Abstract
In all multicellular organisms, the fundamental processes of cell proliferation and cell death are crucial for growth regulation during organogenesis. Strict regulation of cell death is important to maintain tissue homeostasis by affecting processes like regulation of cell number, and elimination of unwanted/unfit cells. The developing Drosophila eye is a versatile model to study patterning and growth, where complex signaling pathways regulate growth and cell survival. However, the molecular mechanisms underlying regulation of these processes is not fully understood. In a gain-of-function screen, we found that misexpression of cullin-4 (cul-4), an ubiquitin ligase, can rescue reduced eye mutant phenotypes. Previously, cul-4 has been shown to regulate chromatin remodeling, cell cycle and cell division. Genetic characterization of cul-4 in the developing eye revealed that loss-of-function of cul-4 exhibits a reduced eye phenotype. Analysis of twin-spots showed that in comparison with their wild-type counterparts, the cul-4 loss-of-function clones fail to survive. Here we show that cul-4 clones are eliminated by induction of cell death due to activation of caspases. Aberrant activation of signaling pathways is known to trigger cell death in the developing eye. We found that Wingless (Wg) and c-Jun-amino-terminal-(NH2)-Kinase (JNK) signaling are ectopically induced in cul-4 mutant clones, and these signals co-localize with the dying cells. Modulating levels of Wg and JNK signaling by using agonists and antagonists of these pathways demonstrated that activation of Wg and JNK signaling enhances cul-4 mutant phenotype, whereas downregulation of Wg and JNK signaling rescues the cul-4 mutant phenotypes of reduced eye. Here we present evidences to demonstrate that cul-4 is involved in restricting Wg signaling and downregulation of JNK signaling-mediated cell death during early eye development. Overall, our studies provide insights into a novel role of cul-4 in promoting cell survival in the developing Drosophila eye.
During organogenesis, regulation of conserved processes like cell proliferation, cell survival and cell death is crucial for organ growth and differentiation. A fine balance between control of cell death and cell survival is responsible for final organ shape and size during development. We used Drosophila eye model to identify genes involved in promoting growth and cell survival. The Drosophila adult eye contains 750–800 differentiated ommatidia, and develops from a sac-like epithelial structure called the imaginal disc housed in the larva. The ommatidia differentiate in the wake of a synchronous wave of retinal differentiation called the Morphogenetic Furrow (MF).1 The MF originates at the posterior eye margin, and the Wingless (Wg) signaling pathway negatively regulates the anterior movement of the MF.2, 3, 4 Wg, a secreted morphogen, initiates an intracellular signaling cascade by binding to its receptors Arrow (Arr) and Frizzled (Fz), which triggers downstream events to control the nuclear localization of the Drosophila beta-catenin Arm, and the spatial expression of Wg target genes.5, 6, 7 In Drosophila eye, Wg is also known to induce the proapoptotic genes, head involution defective (hid), reaper (rpr) and grim (together referred as HRG), to trigger programmed cell death to remove extra cells from the periphery of the pupal retina.8, 9, 10, 11, 12, 13 Further, aberrant signaling during development, e.g., abnormal Wg signaling, also causes stress-induced apoptosis.14
The intrinsic caspase-dependent cell death involves activation of HRG,15, 16, 17 which are negatively regulated by Drosophila inhibitor of apoptosis (DIAPs).18, 19 Inactivation of DIAP-1 can trigger cell death by the activation of cysteine proteases Dronc and Drice, the Drosophila homolog of Caspase-9 and Caspase-3, respectively.20, 21, 22, 23 In Drosophila expression of baculo-virus protein, P35 can block caspase-dependent cell death.24 Besides Wg, activation of c-Jun amino-terminal (NH2) Kinase (JNK) signaling triggers cell death through the activation of caspases.11, 14, 25, 26, 27 JNK belongs to a conserved MAP kinase super-family, which is involved in cell proliferation and cell survival, and is activated through a cascade of phosphorylation by MAP kinases.26, 28, 29, 30 In Drosophila, JNK signaling is activated by binding of the tumor necrosis factor (TNF) Eiger (Egr) to its receptor Wengen (Wgn), and a conserved signaling cascade of Tak 1 (TGFβ activating kinase 1, a Jun kinase kinase kinase (JNKKK), hemipterous (hep) (Jun kinase kinase), basket (bsk) (Jun kinase) and jun. Activation of the pathway leads to expression of the downstream target puckered (puc), a dual phosphatase, which participates in a negative feedback loop by downregulating JNK activity.29, 30
We argued that during early eye development, Wg or JNK levels must be tightly regulated to allow differentiation to proceed, and to prevent premature cell death that results in small or reduced eye phenotype. In a genetic screen, we identified cullin4 (cul-4) as a modifier that rescues the reduced eye phenotype.31 During development cul-4 is globally required. Analysis of cul-4 function revealed its new role in promoting cell survival during early eye development. The cul-4 gene belongs to an evolutionary conserved class of Cullin-family E3 ubiquitin ligases.32 Earlier studies showed that cul-4 is involved in maintenance of genomic integrity by promoting the ubiquitylation and subsequent degradation of key regulators of cell cycle progression.33, 34, 35, 36 Here, we report that cul-4 promotes cell survival by preventing Wg and JNK signaling-mediated cell death in the developing eye.
Results
Gain-of-function of cul-4 rescues reduced eye mutant phenotype
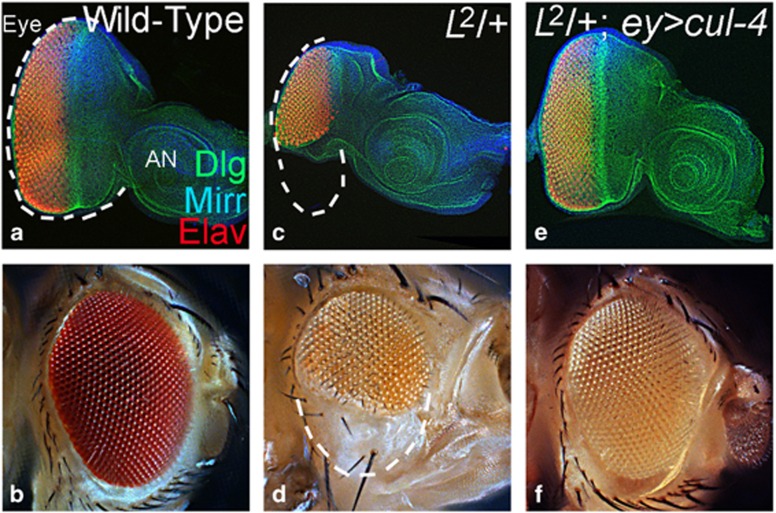
In comparison with the wild-type larval eye disc and the adult eye (Figures 1a and b), L mutant exhibits reduced eye phenotype in larval eye disc (Figure 1c) and adults (Figure 1d).11, 37 Misexpression of cul-4 using Gal4/UAS approach38 (L2; ey>cul-4) resulted in the rescue of L2-reduced eye phenotype (Figures 1e and f). Misexpression of cul-4 (ey>cul-4) does not affect the eye size (not shown) suggesting that cul-4 may not promote cell proliferation. We analyzed loss-of-function phenotype of cul-4 to understand its role during eye development.
