molecular basis for the linkage-specificity in OTU domains is unclear, and structural information is only available for OTUB1 20, OTUB2 22, A20 23,24 in absence of Ub, and for yeast Otu1 and a viral OTU in complex with a Ub suicide inhibitor 25–28. In addition to their catalytic domain, many OTU members have additional Ub binding domains (UBDs) 15. At least 20 different UBD families have been described, and linkage-specific UBDs have provided the key to understand roles of different Ub linkages in cells 29.
A20 is the best-studied OTU family DUB with important roles as a negative feedback regulator in nuclear factor-κB (NF-κB) signaling 30. Other OTU enzymes play roles in interferon signaling 31,32, and in p97/cdc48-mediated processes 33,34, but cellular roles for most OTU enzymes remain to be discovered.
TRABID (ZRANB1) is an OTU family member comprising a catalytic domain closely related to that of A20 22,36, which had previously been linked to Wnt/β-catenin signaling, as a positive regulator of β-catenin-mediated transcription 35. We further showed recently that the TRABID OTU domain hydrolyzes Lys29-linked diUb with 40-fold higher efficiency compared to Lys63-linked diUb, while Lys6-, Lys11-, Lys48-linked diUb or linear chains were not cleaved, identifying TRABID as the first Lys29-specific OTU domain enzyme 11. This implicated atypical chains in Wnt/β-catenin signaling, although the molecular step(s) in the signaling cascade have not yet been identified. Also, TRABID activity towards Lys27- and Lys33-linkages has not yet been tested, as these chain linkages had not been available previously.
Here, we set out to determine how TRABID DUB activity and specificity is achieved. We tested the DUB activity of TRABID against the complete panel of eight different Ub linkages, and show that it cleaves Lys29- and Lys33-linkages with marked preference over Lys63-linkages, but no other chain type. A crystal structure of the N-terminally extended TRABID OTU domain revealed a catalytic fold similar to that of A20, which is extended by two Ankyrin (Ank) repeats positioned such that they could form a proximal Ub binding site. Indeed, the isolated Ank domain binds to Ub, and NMR experiments map the interaction interfaces to a conserved hydrophobic surface of the Ank module and to the hydrophobic Ile44 patch of Ub. We provide evidence that the Ankyrin repeat Ub binding domain (AnkUBD) contributes to enzymatic efficiency and linkage-specificity in vitro and in vivo.
Results
Specificity of the extended OTU domain of TRABID
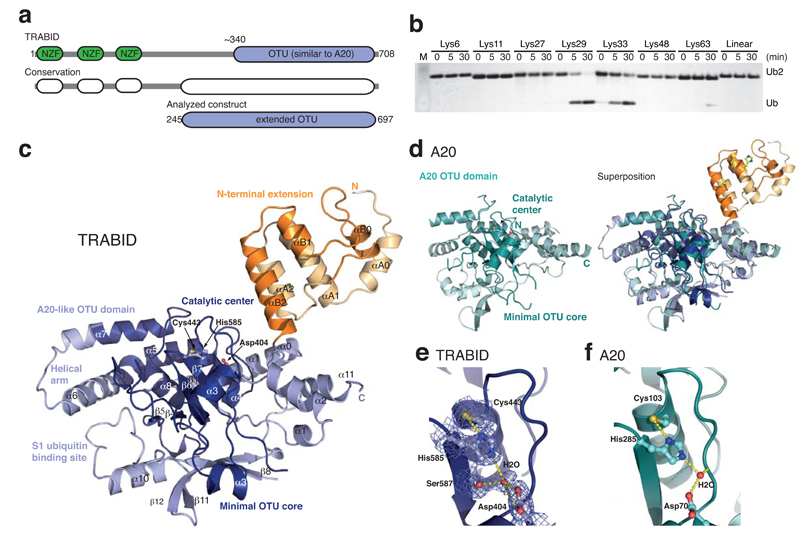
The human TRABID protein spans 708 amino acids (aa) and comprises three N-terminal Npl4-like zinc finger (NZF) domains (aa 1-200), and a C-terminal OTU domain (aa 340-700) (Fig. 1a). The C-terminal OTU domain of TRABID is closely related to the previously characterized OTU domain of A20 23,36. However, a stretch of ~100 highly conserved residues upstream of the TRABID OTU domain (aa 245-340; see below) indicated an extension of the catalytic fold not present in A20.
Figure 1.
Structure and specificity of an extended TRABID OTU domain. (a) Schematic representation of the functional domains of TRABID (top) and species conservation derived from a multiple sequence alignment (http://www.ensembl.org) (middle). An extended catalytic OTU domain was analyzed (aa 245-697, bottom). (b) Linkage specificity of the extended catalytic OTU domain of TRABID using diUb molecules of all eight linkage types, performed as reported before 26. TRABID was incubated with diUb for the indicated times, the reactions resolved on an SDS-PAGE gel and silver stained. (c) Structure of the extended TRABID OTU domain. The catalytic core is colored in shades of blue where dark-blue indicates the minimal OTU core domain, and lighter-blue additional secondary structure elements found in the A20-like subfamily of OTU DUBs. The Ank-repeats are shown in two shades of orange. The catalytic triad residues are indicated in ball-and-stick representation. (d) Structure of A20 (left, pdb-id 2vfj, 23) and superposition with TRABID (right). (e) Catalytic triad residues of TRABID are shown in ball-and stick representation with yellow sulfur, red oxygen and blue nitrogen atoms. A red sphere indicates a water molecule, and yellow dotted lines indicate hydrogen bonds. A 2|Fo|-|Fc| electron density map contoured at 1σ covers relevant residues. (f) The A20 catalytic triad shown as in e.
The extended fragment of the TRABID OTU domain (aa 245-697) was tested against the complete panel of eight differently linked diUb molecules. In this qualitative analysis, we defined DUB specificity as the enzymatic concentration at which the preferred linkage type is cleaved completely at a defined timepoint. TRABID cleaved Lys29- and Lys33-linked diUb with higher activity compared to Lys63-linkages (Fig. 1b), consistent with previous quantitative data 11. The remaining diUb molecules linked through Lys6, Lys11, Lys27 or Lys48, or linear diUb, were not cleaved (Fig. 1b). Hence, TRABID has a dual specificity against two atypical Ub chains linked via Lys29 and Lys33, in marked preference over Lys63-linkages.
Crystal structure of the extended OTU domain
To understand the molecular basis for the unique DUB specificity of TRABID, the extended OTU domain was purified to homogeneity and crystallized. Needle crystals diffracted to 2.23 Å on the ID23-2 beamline at ESRF (Grenoble). Phase information was obtained from a SIRAS experiment using crystals derivatized with gold cyanide. The resulting electron density maps were of high quality (Supplementary Fig. 1a), and the structure was built and refined to final statistics shown in Table 1.
Table 1. Data collection statistics.
