Abstract
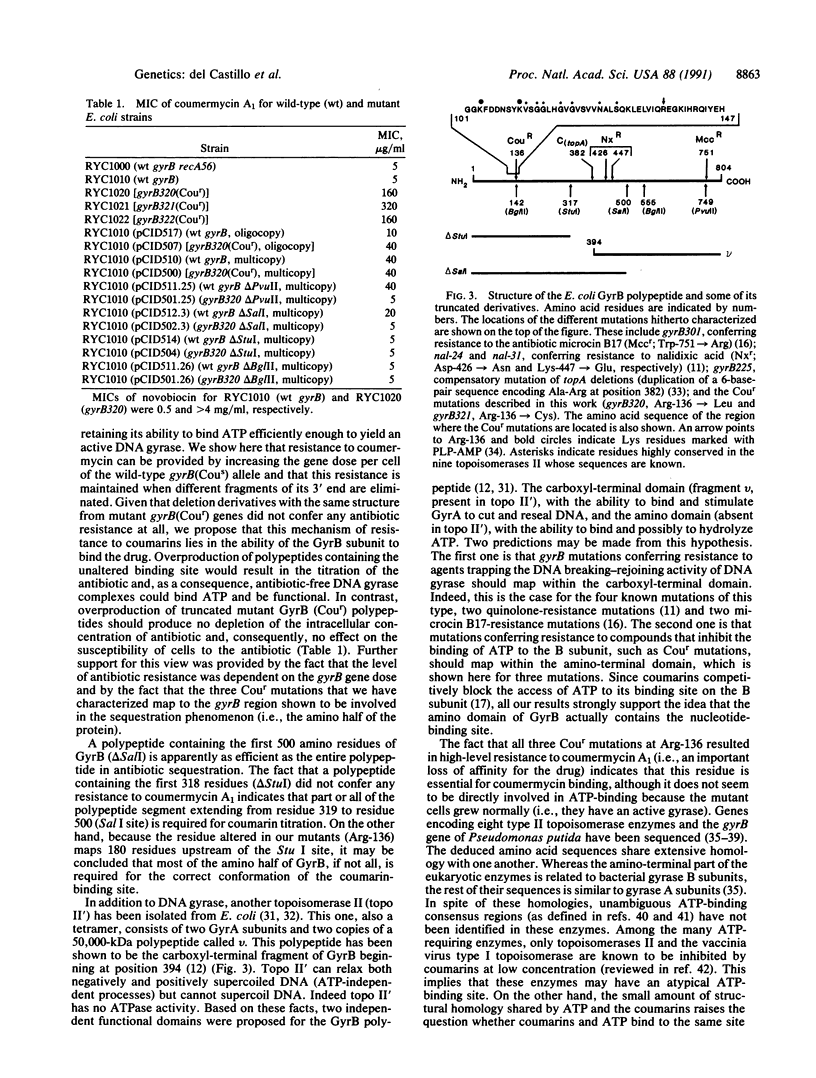
Bacterial DNA gyrases are type II topoisomerases made up of two A subunits and two B subunits. Coumarins are carbohydrate-containing antibiotics that inhibit topoisomerases II by competing with ATP for binding to the enzymes. High resistance to coumarins is produced in bacterial species by mutations in gyrB, the gene encoding subunit B. We have found an unusual mechanism of resistance to coumarins in Escherichia coli. This mechanism is exhibited by cells containing the wild-type gyrB, or its 5' half, in high copy number. Since homologous mutant gyrB (coumermycin resistant) truncated genes did not confer drug resistance at all under the same conditions, we propose that this mechanism of resistance is due to drug sequestration by the overproduced wild-type GyrB polypeptides. A corollary of this is that the amino half of GyrB is required and sufficient to fashion the ATP-binding domain of DNA gyrase, a conclusion that was further supported by mapping three independent coumarin-resistant mutations at Arg-136 of GyrB. Just upstream of this residue there is a glycine-rich sequence highly conserved in all topoisomerases II, which seems to be a good candidate for the actual ATP-binding site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Mizuuchi M., Robinson E. A., Appella E., O'Dea M. H., Gellert M., Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987 Jan 26;15(2):771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Peebles C. L., Cozzarelli N. R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. T., Goff S. A., Webster T., Smith T., Goldberg A. L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988 Aug 25;263(24):11718–11728. [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Dombroski A. J., LaDine J. R., Cross R. L., Platt T. The ATP binding site on rho protein. Affinity labeling of Lys181 by pyridoxal 5'-diphospho-5'-adenosine. J Biol Chem. 1988 Dec 15;263(35):18810–18815. [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Franco R. J. Inhibitors of DNA topoisomerases. Biochemistry. 1988 Apr 5;27(7):2253–2259. doi: 10.1021/bi00407a001. [DOI] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., O'Dea M. H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M. L., Dyall-Smith M. L. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J Bacteriol. 1991 Jan;173(2):642–648. doi: 10.1128/jb.173.2.642-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Wang J. C. Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an alpha 2 beta 2 protein core. Biochemistry. 1980 Nov 11;19(23):5229–5234. doi: 10.1021/bi00564a012. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J Biol Chem. 1984 Dec 10;259(23):14472–14480. [PubMed] [Google Scholar]
- McEachern F., Fisher L. M. Regulation of DNA supercoiling in Escherichia coli: genetic basis of a compensatory mutation in DNA gyrase. FEBS Lett. 1989 Aug 14;253(1-2):67–70. doi: 10.1016/0014-5793(89)80931-7. [DOI] [PubMed] [Google Scholar]