Figure 1.
Gain-of-function of cul-4 can rescue reduced eye mutant phenotype. (a and b) Wild-type (a) eye-antennal imaginal disc and (b) adult eye. The eye disc is stained for a membrane specific marker, Dlg (green); dorsal eye fate marker, Mirr (blue); and pan-neural marker, Elav (red). The white dotted line marks the boundary of eye field. (c and d) L2 mutant exhibits reduced eye phenotype in (c) eye imaginal disc and (d) adult eye. (e and f) Misexpression of cul-4 in the L2 mutant eye background (L2; ey> cul-4) results in a significant rescue to a near wild-type eye as seen in (e) the eye disc and (f) the adult eye
Loss-of-function of cul-4 fail to survive and exhibit reduced eye phenotype
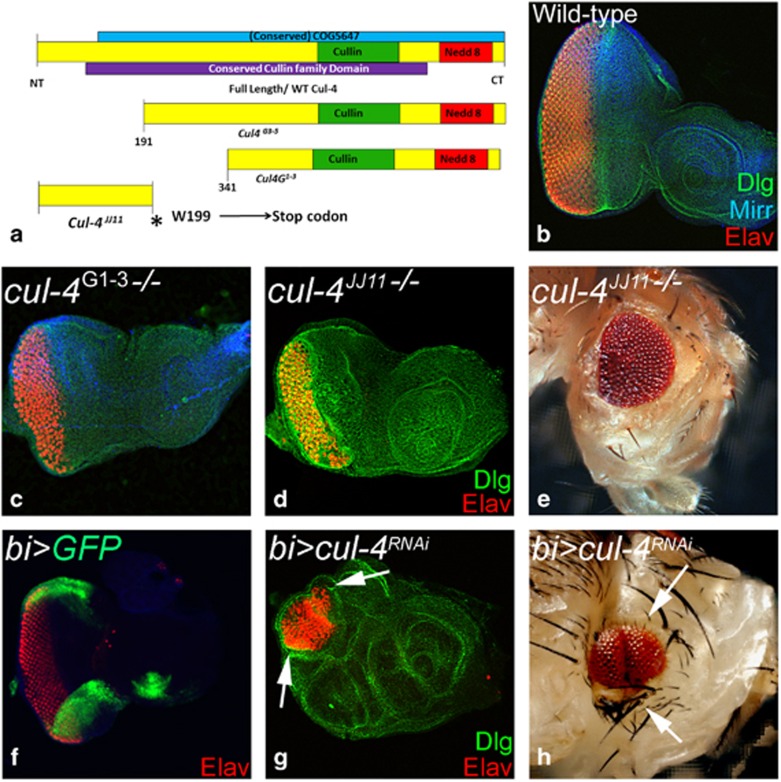
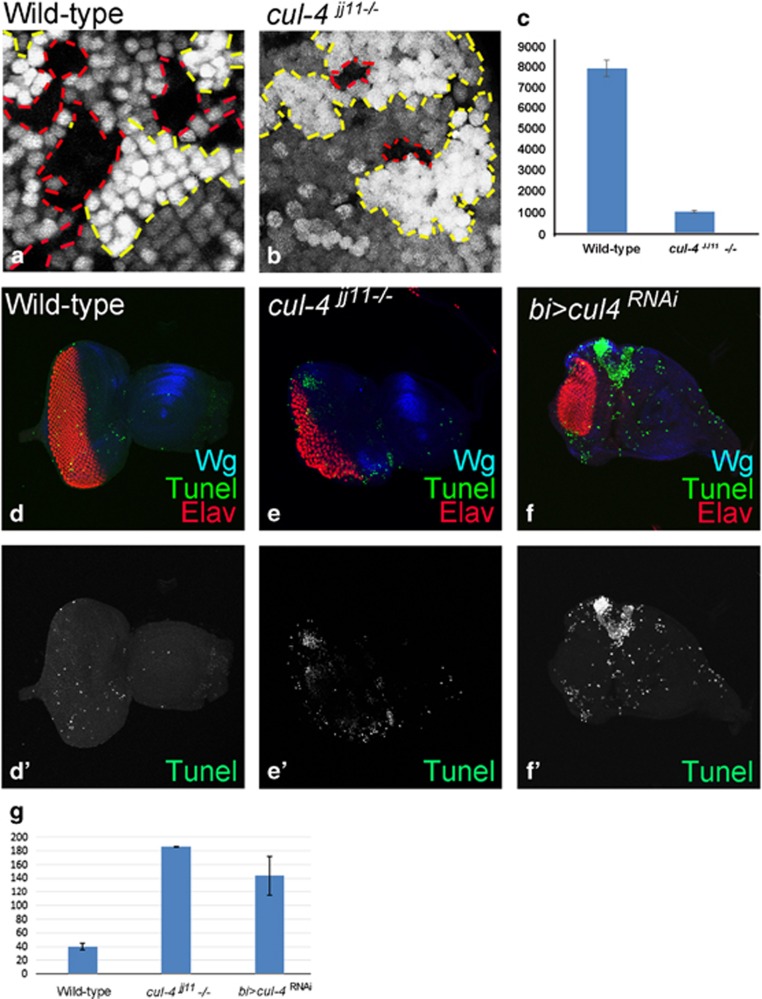
We generated cul-4 loss-of-function clones by cell lethal (cl) approach, which results in an eye disc comprising of nearly 80% mutant cells due to elimination of the wild-type twin spot by a cl mutation.39 Loss-of-function clones of cul-4 alleles (Figure 2a)40 like cul-4ExG1−3 (Figure 2c) or cul-4JJ11 (Figures 2d and e) resulted in a small eye phenotype as compared with the wild-type eye (Figure 2b). It is to be noted that both cul-4JJ11 and cul-4ExG1−3 loss-of-function phenotypes were similar in the eye. Downregulation of cul-4 expression in the dorso-ventral (DV) margins of developing eye disc by using bi-Gal4 driver (Figure 2f; bi>GFP) resulted in reduction of eye field on DV margins (Figures 2g and h, arrows).41, 42 It suggests that there is no domain constraint in cul-4 function in the eye. It is possible that reduced eye phenotype may be due to induction of cell death. To test this, we used ey-Flippase (ey-Flp) to induce somatic ‘twin clones' using Ubi-GFP (1XGFP), where cul-4−/− mutant cells (GFP-negative) were adjacent to their wild-type twin spot (2XGFP). However, we found that only the wild-type twin clone (2XGFP) and the heterozygous cells (express 1XGFP) could be identified. However, we found wild-type twin clones (2XGFP) but no cul-4 mutant clones (GFP-negative) in the third-instar eye disc (not shown), suggesting that the cul-4 mutant cells failed to survive. We performed a 'twin spot' analyses in the heat-shock-Flippase (hs-FLP) -induced clones, to test survival profiles of cul-4 mutant cells. The heat shock was administered in the first instar larva and the resultant clones were examined in the second- (24 h after clone formation (ACF)) and third-instar (48–72 h after clone formation) disc to determine whether or not these clones could survive. Very small clones were detected at second instar stage (within 24 h after clone formation). However, within 48 h after clone formation, the mutant clones were lost. In comparison with the wild-type clones (Figure 3a), the mutant clones generated at later time points (second instar (48 h) or early third instar (60 h)) and examined at late third-instar stage (within 24 h after clone formation), showed smaller cul-4−/− clones (Figures 3b and c; clone boundary marked by red dotted lines). These cul-4−/− clones failed to survive until 120 h of development. Quantification of clone size showed that cul-4 mutant clones are significantly smaller than their wild-type twin clones (Figure 3c), suggesting that cul-4 mutant clones either fail to survive or are slow growing compared with wild-type cells. We tested if cul-4 mutant cells are eliminated by cell death using TUNEL labeling.11, 17 Wild-type eye disc showed few TUNEL-positive dying cells (Figures 3d and d'), whereas eye disc lacking cul-4 function in the entire eye disc (cul-4jj11-/-, Figures 3e and e') or on DV margins (bi>cul4RNAi, Figures 3f and f') showed a threefold increase in TUNEL-positive cells suggesting that cul-4 mutant cells are eliminated by cell death.