Values in parentheses are for the highest resolution shell. A single crystal was used for each data set.
| TRABID 245-697 AuCN | TRABID 245-697 | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| a, b, c (Å) | 60.29, 72.24, 132.55 | 60.40, 72.15, 133.01 |
| α, β, γ (º) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 72.2-3.50 (3.69-3.50) | 44.7 - 2.23 (2.35-2.23) |
| Rmerge | 0.193 (0.513) | 0.077 (0.494) |
| l / σl | 10.2 (5.4) | 12.1 (2.7) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy | 9.0 (9.2) | 4.1 (4.1) |
| Refinement | ||
| Resolution (Å) | 44.7-2.23 | |
| No. reflections | 54670 | |
| Rwork / Rfree | 0.202 / 0.246 | |
| No. atoms | ||
| Protein | 3433 | |
| Ligand/ion | 9 | |
| Water | 188 | |
| B-factors | ||
| Protein | 43.5 | |
| Ligand/ion | 61.5 | |
| Water | 43.2 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.013 | |
| Bond angles (º) | 1.38 |
The crystallized fragment of TRABID (Fig. 1c) comprised an OTU domain (aa 339-693) with the characteristic triangular shape similar to that of A20 (Fig. 1c, d) 23,24 that was preceded by an α-helical domain of 96 aa (see below). The two independent domains are connected by the last helix of the helical domain (αB2) projecting away from the OTU fold (Fig. 1c). A flexible linker between the domains, and no notable additional interactions at the small interface (313 Å2) indicate potential conformational freedom of the α-helical domain with respect to the OTU domain.
The OTU domain folds of TRABID and A20 superpose well (RMSD of 1.5 Å over 275 aa), and all secondary structure elements are conserved (Fig. 1c, d, Supplementary Fig. 1b). The catalytic Cys443 and His585 of TRABID superpose well with the A20 catalytic center (Fig. 1e, f).
Two Ankyrin repeats extend the catalytic domain
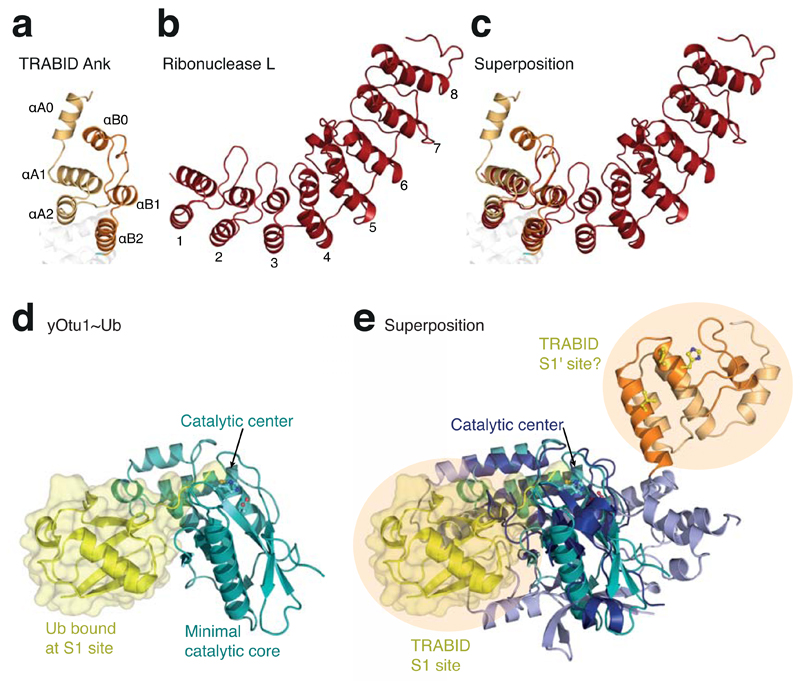
Fold analysis of the adjoining α-helical domain (aa 245-340) with the Dali server 37 revealed the presence of two Ankyrin (Ank) repeats (Fig. 2a). Ank repeats comprise ~30 aa and consist of two interacting helices connected by short loops. Multiple repeats ”stack” via a conserved Ank motif, forming arc-shaped structures 38 (Fig. 2b). In TRABID, the first Ank repeat spans aa 260-290 and is connected to the second repeat (aa 313-340) by a long linker that packs against what would correspond to the concave surface in an extended Ank repeat structure (Fig. 2c). However, a conserved N-terminal helix (αA0, aa 245-259) that packs against the first Ank repeat, and the C-terminal OTU domain that directly extends from the second repeat, define the boundaries for the two Ank repeats in TRABID (Fig. 1c). The primary sequence of these terminal repeats is divergent from the easily identifiable Ank motif in internal repeats, explaining why the Ank domain of TRABID had not been annotated.
Figure 2.
TRABID comprises two Ankyrin repeats with roles in Ub binding. (a) Structure of the Ank domain in TRABID showing the two repeats. (b) Structure of ribonuclease L (pdb-id 1wdy, 47), the Ank repeat protein with highest similarity to the TRABID Ank domain in a DALI search (Z-score 8.4). The eight Ank repeats are numbered. (c) Superposition of the TRABID Ank domain and ribonuclease L. (d) The minimal OTU domain of yOtu1 (green) with Ub (yellow) bound at the S1 site (pdb-id 3by4, 25). The orientation matches that of the minimal OTU domain core indicated in Fig. 1c. (e) Superposition of TRABID and yOtu1 reveals the relative position of the S1 Ub binding site on TRABID, and suggests that the Ank domain may constitute an S1' Ub binding site.
Ub binding sites in TRABID
OTU domain enzymes comprise a high-affinity S1 Ub binding site that binds Ub (termed the ”distal” Ub in a diUb) and coordinates its C-terminus towards the active site independently of the linkage type 15. OTU domain complex structures with Ub bound at the S1 site have been reported for yeast Otu1 (yOtu1, Fig. 2d) 25 and for a viral OTU domain (vOTU, Supplementary Fig. 1c) 26–28. Both OTU domains comprise only ~180 aa and define the minimal OTU domain core conserved in TRABID (Fig. 1c, 2d, 2e). TRABID superposes with yOtu1 with an RMSD of 2.0 Å over 99 aa (Fig. 2e) and with vOTU with an RMSD of 2.5 Å over 105 aa (Supplementary Fig. 1c).
The complex structures indicate the position of the distal Ub bound to the S1 site of the enzyme (Fig. 2e). All residues interacting with the C-terminal tail of the distal Ub are conserved amongst OTU domains, indicating identical binding modes at the catalytic center itself, and a similar position of the S1 Ub binding site. However, Ub binds to yOtu1 and vOTU in different orientations, and interacts with distinct OTU surfaces in the two structures 26. Compared to yOtu1 and vOTU, TRABID contains additional secondary structure elements that would require the distal Ub to rotate and or shift in order to bind the S1 site (Fig. 2e). Hence, while the position of the S1 Ub binding site can be confidently assigned, the orientation of the S1 Ub is difficult to predict in absence of a complex structure.
In contrast, the existence or location of an S1' Ub binding site (coordinating the ‘proximal’ Ub in diUb, which provides the Lys residue to the isopeptide bond) is not known for any OTU domain. Interestingly, in TRABID, the adjacent Ank module preceding the OTU fold is located such that it could serve as an S1' Ub binding site (Fig. 2e). Ank repeat domains mediate protein interactions through a variety of surfaces 38, but Ub has not been reported to bind Ank repeats.