- Mildvan A. S., Fry D. C. NMR studies of the mechanism of enzyme action. Adv Enzymol Relat Areas Mol Biol. 1987;59:241–313. doi: 10.1002/9780470123058.ch6. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., O'Dea M. H., Gellert M. Cloning and simplified purification of Escherichia coli DNA gyrase A and B proteins. J Biol Chem. 1984 Jul 25;259(14):9199–9201. [PubMed] [Google Scholar]
- Ohmi N., Hoshino M., Tagaya M., Fukui T., Kawakita M., Hattori S. Affinity labeling of ras oncogene product p21 with guanosine diphospho- and triphosphopyridoxals. J Biol Chem. 1988 Oct 5;263(28):14261–14266. [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Parales R. E., Harwood C. S. Nucleotide sequence of the gyrB gene of Pseudomonas putida. Nucleic Acids Res. 1990 Oct 11;18(19):5880–5880. doi: 10.1093/nar/18.19.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J Biol Chem. 1989 Nov 25;264(33):19648–19653. [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Strauss P. R., Wang J. C. The TOP2 gene of Trypanosoma brucei: a single-copy gene that shares extensive homology with other TOP2 genes encoding eukaryotic DNA topoisomerase II. Mol Biochem Parasitol. 1990 Jan 1;38(1):141–150. doi: 10.1016/0166-6851(90)90214-7. [DOI] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya M., Noumi T., Nakano K., Futai M., Fukui T. Identification of alpha-subunit Lys201 and beta-subunit Lys155 at the ATP-binding sites in Escherichia coli F1-ATPase. FEBS Lett. 1988 Jun 20;233(2):347–351. doi: 10.1016/0014-5793(88)80457-5. [DOI] [PubMed] [Google Scholar]
- Tagaya M., Yagami T., Fukui T. Affinity labeling of adenylate kinase with adenosine diphosphopyridoxal. Presence of Lys21 in the ATP-binding site. J Biol Chem. 1987 Jun 15;262(17):8257–8261. [PubMed] [Google Scholar]
- Tamura J. K., Gellert M. Characterization of the ATP binding site on Escherichia coli DNA gyrase. Affinity labeling of Lys-103 and Lys-110 of the B subunit by pyridoxal 5'-diphospho-5'-adenosine. J Biol Chem. 1990 Dec 5;265(34):21342–21349. [PubMed] [Google Scholar]
- Tsai-Pflugfelder M., Liu L. F., Liu A. A., Tewey K. M., Whang-Peng J., Knutsen T., Huebner K., Croce C. M., Wang J. C. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizán J. L., Hernández-Chico C., del Castillo I., Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991 Feb;10(2):467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Recent studies of DNA topoisomerases. Biochim Biophys Acta. 1987 Jun 6;909(1):1–9. doi: 10.1016/0167-4781(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Wyckoff E., Natalie D., Nolan J. M., Lee M., Hsieh T. Structure of the Drosophila DNA topoisomerase II gene. Nucleotide sequence and homology among topoisomerases II. J Mol Biol. 1989 Jan 5;205(1):1–13. doi: 10.1016/0022-2836(89)90361-6. [DOI] [PubMed] [Google Scholar]
- Yamagishi J., Yoshida H., Yamayoshi M., Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986 Sep;204(3):367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Kojima T., Yamagishi J., Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988 Jan;211(1):1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]