Figure 2.
Loss-of-function of cul-4 results in reduced eye phenotype. (a) Schematic representation of wild-type and truncated Cul-4 protein encoded by different mutant alleles of cul-4 used in the loss-of-function studies.40 (b) Wild-type eye disc stained for dorsal fate marker Mirr (blue), Dlg (green) and Elav (red). (c–e) Loss-of-function clones of cul-4 (e) cul-4G1−3 (d and e) cul-4JJ11 generated by using 'cell-lethal' approach39 results in reduced eye phenotype as seen in (c and d) eye imaginal disc and (e) adult eye. (f) bi-Gal4 drives expression of GFP reporter on DV of the eye imaginal disc. (g and h) Misexpression of cul-4 RNAi on DV margins of the eye using bi-Gal4 driver (bi>cul4RNAi) exhibits eye fate suppression on both DV margins as seen in (h) eye disc and the (i) adult eye
Figure 3.
Loss-of-function clones of cul-4 fail to survive. (a and b) Genetic mosaics generated by using a Flp-FRT system in the eye results in GFP-positive and GFP-negative patches of cells. (a) Note that in wild-type controls, clonal areas marked by the absence of GFP (red dotted lines) are comparable in terms of size to the wild-type clones (with strong (2X) GFP-positive areas, marked by red dotted lines). (b) Loss-of-function clones of cul-4JJ11 mutant in the eye imaginal disc (no GFP, marked by red dotted line) are smaller compared with the wild-type twin spot (2XGFP, marked by yellow dotted line). Note that these cul-4JJ11 mutant clones fail to survive 24 h after they are formed. Only GFP-positive (wild-type) cells were seen. (c)The cell number of cul-4 clones was less than eightfold as compared with the wild-type clones based on counting five eye discs for each. (d–f') TUNEL staining was used to mark the dying cells nuclei in (d and d') wild-type, (e and e') cul-4jj11−/− clones and (f and f') bi>cul4RNAi eye imaginal disc. (g) The dying nuclei were counted from five imaginal discs from each of these category. There is more than fourfold increase in dying cell population in cul-4 mutant eye disc as compared with wild-type
cul-4 prevents cell death in the developing eye
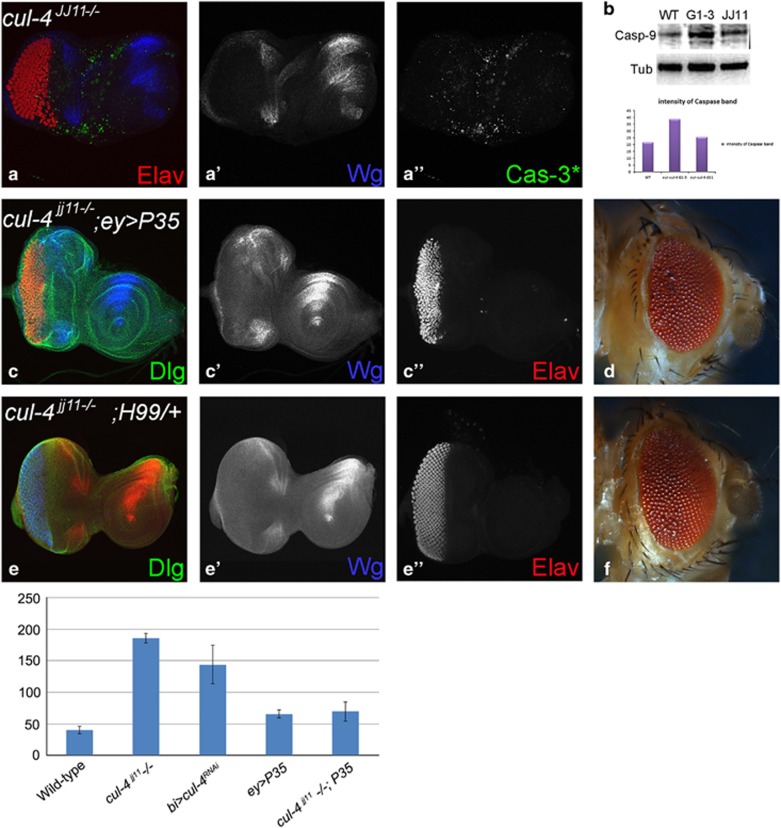
We, therefore, tested if Drosophila Caspases- Dronc and Drice activation is part of the mechanism. In the eye disc, cul-4JJ11loss-of-function clones generated by 'cell-lethal' clonal approach,39 exhibited robust induction of activated Caspase-3* (Cas-3*) and a signaling molecule Wg (Figures 4a and a''). Similarly, in semi-quantitative western blots, Dronc levels were nearly two fold higher in cul-4 mutant as compared with the wild-type eye disc (Figure 4b). Thus, both Dronc and Drice are induced in cul-4 mutant cells. Misexpression of baculo-virus P35 in the cul-4 loss-of-function clones (cul-4JJ11−/−, ey>P35) resulted in a significant rescue of the reduced eye phenotype (Figures 4c and c”). In cul-4 loss-of-function background reducing level of proapoptotic genes using H99 deficiency43 (cul-4JJ11−/−, H99−/+) resulted in significant rescue of the cul-4 mutant phenotype (Figures 4e and e”). It suggests that activation of caspases triggers apoptosis in cul-4 mutant cells. Aberrant signaling from key developmental and signaling pathways, like Wg/Wnt, can induce apoptosis to prevent defective development.10, 14
Figure 4.