TRABID Ank domain mediates Ub interactions
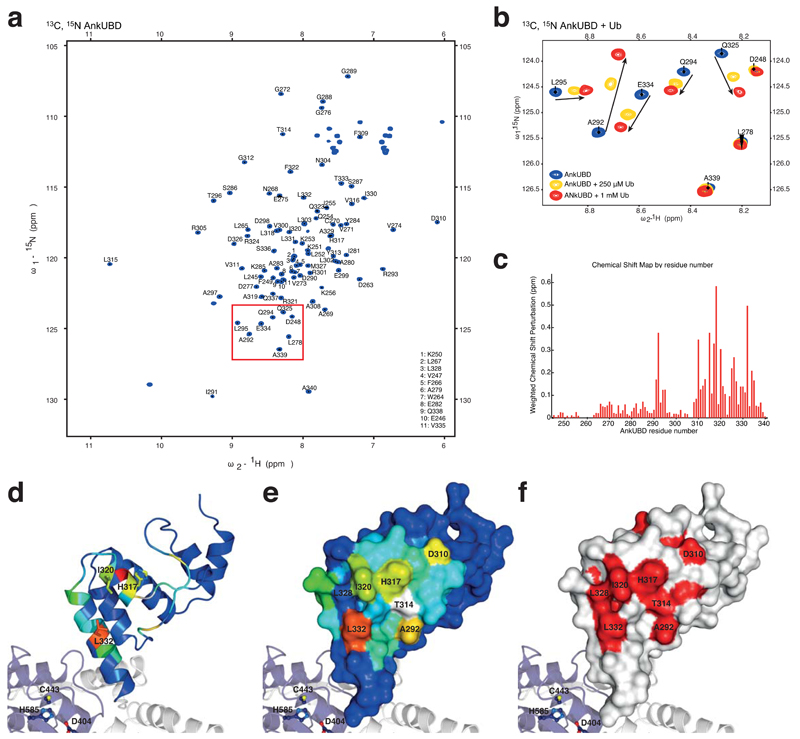
To understand whether and how the Ank domain interacts with Ub, nuclear magnetic resonance (NMR) studies were performed with a 13C, 15N-doubly labeled Ank domain. Full sequence specific backbone resonance assignments were obtained from triple resonance experiments (Fig. 3a, Supplementary Fig. 2a). Addition of 250 μM unlabeled Ub resulted in marked perturbations of a subset of Ank domain resonances in 1H,15N-HSQC spectra. A further increase of Ub concentration (1 mM) allowed unambiguous assignment of the perturbed resonances (Fig. 3b, Supplementary Fig. 2a). This confirmed that the Ank domain of TRABID interacts with Ub. To our knowledge, this is the first description of an Ank repeat as a Ub binding fold, which we shall refer to as the Ank Ub binding domain (AnkUBD).
Figure 3.
A conserved hydrophobic surface on AnkUBD binds Ub. (a) 1H,15N-HSQC spectrum of 13C, 15N-labelled TRABID Ank domain. (b) Close-up of a region boxed in a, showing resonances of the doubly labeled Ank domain (blue) and their shifts upon addition of 250 μM (yellow) or 1 mM (red) unlabeled Ub. Arrows indicate the shift of individual resonances. (c) Weighted chemical shift perturbation map of the AnkUBD binding to Ub. (d) AnkUBD residues are colored according to the degree of perturbation from blue (unperturbed) to red (strongly perturbed) and key residues are shown in stick representation. (e) The AnkUBD surface is shown colored as in d and key residues are labeled. (f) Invariant residues derived from a species sequence alignment (Supplementary Fig. 2b) are shown in red on a white AnkUBD surface.
The resulting chemical shift map showed that the entire second Ank repeat (aa 313-340) and in addition a small loop (aa 291-295) showed strong perturbations upon addition of Ub (Fig. 3c). The four residues that show the strongest perturbation, Ala292, His317, Ile320 and Leu332 form a hydrophobic patch on the AnkUBD (Fig. 3d, e). Furthermore, Thr314 and Leu332 showed exchange broadening upon addition of Ub, indicating that they may also be involved in Ub interactions (Supplementary Fig. 2a).
The AnkUBD extension of the TRABID OTU domain is an evolutionarily conserved feature. A sequence alignment derived from all TRABID sequences annotated in the Ensembl database (www.ensembl.org) reveals that the Ub-interacting residues on the AnkUBD are evolutionarily invariant amongst TRABID orthologues, indicating their functional importance (Fig. 3f, Supplementary Fig. 2b).
AnkUBD interacts with Ub hydrophobic patch
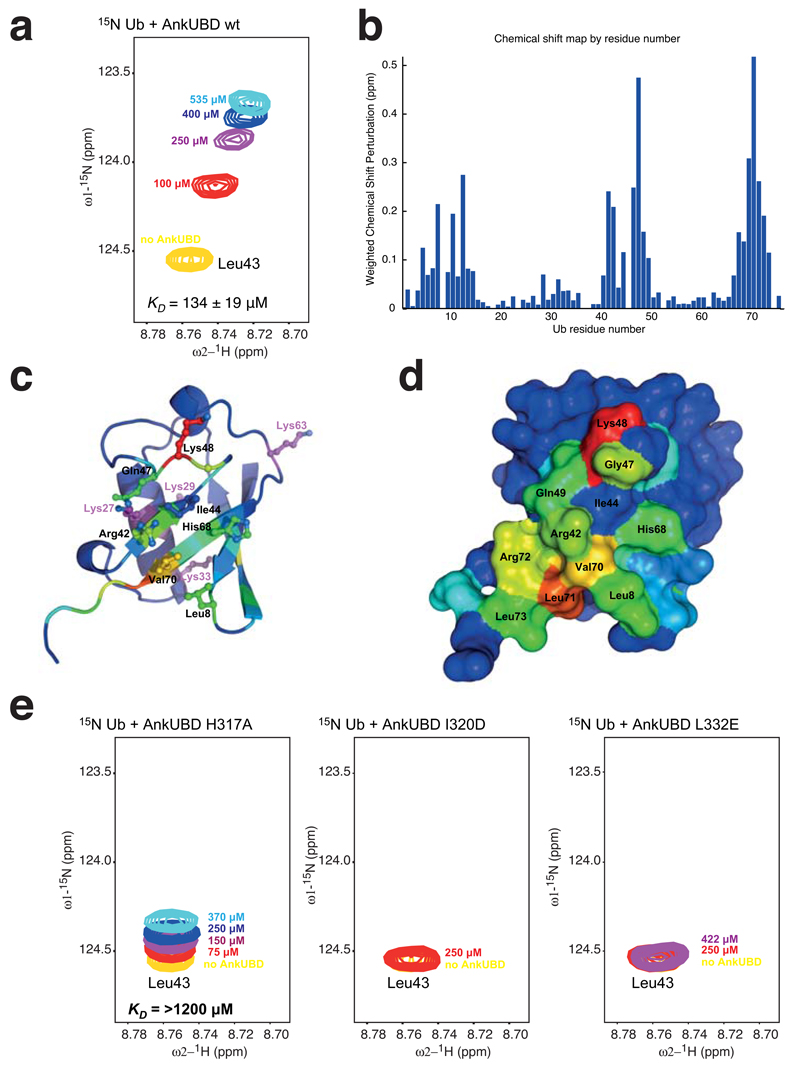
We next confirmed the interaction of the isolated AnkUBD with 15N-labelled Ub using chemical shift analysis of 1H,15N-HSQC spectra. Ub resonances that are perturbed upon addition of increasing AnkUBD concentrations (from 100-535 μM, Fig. 4a) were found in three regions of Ub, surrounding Leu8, Ile44 and Val70, which constitute the most common Ub binding site for protein interactions, the Ile44 hydrophobic patch (Fig. 4b-d). NMR-based affinity measurements indicated a KD of 134 ± 19 µM for the AnkUBD-Ub interaction (Fig. 4a), which is comparable to other UBD-Ub interactions 29. Hence, the hydrophobic surface of the AnkUBD interacts with the hydrophobic Ile44 patch of Ub.