cul-4 mutant cells are eliminated by activation of caspases. (a, a' and a'') Loss-of-function of cul-4 in eye results in enhanced caspase-3* (cas-3*, green) levels along with Wg (blue) upregulation and suppression of eye fate based on Elav (red) expression. (b) In comparison with the wild-type controls, nearly twofold increase in activator caspase-9 protein are seen in total protein extracted from eye imaginal discs of cul-4 loss-of-function background. Caspase band staining intensity calculated by Image –J. (c and d) Loss-of-function phenotype of cul-4 can be rescued by misexpression of baculo-virus protein, P35 (cul-4−/−; ey>P35) as seen in (c, c' and c”) eye imaginal disc and (d) adult eye. (e and f) Reduction in the levels of Hid-Reaper-Grim (HRG) complex by using deficiency of H99 can rescue the loss-of-function phenotype of cul-4, as seen in (e, e' and e”) eye imaginal discs and (f) adult eye. (g) The dying nuclei were counted from five imaginal discs from each of these category. Misexpression of P35 can significantly reduce the number of dying cell nuclei in cul-4 loss-of-function eye disc as compared with wild-type
cul-4 represses Wg levels in the developing eye
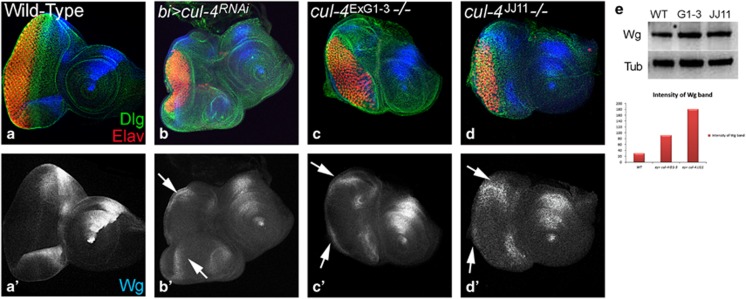
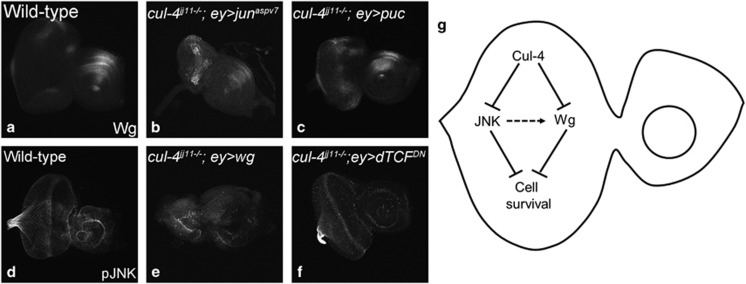
In the third-instar stage, Wg is expressed along the antero-lateral margins of the wild-type eye disc (Figures 5a and a'). Reducing cul-4 function on DV margin of eye disc using bi-Gal4 driver (bi>cul-4RNAi) resulted in strong ectopic induction of Wg in DV domain of the eye (Figures 5b and b'' white arrows). Loss-of-function clones of cul-4 using the cul-4ExG1−3(Figures 5c and c') and cul-4JJ11(Figures 5d and d') alleles showed a robust ectopic induction of Wg in the eye disc (Figures 5c', white arrows). Wg levels were significantly upregulated in semi-quantitative western blots on total protein isolated from eye imaginal discs from wild-type, and cul-4ExG1−3and cul-4JJ11 (Figure 5e). These data suggest that cul-4 may downregulate Wg in the eye imaginal disc. Misexpression of wg on DV margins of eye disc bi>wg results in ectopic wg transcription suggesting that wg is a target of Wg pathway in developing eye (Supplementary Fig. S1). We then tested if aberrant Wg signaling is responsible for cul-4 mutant phenotypes.
Figure 5.
Wg is ectopically induced in cul-4 mutant background. Expression of Wg (blue) in (a,a') Wild-type, (b and b') bi>cul4RNAi, (cul-4 RNAi is misexpressed on DV margin using bi-Gal4), (c and c') cul-4ExG1−3and (d and d') cul-4JJ11 loss-of-function clones. Note robust ectopic Wg (blue) expression on (b') DV margin (marked by white arrows) along with suppression of eye fate. (c and d) The reduced eye phenotype of cul-4 loss-of-function clones generated by cell-lethal approach is accompanied by ectopic upregulation of Wg (blue, marked by white arrow). (a'–d') Shown is the split channel of Wg expression. (e) In western blot analysis, the Wg protein levels are more than twofolds in eye discs with cul-4 loss-of-function clones as compared with wild-type eye disc. Wg band staining intensity calculated by Image-J
Wg signaling pathway alters the cul-4 mutant phenotype
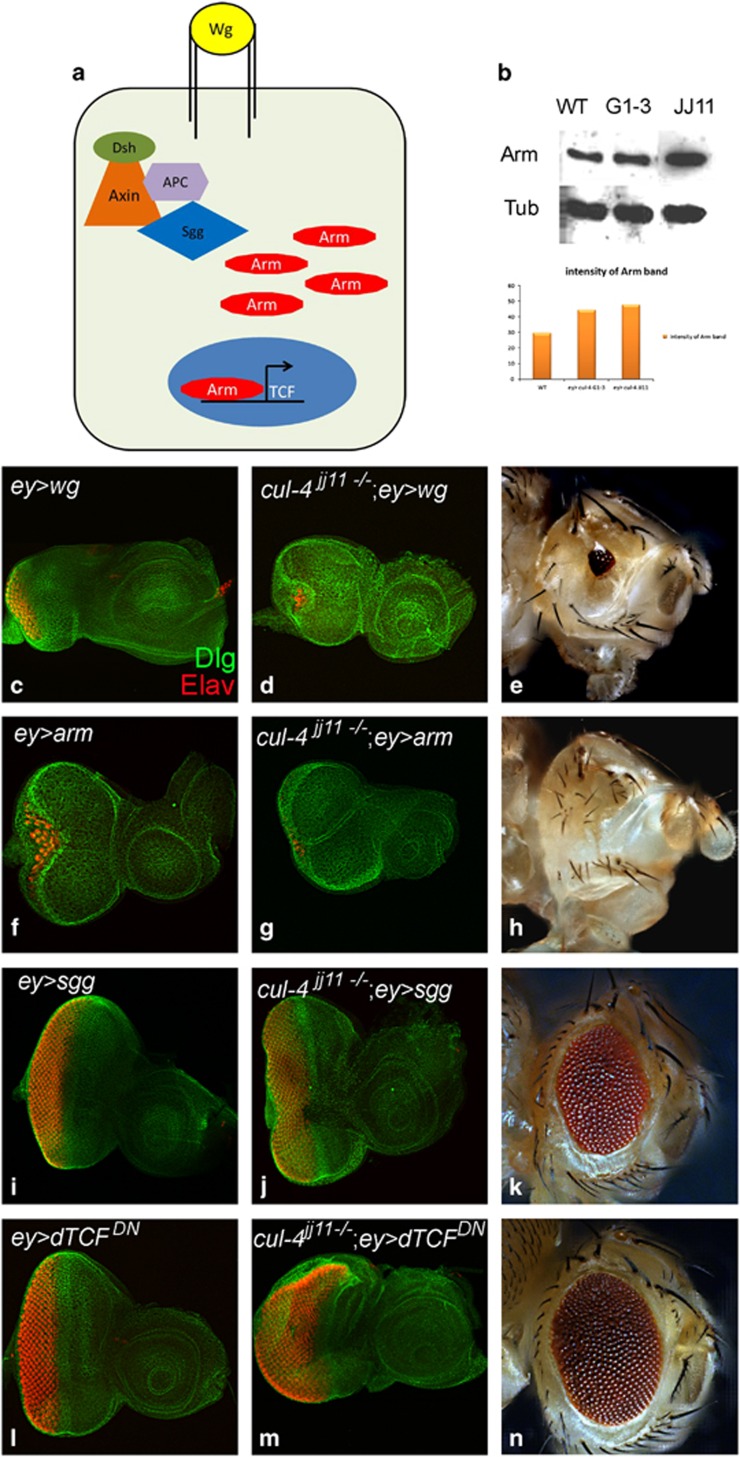
Arm translocates to the nucleus in response to Wg signaling and binds with the transcription factor dTCF (LET/TCF family protein) to turn on the transcription of Wg target genes (Figure 6a).44 In western blots a two fold increase in Arm protein levels was observed in cul-4 mutant eye discs as compared with wild-type (Figure 6b). We tested if modulating Wg signaling levels can affect the cul-4 mutant phenotype. In the eye imaginal discs, activation of Wg signaling by misexpression of wg (ey>wg) (Figure 6c) or arm (ey>arm) (Figure 6f) resulted in reduced eye phenotypes.11 Misexpression of wg (cul-4JJ11−/−, ey>wg) (Figures 6d and e) or arm (cul-4JJ11, ey>arm) (Figures 6g and h) in cul-4 loss-of-function background, resulted in near complete loss of eye. Blocking Wg signaling by misexpression of a constitutive active form of Shaggy/Zeste-White-3/GSK-3 (Sgg), a negative regulator of the Wg signaling pathway4, 45 (ey>sggS9A) (Figure 6i),11 or dominant negative form of TCF (dTCFDN)44 (ey>dTCFDN) (Figure 6l) does not affect the size of the eye field. However, in cul-4 loss-of-function background, misexpression of sgg (cul-4JJ11−/−, ey>sgg) (Figures 6j and k) or dTCFDN (cul-4JJ11−/−, ey>dTCFDN), (Figures 6m and n) resulted in a significant rescue of the cul-4 loss-of-function phenotype to a near wild-type eye. It suggests that cul-4 is involved in downregulation of Wg signaling in the eye. JNK is known to work in conjunction with Wg in multiple contexts including correction of morphogen gradient discontinuities;26 and differential levels of JNK signaling are associated with cell survival.26, 28
Figure 6.
Activation of Wg pathway results in the cul-4 mutant's reduced eye phenotype. (a) Cartoon showing Wg signaling pathway. (b) In the western blot performed by using protein extracts prepared from the wild-type and cul-4 mutant eye imaginal discs, the Armadillo (Arm) (a downstream cytoplasmic target of Wg signaling), protein levels are enhanced (twofold) in cul-4 loss-of-function background as compared with the wild-type (WT) control. Activation of Wg signaling by misexpression of (d and e) wg (cul-4−/−; ey>wg), (g and h) arm (cul-4−/−; ey>arm) results in enhancement of loss-of-function phenotype of cul-4 as seen in (d and g) eye disc as well as (e and h) adult eye. (c and f) Misexpression of (c) wg (ey>wg) and (f) arm (ey>arm) results in small eye. Blocking Wg signaling by misexpression of negative regulators/ antagonists like (j and k) sggS9A (cul-4−/−; ey> sggS9A) and (m and n) dTCFDN(cul-4−/−; ey> dTCFDN) suppresses the reduced eye phenotype of cul-4 loss-of-function to near normal as seen in (j and m) eye disc and (k and n) adult eye. Misexpression of (i) sgg (ey>sggS9A) and (l) dTCFDN(ey>dTCFDN) results in normal eye sizes
cul-4 prevents JNK-mediated cell death in the developing eye
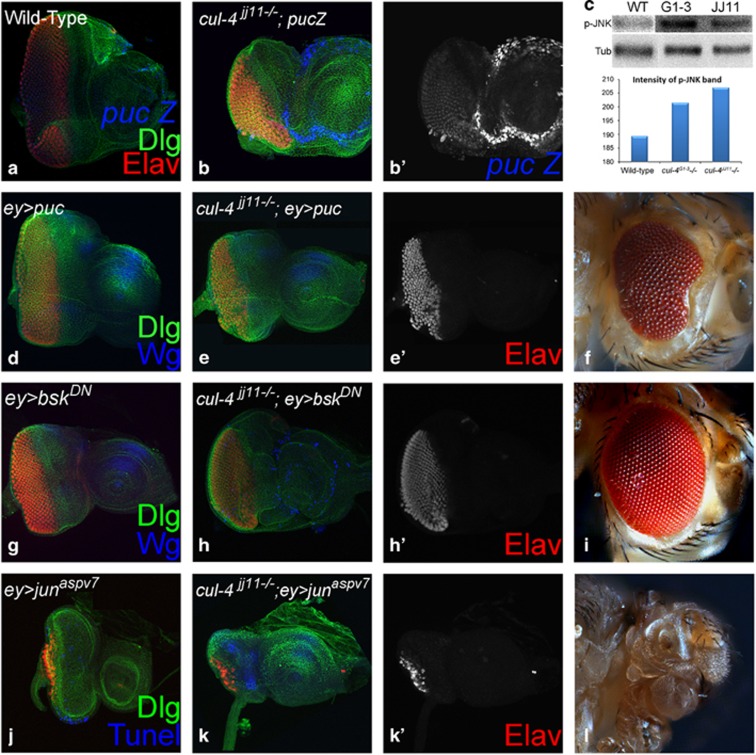
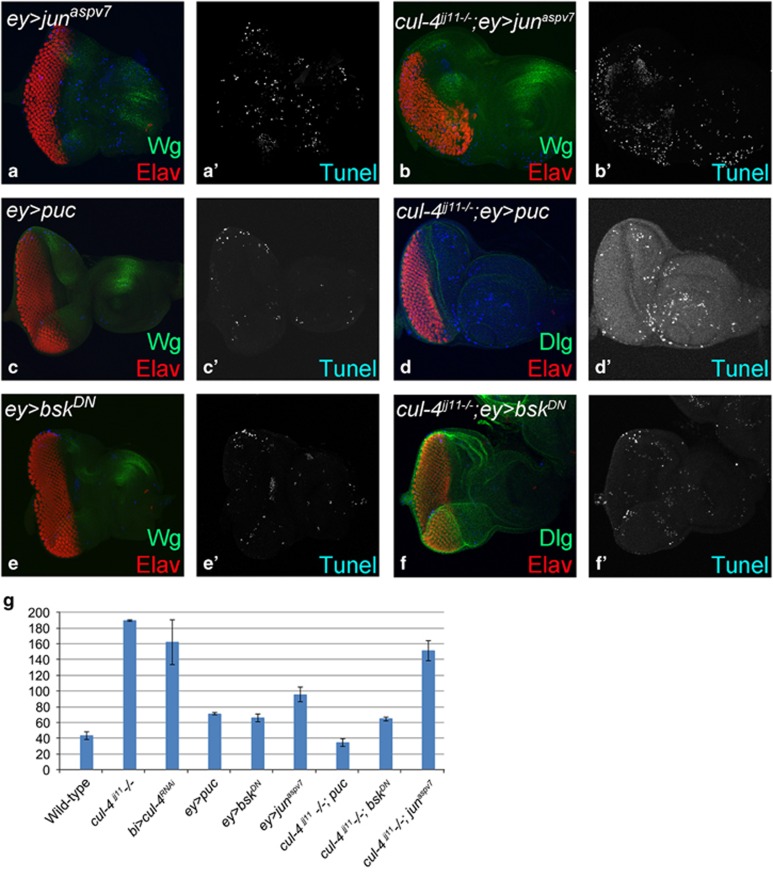