Figure 4.
AnkUBD binds the Ub hydrophobic patch. Ub binding to the AnkUBD was confirmed by NMR shift mapping experiments, for which spectra of 15N Ub were recorded in absence and presence of AnkUBD (Supplementary Fig. 3). (a) Perturbation of a selected resonance (that of Ub Leu43, yellow) by titration of increasing concentrations of AnkUBD (colored from red to cyan) is shown as an example. The complete spectra can be found in Supplementary Fig. 3. (b) The resulting weighted chemical shift perturbation map reveals a familiar pattern seen when proteins bind to the Ub hydrophobic patch. (c) Ub residues are colored according to the degree of perturbation from blue (unperturbed) to red (strongly perturbed) and key residues are shown in stick representation. (d) The Ub surface is shown colored as in d and key residues are labeled. (e) Titration experiments with indicated concentrations of AnkUBD mutants H317A (left), I320D (middle), and L332E (right) were performed. The same resonance as in a is shown. Mutant L332E did not perturb any resonances, while H317A and I320D perturbed few resonances to a weaker degree (Supplementary Fig. 3).
To probe the interaction between AnkUBD and Ub further, three AnkUBD mutants, His317 to Ala (H317A), Ile320 to Asp (I320D) and Leu332 to Glu (L332E), were analyzed for their ability to interact with 15N-labeled Ub. The correct folding of all mutants was confirmed by NMR and circular dichroism (not shown). No chemical shift perturbation in 15N-labeled Ub was observed with AnkUBD L332E, while fewer and greatly attenuated shift perturbations were detected with H317A and I320D AnkUBD mutants, indicating some residual Ub binding (Fig. 4e, Supplementary Fig. 3). KD measurements confirmed this and indicated an at least 10x weaker affinity (>1200 µM) for the AnkUBD H317A mutant (no KD values could be determined for the I320D or L332E). This mutagenesis analysis therefore confirmed the presence of a single Ub binding surface on the AnkUBD of TRABID.
AnkUBD contributes to linkage-specificity of TRABID
The presence of the AnkUBD as a Ub binding fold in close proximity to the catalytic site suggested that it may serve as an enzymatic S1' Ub binding site, directly affecting TRABID’s DUB efficiency. Moreover, the S1' site may impact on TRABID specificity, as it may preferentially present a subset of Ub Lys residues to the catalytic site.
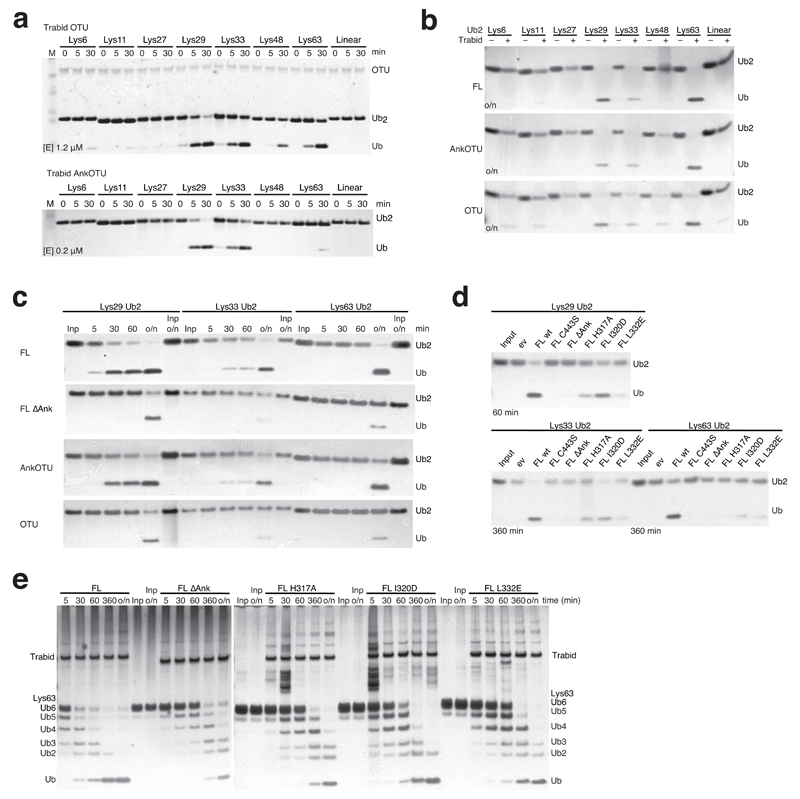
Indeed, the crystallized fragment including AnkUBD and OTU domain (AnkOTU) displayed higher DUB activity compared to the isolated OTU domain (OTU, aa 339-697), which was less active at similar concentration (Supplementary Fig. 4a) and was hence used at 5-fold higher concentration to allow detection of enzymatic activity (Fig. 5a). The OTU fragment hydrolyzed Lys29, Lys33 and Lys63 with similar efficiency, but in addition also Lys48-linkages and with low efficiency Lys6- and Lys11-linked diUb (Fig. 5a). This contrasted with the AnkOTU fragment that cleaved Lys29-, Lys33- and less efficiently Lys63-linked diUb (Fig. 5a).
Figure 5.
Analysis of TRABID DUB activity. Bacterial (a) and mammalian (b-e) TRABID variants were incubated with polyUb substrates for indicated times and visualized by silver staining. (a) Comparison of activity and specificity of the isolated OTU domain (above, [E] 1.2 µM) with TRABID AnkOTU (below, [E] 0.2 µM, reproduced from Fig. 1b to allow comparison). Input enzyme levels are shown in OTU panel. Also see Supplementary Fig. 4a. (b-e) FLAG-tagged TRABID variants were purified from HEK293 cells, and used in DUB assays. (b) Specificity of mammalian full length (FL), AnkOTU and OTU TRABID against the diUb panel after over night (o/n) incubation. (c) Time course analysis of mammalian TRABID variants against its substrate linkages. FL ∆Ank, FL lacking AnkUBD. (d) Activity of FL TRABID with point mutations in the AnkUBD against its preferred diUb substrates. FL C443S, catalytic mutant. (e) Time course activity of TRABID variants against Lys63-linked hexaUb.