We tested if JNK pathway is associated with the cul-4 loss-of-function phenotypes (Figure 7). We tested JNK levels in cul-4 loss-of-function background using puc (puc-lacZ) the downstream target, which serves as the functional readout for JNK pathway activation.30 In wild-type eye disc, puc is expressed in differentiated photoreceptor neurons of eye disc (Figure 7a). In cul-4 loss-of-function background, ectopic induction of puc was seen in the eye as well antenna disc (Figures 7b and b'), suggesting that JNK signaling is activated in cul-4 mutant cells. To confirm, we checked levels of p-JNK, a reporter for activated JNK signaling, in western blots and found significant upregulation of p-JNK levels in cul-4 mutants compared with wild-type eye disc (Figure 7c). Blocking JNK signaling in the developing eye by misexpression of puc (ey>puc, Figure 7d) or bsk dominant negative (bskDN) (ey>bskDN, Figure 7g)25 did not affect the eye size. However, in cul-4 loss-of-function background misexpression of puc (cul-4JJ11−/−, ey>puc, Figures 7e and e') or bskDN (cul-4JJ11−/−, ey> bskDN, Figures 7h and h'), resulted in a significant rescue of cul-4 loss-of-function phenotype of reduced eye (Figure 2). Conversely, in cul-4 loss-of-function background activation of the JNK signaling pathway by misexpression of activated Jun (junaspv7) (cul-4JJ11−/−, ey>junaspv7) in the eye disc, strongly enhanced the reduced eye- to a 'no-eye' phenotype (Figures 7k and k'). Misexpression of junaspv7(ey>junaspv7) alone in the eye results in a highly reduced eye field (Figure 7j). It suggests that loss of cul-4 leads to activation of JNK signaling in the eye.
Figure 7.
Aberrant JNK signaling in cul-4 loss-of-function background triggers cell death. (a) A functional readout of JNK signaling pathway, puc-lacZ (blue) is expressed in differentiated photoreceptor neurons (marked by Elav, red) in the wild-type eye discs.60 (b and b') Loss-of-function of cul-4 causes ectopic induction of puc-lacZ reporter in the eye imaginal disc. (c) Activation of JNK signaling was detected by analyzing phospho-Jun (p-JNK) levels in western blots. Twofold increase in the levels of JNK signaling pathway was detected in cul-4 loss-of-function backgrounds in comparison to the wild-type eye imaginal disc. (d–i) Blocking JNK signaling by misexpression of (e, e' and f) puc (cul-4−/−; ey>puc) or (h, h' and i) dominant negative bskDN(cul-4−/−; ey>bskDN) in loss-of-function clones of cul-4 significantly restores their reduced eye phenotype as seen in the (e, e', h and h') eye imaginal disc and the (f and i) adult flies. Misexpression of (d) puc (ey>puc) or (g) bsk dominant negative (ey>bskDN) does not affect the eye size. (j–l) Misexpression of activated form of (j) jun (ey>junaspv7) in the developing eye results in reduced eye size, whereas (k, k' and l) misexpression of activated jun (cul-4−/−; ey>junaspv7) results in further enhancement of cul-4 loss-of-function phenotype of reduced eye
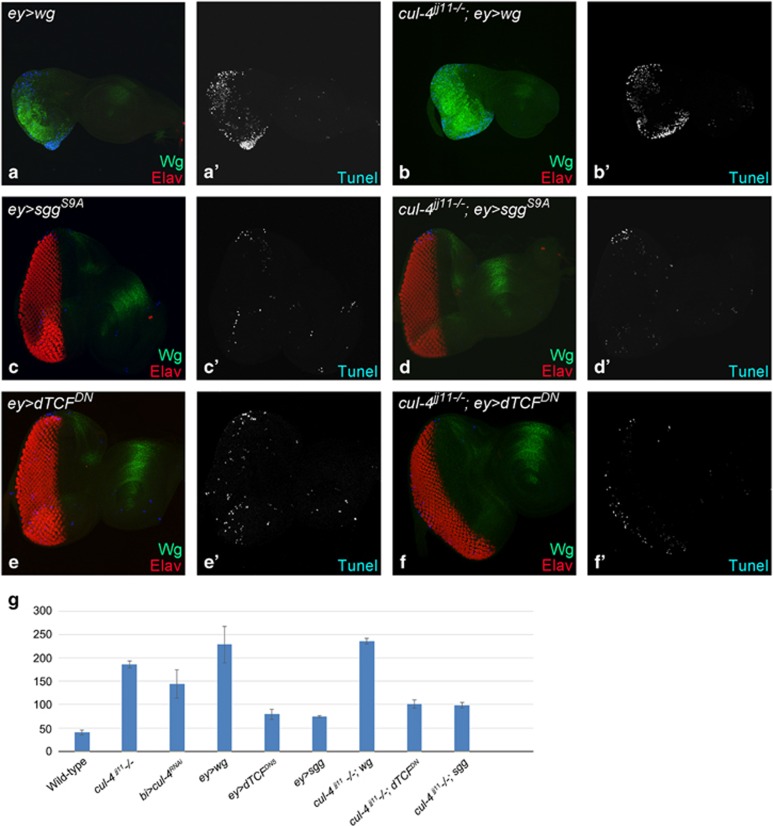
To confirm that activation of Wg /JNK signaling pathway are both associated with the induction of cell death observed in cul-4 mutant cells, we monitored cell death using TUNEL assay when Wg (Figure 8) and JNK (Figure 9) levels are modulated in the wild-type, and in cul-4 mutant eye discs. We found that cell death is reduced when Wg or JNK signaling is downregulated in cul-4 mutant background. However, cell death is elevated when Wg/ JNK signaling is activated. Thus, cul-4 may be involved in limiting JNK as well as Wg activation in the developing eye disc, and thereby promote cell survival during development.
Figure 8.
Aberrant Wg signaling triggers cell death in cul-4 loss-of-function background. Eye imaginal discs showing TUNEL (blue) labeling in (a and a') ey>wg, (b and b') cul-4−/−; ey>wg (c and c') ey>sggS9A, (d and d') cul-4−/−; ey>sggS9A, (e and e') ey>dTCFDN and (f and f') cul-4−/−; ey> dTCFDN. (g) Graph shows quantification of the number of dying cells in the wild-type, cul-4 loss-of-function and genotypes shown in (a–f). Note that in cul-4 loss-of-function background cell death is enhanced when Wg is activated. Conversely, cell death is suppressed to near wild-type when Wg signaling is blocked in cul-4 loss-of-function background
Figure 9.
Aberrant JNK signaling triggers cell death in cul-4 loss-of-function background. Panels show TUNEL (blue) staining marking the dying cells nuclei in eye discs from (a and a') ey>junaspv7, (c and c') ey>puc, (e and e') ey>bskDN, (b and b') cul4−/−; ey>junaspv7, (d and d') cul4−/−; ey>puc or (f and f') cul4−/−; ey> bskDN. (g) Graph shows a comparison of the number of dying cells in wild-type, cul-4 loss-of-function and the genotypes shown in a–f. Note that rate of cell death is enhanced when JNK signaling is activated whereas the rate of cell death is suppressed when JNK signaling is blocked in cul-4 loss-of-function background
Discussion
Cul-4, an E3 ligase, is involved in regulation of chromatin function through heterochromatin gene silencing, maintenance of genomic integrity by promoting the ubiquitylation and degradation of key cell cycle regulators.46, 47, 48, 49 A number of ligases work in concert with the signaling pathways (Notch (N), Hedgehog (Hh), Wg and so on) for regulating gene expression. For example, Slimb, is involved in regulating Wg and Hh signaling during eye development,50 Neuralized (Neu)51 and Mind Bomb (Mib), are E3 ligases that are components of N signaling pathway; and are required for Drosophila eye development.52 Recently other functions for E3 ligases are being recognized. For example, DIAP1 regulates Dronc/Hid caspases,20, 53 and is transcriptionally regulated by yorkie (yki) for survival function.54 DIAP-1 in turn, is regulated by Cul-3 in the developing eye to regulate apoptosis.71 Our studies provide evidences for a new function for cul-4 in cell survival during eye development.
Homozygous larvae of some cul-4 alleles are larval lethal that can survive until early third instar and produce smaller imaginal discs than wild-type discs at comparable developmental age.40 These phenotypes were attributed to problems with cell division. Our twin spot analysis revealed an interesting result that cul-4 mutant tissues in the developing eye imaginal disc failed to survive (Figure 2), and are eliminated by activation of caspases (Figure 3). Generating cul-4 mutant clones by using multiple approaches (for example, eyeless and heat-shock flippase) validated that cul-4 mutant cells failed to survive when generated in early embryonic or larval stages. Blocking caspase-mediated cell death led to significant rescue of reduced eye phenotypes of cul-4 loss-of-function (Figures 4c and d), supporting a role for cul-4 in cell survival.
We tested several cell signaling pathways in cul-4 loss-of-function background and found aberrant activation of Wg and JNK signaling (Figure 10). Wg is required for patterning, growth regulation and cell survival in multiple tissues including the eye discs. Ectopic induction of Wg induces cell death.9, 10, 11, 55 We found that cells lacking cul-4 function also undergo cell death and they express high levels of Wg. Arm, the nuclear effector of the Wg signaling pathway, is a target of E3 ubiquitin ligase-mediated degradation.56 Loss-of-function phenotype of cul-4 mutants could be modified by modulating the levels of canonical Wg signaling (Figures 6 and 7). Our data suggests that cul-4 may downregulate Wg signaling in the eye to promote cell survival in the eye disc. Since the cul-4 mutant phenotype was not completely rescued by blocking Wg signaling, we also tested the JNK signaling in the cul-4 mutant clones. The possibility of indirect consequences responsible for the mutant phenotype can be refuted because these phenotypes can be rescued by blocking Wg as well as JNK-mediated cell death. We found that Wg levels were affected when JNK signaling was modulated in cul-4 mutant background (Figure 10a–c). However, the converse did not show effect on phospho-JNK levels (Figure 10d–f). Our studies generate insights into genetic mechanisms that regulate cell survival during normal development by demonstrating the role of cul-4 in preventing inappropriate upregulation of Wg and JNK signaling in the developing Drosophila eye during early stages (Figure 10). A recent study showed that loss of Godzilla, a member of the RNF family of membrane-anchored E3 ubiquitin ligases regulates Wg levels on the basolateral surface of the tissues through dynamin-dependent endocytosis from the apical surface and subsequent trafficking from early apical endosomes to the basolateral surface.57 Our studies also generate mechanistic insights into genetic mechanisms that regulate cell survival during normal development.
Figure 10.
cul-4 promotes cell survival by regulating wg and JNk signaling in the developing eye. (a–c) Wg expression in (a) wild-type, (b) cul-4−/−; ey> junaspv7; and (c) cul-4−/−; ey>puc eye imaginal disc. Note that Wg levels are upregulated when JNK signaling is activated, whereas Wg levels are not much affected when JNK signaling is downregulated. p-JNK expression in (a) wild-type, (b) cul-4−/−; ey> wg; and (c) cul-4−/−; ey>dTCFDN eye imaginal disc. Note that pJNK levels are not affected. (g) During eye development cul-4 is involved in (i) downregulating Wg signaling and (ii) inhibiting JNK signaling to promote cell survival. Activated JNK signaling can trigger Wg induction
Numerous studies have shown the role of ubiquitin-mediated proteolysis in a broad array of cellular processes like defects in organogenesis, growth, differentiation, metabolism and aging in all organisms.58 Abnormal protein homeostasis underlies various disorders ranging from growth defects to neurodegenerative disorders.59 Our studies introduced new role of cul-4 in cell survival in the developing Drosophila eye. Since Drosophila serves as an excellent model to study development and human disease,60 these studies may shed light on understanding genetic basis of neurodegenerative orders in future.