This result was reflected in a second analysis, in which TRABID variants were purified by virtue of an N-terminal 3xFLAG-tag from HEK293 cells and used in endpoint DUB assays. Full-length (FL) TRABID, like AnkOTU, only cleaved Lys29-, Lys33- and Lys63-linked diUb over night, while an OTU construct was more promiscuous, and cleaved Lys48-linked diUb (as well as weakly Lys6- and Lys11-linkages) (Fig. 5b). The specificity of HEK293 expressed TRABID variants for Lys29/Lys33-linkages was confirmed by time course analysis, in which these linkages were cleaved within 60 min by FL and AnkOTU TRABID, while Lys63-linked diUb was not cleaved during the first hour but only after over night incubation (Fig. 5c, panel 1 and 3). Whilst this is not a quantitative analysis, we note slightly higher activity of TRABID against Lys29- compared to Lys33-linkages in our qualitative time course assays (Fig. 5a, c). Together, these data show that the AnkUBD restricts activity of the OTU domain, which cleaves (at least) four linkage types, to make TRABID most efficient for Lys29 and Lys33 linkages.
Removal of the AnkUBD from either construct (i.e. FL ΔAnk, and OTU), also resulted in less active protein without noticeable activity in the first 60 min (Fig. 5c, panel 2 and 4), confirming the role of the AnkUBD in TRABID efficiency.
We next analyzed HEK293 expressed TRABID variants with point mutations in the AnkUBD that affect Ub binding (Fig. 3). Mutation of H317A, L332E and I320D in the AnkUBD reduced TRABID activity against diUb substrates, either in context of the FL protein (Fig. 5d) or in context of bacterially produced AnkOTU (Supplementary Fig. 4b). The observed reduced activity of TRABID point mutants (in particular H317A, L332E) was similar to removal of the AnkUBD from either construct.
While Lys29 and Lys33 linkages are available only as diUb, Lys63-linked Ub chains are available as longer polymers, and we next studied activity of TRABID variants against Lys63-linked hexaUb (Fig. 5e). FL TRABID cleaved hexaUb efficiently, hydrolyzing most input material within 60 min. Consistent with diUb assays, removal or mutation of the AnkUBD decreased activity against K63-linked hexaUb (Fig. 5e), suggesting that the AnkUBD is essential also for hydrolysis of longer polyUb chains.
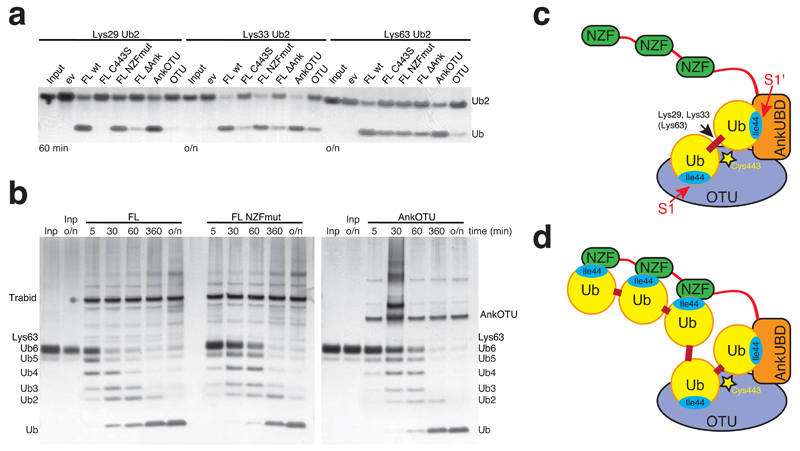
NZF domains contribute to hydrolysis of longer Ub chains
TRABID contains three N-terminal NZF domains, which may also impact on TRABID activity and specificity. A TRABID variant with mutations in all three NZF domains (changing the Thr-Phe motif with Ala-Ala 35, FL NZFmut) was as active against diUb as FL or AnkOTU (Fig. 6a). However FL NZFmut TRABID is less active compared to FL TRABID when longer Lys63 chains were used as a substrate (Fig. 6b), suggesting that the NZF domains contribute to cleaving longer Ub chains by providing additional binding sites. The combined S1-S1' site in TRABID may constitute the highest affinity diUb binding module (Fig. 6c), since otherwise the NZF domains would compete for binding to the hydrophobic patches in diUb and render TRABID less active towards diUb substrates, which is not the case (Fig. 5c). The NZF domains however provide increased affinity for longer TRABID substrates that can bind simultaneously to AnkOTU and NZF domains (Fig. 6d).
Figure 6.
Role of the NZF domains in cleaving longer Ub chains. Mammalian TRABID variants were incubated with polyUb substrates for indicated times and visualized by silver staining. (a) Activity of TRABID variants against Lys29, Lys33 and Lys63-linked diUb at indicated time point. FL C443S, catalytic mutant; FL NZFmut, FL with mutations in all three NZF domains; FL ∆Ank, FL lacking AnkUBD; AnkOTU, crystallized fragment; OTU, OTU domain. (b) Time course analysis of mammalian TRABID FL, FL NZFmut and AnkOTU activity towards Lys63-linked hexaUb. (c) Model for the role of the AnkUBD as an S1' Ub binding site in TRABID. (d) Model for the additional contribution of the NZF domains in cleaving longer polyUb chains.
Overall, our in vitro analysis showed that i) the TRABID OTU domain cleaved four out of eight linkage types, ii) addition of the AnkUBD increases activity of TRABID for Lys29 and Lys33 linkages, effectively making it specific for these linkages (Fig. 6c) and iii) the N-terminal NZF domain does not affect TRABID specificity against diUb, but may increase efficiency of the enzyme against longer polymers (Fig. 6d). This confirmed the importance of the AnkUBD as a key determinant of TRABID DUB efficiency and specificity in vitro.
Inactive TRABID forms puncta enriched in atypical polyUb
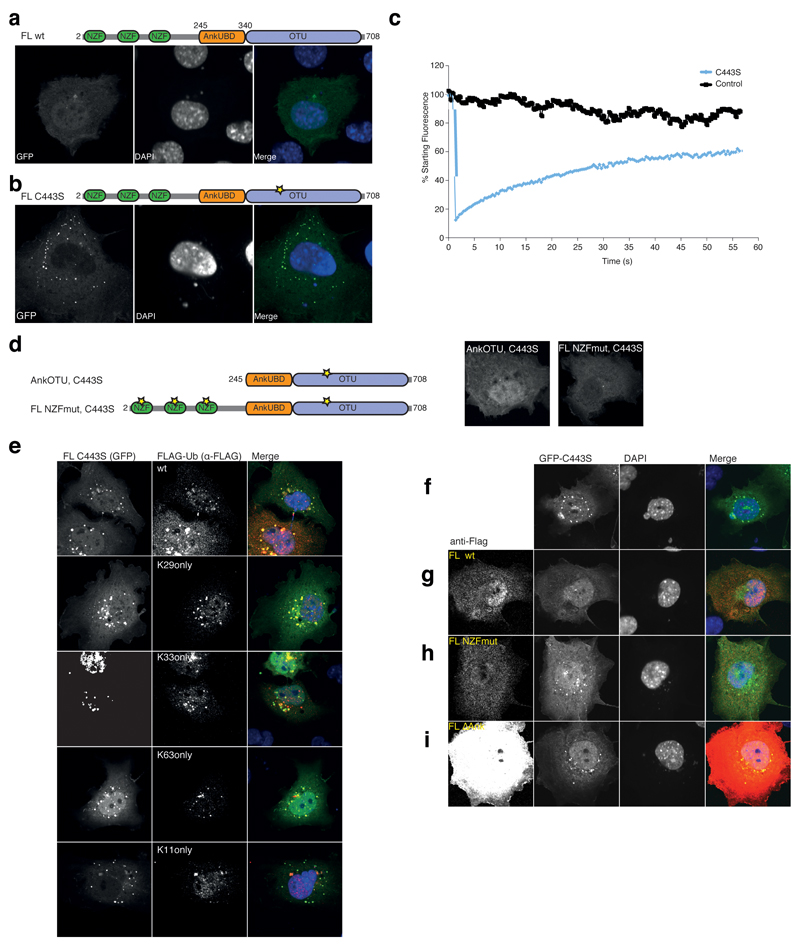
We next set out to provide evidence for the mechanistic models shown in Fig. 6 in a physiological setting, for which we had to establish DUB assays in vivo. It had been reported that HA-tagged catalytically inactive FL TRABID C443S (i.e. mutation of the catalytic Cys443 to Ser) forms distinct cytoplasmic puncta upon overexpression in various human cell lines, while wild-type (wt) TRABID is distributed diffusely throughout the cytoplasm and nucleus 35. An equivalent localization was observed with green fluorescent protein (GFP)-tagged TRABID FL wt and FL C443S (Fig. 7a, b). The latter was used for fluorescence recovery after photobleaching (FRAP) experiments, revealing that the FL C443S puncta are highly dynamic (i.e. 50% of puncta fluorescence recovers within 30 s) and in rapid equilibrium exchange with diffuse protein (Fig. 7c). Thus, inactive TRABID forms dynamic protein assemblies rather than stable aggregates. An equivalent catalytically inactive mutant of the closely related DUB Cezanne does not form puncta upon overexpression (Supplementary Fig. 5a), suggesting that the propensity to form dynamic protein assemblies is not a general property of catalytically inactive OTU family proteins.
Figure 7.
In vivo DUB assay NZF and AnkUBD are essential for TRABID puncta. (a) Localization studies with GFP-TRABID in COS-7 cells 18 h after transfection (left). Nuclei are stained using DAPI (middle). The right image is a merge of the channels. The domain structure of TRABID is shown above. (b) A GFP-tagged full-length TRABID catalytic mutant (C443S, a yellow star in the domain representation indicates the mutation) adopts a punctate localization in COS-7 (shown) and other cell types 35. (c) Fluorescence recovery after photobleaching (FRAP) experiments on a control (black) or puncta containing volume (C443S, blue). Fluorescent recovery is recorded over time. (d) Localization studies of TRABID GFP-AnkOTU C443S and GFP-FL NZFmut C443S as in a. The domain structure is shown (left), and the GFP fluorescence of either construct (right) shows that no puncta are formed. (e) Puncta-forming GFP-C443S (left image) was co-expressed with FLAG-Ub or single-Lys (Konly) Ub mutants (middle image). The merged image is shown to the right. Further Ub mutants are shown in Supplementary Fig. 6a. (f-i) Dissolving TRABID assemblies requires the AnkUBD. (f) Puncta-forming GFP-tagged full-length TRABID C443S (left) was expressed in COS-7 cells and TRABID assemblies were visualized (left). Nuclei are stained using DAPI (middle). The merged image is shown to the right. (g-i) As in f, but in addition, FLAG-tagged TRABID constructs FL wt (g), FL NZFmut (h) or FL ∆Ank (i) were co-expressed (far left) and the presence of GFP puncta was assessed. Additional data is shown in Supplementary Fig. 6b.
Ubiquitin binding via the N-terminal NZF domains of TRABID is crucial for puncta formation (since AnkOTU C443S or FL NZFmut C443S do not form puncta 35, Fig. 7d), indicating that FL C443S puncta contain Ub. Indeed, co-expression of GFP-FL C443S with FLAG-Ub revealed that the FL C443S puncta were positive for FLAG-Ub (Fig. 7e). We then tested the panel of seven FLAG-Ub ‘Konly’ mutants (in which six out of seven Lys residues are mutated to R) for co-localization with TRABID puncta. We found that the FL C443S puncta were positive for K29only, K33only, K27only and K63only Ub mutants, while there was no detectable co-localization with K6only, K11only or K48only Ub mutants (Fig. 7e and Supplementary Fig. 6a). This indicated that protein assemblies comprising inactive TRABID contain the defined set of atypical Ub chains that TRABID preferentially hydrolyzes (with the exception of Lys27 linkages, see above).
All Ub mutants were expressed at similar levels and did not form punctate patterns when transfected alone, in absence or presence of the proteasome inhibitor MG132 (Supplementary Fig. 5b). Interestingly, the number or size of TRABID puncta was not affected when cells were pre-treated with MG132 (Supplementary Fig. 5c). It is possible that the puncta reflect TRABID self-assembly mediated by NZF domain binding to Ub chains on TRABID itself. Co-immunoprecipitation of Flag-tagged TRABID C443S with HA-tagged Ub after stringent washing under denaturing conditions revealed that TRABID is indeed ubiquitinated (data not shown). However, the puncta may also contain TRABID substrates bearing Lys29-, Lys33- and or Lys63-linked Ub chains.
In vivo DUB assay based on TRABID puncta
Given that atypical Ub chains are enriched in TRABID puncta, we explored whether we could exploit these as substrates for in vivo DUB assays, to validate the mechanistic models derived from our structural analysis (Fig. 7f-i and Supplementary Fig. 6b). Co-expression of puncta forming GFP-FL C443S with FLAG-tagged TRABID FL results in disappearance of GFP puncta, and instead produces a diffuse GFP fluorescence throughout the cell (Fig. 7g); the levels of GFP-FL C443S do not change under these conditions, as judged by Western blot analysis; data not shown). In contrast, co-expression with FL NZFmut (Fig. 7h) or with FL C443S (not shown) did not alter the punctate pattern. AnkOTU resulted in complete loss of puncta formation upon high but not moderate expression (Supplementary Fig. 6b) Similarly, highly overexpressed TRABID OTU domain did not result in loss of TRABID puncta (Supplementary Fig. 6b).
Importantly, the punctate pattern was also unchanged upon co-expression of FL ΔAnk TRABID, even after high levels of overexpression (Fig. 7i). This construct still comprises functional NZF domains, and localizes to FL C443S puncta (Supplementary Fig. 5d). This highlights the important functional role of the AnkUBD in TRABID activity and efficiency, which is essential to antagonize C443S puncta in vivo.
Discussion
Availability of all Ub chain types now allows a comprehensive linkage-specificity analysis of DUBs, and in the case of TRABID reveals its dual specificity for Lys29- and Lys33-linked Ub chains, two previously unstudied atypical Ub chain types. We achieved a molecular understanding of this unique specificity profile, and identified a novel UBD based on an Ank repeat fold, which serves as a S1' Ub binding site to position a subset of Ub chains across the active site of the OTU domain.
The TRABID OTU domain is similar in sequence and structure to that of A20, however, both domains display distinct specificities. While A20 cleaves preferentially Lys48-linkages (11, TETM and DK, unpublished), the TRABID OTU domain cleaves Lys29, Lys33, Lys63 and Lys48 chains, but not the remaining chain types. Other OTU family members have yet again distinct specificity profiles 21,26. This indicates that OTU domain folds comprise intrinsic specificity, and the molecular basis for this is currently unknown. Linkage specificity in DUBs is so far only understood for the Lys63-specific JAMM-family DUB AMSH-LP, which forms specific interactions with Lys63-flanking sequences 39. However, the sequences flanking Lys29, Lys33, Lys48 and Lys63 is different 15, and hence this is unlikely to explain TRABID OTU domain specificity.
A novel Ub binding fold, the AnkUBD, further restricts the linkage specificity of the TRABID OTU domain and makes TRABID most efficient for Lys29- and Lys33-linkages. Our data are consistent with the AnkUBD serving as an enzymatic S1' site that positions proximal Ub moieties such that Lys29 and Lys33 are juxtaposed to the catalytic triad (Fig. 6c). The AnkUBD binds directly to Lys48 of the proximal Ub, suggesting that a Lys48-linked chain cannot utilize the AnkUBD. This is consistent with in vitro data showing that residual Lys48 cleavage is independent of the AnkUBD. Therefore, the AnkUBD adjacent to the catalytic domain explains TRABID’s DUB specificity profile.
It is not clear how the AnkUBD orients a proximal Ub towards the active site. Docking of Ub onto the AnkUBD using HADDOCK webserver 40 indicated several potential binding modes (data not shown), however, all models indicated that the AnkUBD has to shift and rotate for any diUb to bind across the active site. The flexible linker between AnkUBD and OTU domain likely permits this required plasticity, but future crystallographic studies of TRABID-diUb complexes will be required to confirm this.
Almost 20,000 Ank repeats have been annotated in >3500 human proteins, making this domain one of the most common protein folds 38. Ank repeats serve as protein binding modules, and our work adds Ub to the list of Ank-interacting proteins. Ub binding by the AnkUBD is mediated by both helices of the C-terminal Ank repeat, via a hydrophobic surface mediated by residues that are conserved amongst Ank repeats. Indeed, these residues are reminiscent of the TPLH motif that is characteristic for internal Ank repeats 41, and it is possible that other Ank repeat-containing proteins may bind Ub in a similar fashion to TRABID. However, terminal Ank repeats suited to bind Ub are not easily identified by sequence analysis alone.
While the AnkUBD is essential for TRABID efficiency and specificity, our analysis has also indicated a functional contribution of the NZF domains (Fig. 6). However, their relative position with respect to the catalytic site is unclear. A linker of ~50 non-conserved and flexible aa separates the NZF domains and the AnkUBD, and individual NZF domains are themselves connected by non-conserved, flexible linkers. Hence, the NZF module could provide additional Ub binding sites towards the distal (S2, S3, S4) or proximal (S2', S3', S4') end (Fig. 6d indicates distal Ub binding sites). The number of Ub binding sites within the three NZF domains is also not clear, as they may comprise two Ub binding interfaces each 42,43. Alternatively, the NZF domains may target TRABID to its polyubiquitinated substrates, increasing DUB efficiency. This is suggested by our findings that the NZF domains are essential for recognition of TRABID puncta (Supplementary Fig. 5d).
Additional UBDs are found in many DUBs, and seven out of the 14 human OTU DUBs contain known UBD folds 15. A Ub interacting motif (UIM) in DUBA was shown to be functionally important for its role in interferon signaling 31, and the A20 ZnF domains bind Ub molecules via several interfaces 44. The USP enzyme USP5/IsoT comprises three UBDs that constitute S1', S2 and S3 sites respectively 45. However, for most DUBs it is not known whether their additional UBDs constitute enzymatic Ub binding sites (S1', S2 etc) or whether they serve as targeting domains. Our work highlights the potential of UBDs to contribute additional levels of linkage-specificity to DUBs, and presumably also to other aspecific enzymes of the Ub cascade, such as E2s and E3s.
Until now, there are no firmly established cellular functions for the atypical Ub chain types cleaved by TRABID. Recent proteomic data in unstimulated HEK293 cells suggests a relatively high abundance of Lys63- and Lys29-linkages, while Lys33-linkages are less abundant 4. A recent study indicated that β-catenin itself is modified with Lys29-linkages by the HECT domain ligase EDD (Lys33 chains were not analyzed). However, whether this leads to β-catenin stabilization 6 or destabilization 46 is not clear. It will be interesting to identify the functionally relevant TRABID substrates within the Wnt pathway that carry Ub chains linked through Lys29 or Lys33, and to determine their functional relevance in signal transduction. This may shed light on the cellular function of these unusual and rare atypical Ub linkages.
Online Methods
Purification and structure determination of TRABID AnkOTU
TRABID AnkOTU was expressed as GST-fusion protein in E. coli and purified by affinity chromatography. After removal of the GST-tag by PreScission protease, anion exchange chromatography and gel filtration produced homogeneous protein that was crystallized at 3.5 mg ml-1. AnkOTU crystals grew from 150 mM NaCl, 100 mM NaOAc, 5 mM MgCl2, 50 mM MES [pH 5.9]. Crystals were soaked in mother liquor containing 27.5 % (v/v) ethylene glycol prior to freezing in liquid nitrogen. To obtain phase information, crystals were soaked in 1 mM KAu(CN)2 for 1 h prior to cryo-protection. Diffraction data of the AnkOTU crystals were collected at ESRF beamline ID23-2 to 2.23 Å (native) and 3.0 Å resolution at the peak-wavelength for Au. Phases were obtained by SIRAS, from an initial set of sites obtained with the SHELX/hkl2map suite 48. Site refinement was performed in SHARP 49, and subsequent density modification resulted in a high-quality map, which was interpreted by WarpNTrace 50 and manually rebuilt in Coot 51. Refinement was performed using PHENIX 52, including simulated annealing and TLS B-factor refinement, resulting in the final statistics reported in Table 1.
NMR analysis
13C, 15N-labeled TRABID AnkUBD (aa 245-339) was expressed from a pOPIN-K vector in Rosetta2 pLacI cells. A 100 ml overnight culture grown in LB medium was pelleted and resuspended in modified K-MOPS minimal media 53, lacking nitrogen and carbon sources, and used to inoculate 3 L modified K-MOPS media supplemented with 13C glucose/15N ammonium chloride. Protein expression was induced after 16 h at 30˚C with 0.4 mM IPTG, and cells were harvested after a further 4 h. The protein was purified as for crystallography (see above) and concentrated to 10 mg ml-1. Unlabeled Ub (from bovine red blood cells, Sigma-Aldrich) was reconstituted in 50 mM Tris [pH 7.6] at 5 mM. 15N-labeled Ub was a kindly provided by Dr. Mark Allen (MRC LMB). Prior to data acquisition, samples were dialyzed against phosphate buffered saline (125 mM Tris [pH 7.2], 16.5 mM Na2HPO4, 8.5 mM NaH2PO4 × 2H2O) in 3 kDa cut-off Slide-A-Lyzer dialysis cassettes (Thermo Scientific). NMR experiments were acquired on Bruker DRX600MHz and AV2+ 700MHz spectrometers equipped with cryogenic triple resonance TCI probes and at a sample temperature of 298 K. All data were processed in Topspin 2.1 (Bruker, Karlsruhe) and analyzed in Sparky (Goddard & Kneller, UCSF). Standard triple resonance experiments (HNCACB, CBCA(CO)NH, HN(CA)CO and HNCO) were used to assign backbone resonances of the 13C, 15N double labeled TRABID AnkUBD, using a sample concentration of 180 μM. All observed correlations in the HSQC were assigned unambiguously, corresponding to 88 out of 95 non-Pro residues in the construct. Titration experiments and KD estimation are detailed in Supplementary Methods online.
Production of full-length TRABID from mammalian cells
500 ng of plasmid was transfected using Lipofectamine 2000 (Invitrogen) in each well of a 6-well plate containing HEK293 cells. 48 h after transfection, cells from the six wells were combined and lysed in 1 ml of lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, Complete protease inhibitor (Roche)), sonicated and centrifuged at 15,000 × g for 20 min. The entire lysate was incubated with 80 μL of 50% (v/v) slurry of ANTI-FLAG® M2 affinity gel (Sigma) for 2 h at 4°C. Beads were then washed five-times in 50 mM Tris (pH 7.4), 500 mM NaCl, 0.5 mM EDTA, 1% (v/v) Triton X-100, 1 mM DTT, complete protease inhibitor; five-times in 50 mM Tris (pH 7.4), 150 mM NaCl, 0.5 mM EDTA, 1% (v/v) Triton X-100, 1 mM DTT, complete protease inhibitor; and twice in 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM DTT. Immunoprecipitated TRABID was finally eluted in two consecutive steps of 100 µl elution buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM DTT) with a final concentration of 100 μg ml-1 3xFLAG® peptide (Sigma). The expression level of all TRABID variants was similar (see Fig. 5e, 6b).
Deubiquitinase assays
Deubiquitination assays using purified bacterial enzymes were performed as reported before 26. DUBs were diluted to 2 × final concentration in 150 mM NaCl, 25 mM Tris (pH 7.5) and 10 mM DTT and activated at 23 °C for 10 min. Subsequently, 10 µl of diluted enzyme were mixed with 1-2 µg diUb and 2 µl of 10 × DUB buffer (500 mM NaCl, 500 mM Tris [pH 7.5] and 50 mM DTT) in a 20 µl reaction. For DUB assays with TRABID produced in mammalian cells, 6 μl of eluted enzyme was used in a 12 μl reaction in DUB buffer containing 200 ng di- or hexaUb. Reactions were stopped by addition of 4 μl LDS sample buffer (containing 100 mM DTT) after 5, 30, 60, 360 min or after o/n incubation at 37 °C. Ub cleavage was detected by silver staining using the Silver Stain Plus kit (BioRad).
Confocal image analysis and in vivo DUB assays
GFP-TRABID constructs (500 ng) were transfected in COS-7 cells. After 18 h the cells were rinsed in PBS and fixed using 1% (w/v) paraformaldehyde, permeabilized in 0.1% (v/v) Triton X-100 and mounted using Vectashield mounting medium with DAPI (Vector Laboratories Inc).
For in vivo DUB assays, cells were co-transfected with 500 ng of GFP-TRABID C443S and 500 ng of various 3xFLAG-TRABID constructs, permeabilized, blocked in PBS supplemented with 3% (w/v) BSA, stained using a mouse monoclonal anti-FLAG® M2 antibody (Sigma) (1:500 dilution) followed by Cy3® goat anti-mouse IgG (Invitrogen) (1:1000 dilution). Slides were finally washed and mounted as described above. Images were obtained using a Zeiss LSM510 (Jena, Germany) confocal microscope. Double-label images of Cy3/GFP conjugates were detected with standard filter-sets and laser lines.
Supplementary Material
Acknowledgements
We would like to thank Roger Williams, Mark Allen, Mark Fiedler, Nicolas Soler, , Stefan Freund, Chris Johnson, Stephen McLaughlin, Yogesh Kulathu, Yu Ye, Anja Bremm, Martin Busch (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK), Dharjath S. Hameed (Netherlands Cancer Institute, Amsterdam, The Netherlands) and Kay Hofmann (Miltenyi Biotec) for reagents, help with experiments and discussions. Crystallographic data was collected at ESRF beamline ID23-2. JDFL was supported by an Association for International Cancer Research grant (no. 07-0040 to MB). DK is an EMBO Young Investigator. This work was supported by the MRC (MC_US_A024_0059, MC_US_A024_0056, MC_US_A024_0051 and MC_US_A021_0013).
Footnotes
Data deposition
Coordinates and structure factors have been submitted to the protein data bank, accession number 3zrh.
Conflict of interest statement
HO and FE are co-founders of UbiQ Bio B.V.
Author contributions
DK, MB and JDFL designed research. JDFL, DK, JM, TETM, TR and MA performed experiments. FE, HO, SV and JWC contributed reagents. DK wrote the manuscript with help from all authors.
References
- 1.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–53. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dammer EB, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–65. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–9. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates {beta}-catenin. Mol Biol Cell. 2011;22:399–411. doi: 10.1091/mbc.E10-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chastagner P, Israël A, Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006;7:1147–53. doi: 10.1038/sj.embor.7400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Hakim AK, et al. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411:249–60. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–56. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 10.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–44. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 11.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–7. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 12.El Oualid F, et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl. 2010;49:10149–53. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Total chemical synthesis of di-ubiquitin chains. Angew Chem Int Ed Engl. 2010;49:9126–31. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 14.Virdee S, et al. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc. 2011;133:10708–11. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 16.Nijman SMB, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCullough J, Clague MJ, Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper EM, et al. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–31. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelmann MJ, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–90. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 21.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanao M, et al. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–8. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 24.Lin SC, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–40. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messick TE, et al. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–49. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci U S A. 2011;108:2228–33. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capodagli GC, et al. Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin. J Virol. 2011;85:3621–30. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James TW, et al. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc Natl Acad Sci U S A. 2011;108:2222–7. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains — from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–41. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 31.Kayagaki N, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–32. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 32.Li S, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–7. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiyama K, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–66. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst R, Mueller B, Ploegh HL, Schlieker C. The Otubain YOD1 Is a Deubiquitinating Enzyme that Associates with p97 to Facilitate Protein Dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–42. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans PC, et al. Isolation and characterization of two novel A20-like proteins. Biochem J. 2001;357:617–23. doi: 10.1042/0264-6021:3570617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–78. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 39.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 40.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–97. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 41.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–48. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28:3903–9. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–30. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 44.Bosanac I, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–57. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–92. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohshima R, et al. Putative tumor suppressor EDD interacts with and up-regulates APC. Genes Cells. 2007;12:1339–45. doi: 10.1111/j.1365-2443.2007.01138.x. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka N, et al. Structural basis for recognition of 2',5'-linked oligoadenylates by human ribonuclease L. EMBO J. 2004;23:3929–38. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pape T, Schneider TR. Hkl2map: a graphical user interface for macromolecular phasing with shelx programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 49.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–30. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 50.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 53.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–47. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.