Materials and methods
Fly stocks
The fly stocks used are described on Flybase (http://flybase.bio.indiana.edu). Cul-4 stocks used are EP 2518 (UAS-cul-4); cul-4 RNAi lines 8711 and 8711-R1 (from NIG).61 The N-terminal deletion mutants used were cul-4 ExG1−3/CyO, which lack 340 amino acids from amino terminal. cul-4JJ11/ twi>GFP, CyO carries a nonsense mutation at Trp199 position.40
Other stocks include Canton-S, y w eyFLP,39 L2/Cyo;37, 62 wg-lacZ/CyO,63 UAS-wg,64 UAS-sggS9A,65 UAS-arm,66 UAS-dTCFDN,44 UAS-P35,24 Df(3L)H99/TM6B,43 pucE69, UAS-puc,30 UAS-bskDN,25 and UAS-DJunaspv7.67 The Gal4/UAS system was used for targeted misexpression studies38 using ey-Gal4,65 and bi-Gal4 (refs 41, 42) lines.
Mosaic analysis
To generate loss-of-function clones68 of cul-4 in the eye, virgins of eyFlp; FRT42D, cl-w+/CyO-GFP were crossed to (i) FRT 42D, cul-4 ExG1−3/CyO, (ii) FRT 42D, cul-4 ExG3−5, (iii) FRT 42D, cul-4 ExL2−1/CyO and (iv) FRT 42D, cul-4 JJ11/twi>GFP, CyO.
Twin spot analysis
We used hsFlp; FRT42D ubi-GFP to generate loss-of-function clones of cul-4 ExG1−3 and cul-4 JJ11in the eye imaginal disc at different larval development stages. Egg laying were collected from synchronous cultures maintained at 25 °C. The cultures were heat shocked at 24 and 48 h after egg laying (AEL) at 37 °C for 50 min in order to induce loss-of-function clones. Eye discs were dissected in second and third-instar stages to analyze/identify cul-4 loss-of-function clones marked by the absence of GFP expression.
Immunohistochemistry
Eye-antennal imaginal discs were dissected from wandering third-instar larvae and stained following the standard protocol.62 Antibodies used were rat anti-Elav (1:100), mouse anti-Wg (1:50), mouse anti-β galactosidase (1:200) (Developmental Studies Hybridoma Bank, DSHB, Iowa City, IA, USA), rabbit anti-Dlg (1:250) (gift from Kyung- Ok Cho), rat anti-Mirror (1:200) (gift from Kwang Wook Choi), rabbit anti-caspase-3* (1:200) and rabbit Phospho-SAPK/JNK (Cell Signaling Thr183/Tyr185) (81E11) (Cell Signaling Technology, Danvers, MA, USA). Secondary antibodies (Jackson Immuno Research Laboratories Inc., West Grove, PA, USA) were goat anti-rat IgG conjugated with Cy5 (1:200), donkey anti-rabbit IgG conjugated to Cy3 (1:250), donkey anti-rabbit IgG conjugated to FITC and donkey anti-mouse IgG conjugated to Cy3 (1:200). The discs were mounted in Vectashield (Vector Laboratories Inc., Burlingame, CA, USA) and imaged using Olympus Fluoview 1000 microscope (Olympus America, Scientific Solutions Group, Center Valley, PA, USA). Images were analyzed using the Adobe Photoshop CS4 (Adobe Systems, San Jose, CA, USA) and image intensity was calculated using the Image J software.
TUNEL assays
Apoptotic cell death was assayed using TUNEL assays in the mutant clones generated via twin spot analysis and cell lethal approach. Eye discs, after secondary antibody staining,62 were blocked in 10% Normal Goat Serum in Phosphate Buffered Saline with 0.2% Triton X-100.TUNEL assays were done using the Cell-death Detection Kit from Roche Diagnostics following the standardized protocol.11, 17 The TUNEL-positive nuclei were counted from five eye imaginal discs for each genotype using Image-J and statistical analysis was performed using Microsoft Excel 2013. The P-values were calculated and the error bars represent Standard Deviation.
Adult eye imaging
Adult Drosophila eye images were taken70 using a Zeiss Apotome Imager.Z1 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). The flies were prepared by freezing them at −20 °C for ~2 h. The legs and wings of the flies were removed and flies were mounted on a dissection needle, and the fly was positioned on a glass slide using mounting putty. Images were captured by using extended depth of focus function of the Axiovision software version 4.6.3 (Carl Zeiss Microscopy GmbH, Jena, Germany) by compiling the individual stacks from the Z-sectioning approach. The final images and figures were prepared using Adobe Photoshop CS4 software.
Western blot analysis
Protein samples were prepared from third-instar eye-antennal imaginal discs of different cul-4 mutants dissected in ice-cold PBS. Samples were transferred to sample buffer containing SDS-β-mercaptoethanol, boiled for 10 min, stored in −80 °C. Protein samples were separated on 10% SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in blocking solution (AMRESCO LLC, Solon, OH, USA) and incubated with primary antibody. The antibodies used were anti- mouse Wg (1:100) (DSHB); anti-mouse arm (1:2000) (DSHB), anti-rabbit p-JNK (1:2000) (Cell signaling Technologies), anti-rabbit Caspase-9 (1:1000) (Cell signaling Technologies) or anti- mouse tubulin (1:5000) (Sigma-Aldrich Corp., St. Louis, MO, USA). Secondary antibodies were horseradish peroxidaseconjugated goat anti-rabbit IgG, and the signal was detected using super-signal chemiluminiscence substrate (Pierce Biotechnology, Thermofisher Scientific, Rockford, IL USA).
Acknowledgments
We thank, Chen Ting-Chen, Robert J Duronio, Tin Tin Su, the Bloomington Stock Center for the Drosophila strains, K Cho and the Developmental Studies Hybridoma Bank (DSHB) for the antibodies. We thank members of the Singh laboratory for critical comments on the manuscript. Confocal microscopy was supported by Biology Department central core facility. MK-S is supported by Knight's Templar Ophthalmology Research Foundation and startup support from UD. This work is supported by start-up support from UD, and Ohio Cancer Research Associates to AS.
Author contributions
MT performed majority of the experiments and the data analysis. AS performed experiments. SB performed experiments. MK-S did the data analysis and the manuscript writing. AS developed the concept and did the data analysis and manuscript writing.
Glossary
- Cul-4
Cullin-4
- Wg
Wingless
- JNK
c-Jun amino-terminal (NH2) Kinase
- MF
Morphogenetic Furrow
- Arm
Armadillo
- dTCF
Drosophila T cell factor
- Arr
Arrow
- Fz
Frizzled
- Sgg
Shaggy
- Hid
head involution defective
- Rpr
reaper
- HRG
hid, reaper, and grim
- DIAP1
Drosophila inhibitor of Apoptosis
- PCD
programmed cell death
- Cas-9
caspase-9
- Cas-3
Caspase-3
- TNF
Tumor necrosis factor
- Egr
Eiger
- Wgn
Wengen
- Tak1
TGFβ activating kinase 1
- Hep
hemipterous
- Bsk
basket
- BskDN
basket dominant negative
- Puc
puckered
- L
Lobe
- Ey
eyeless
- UAS
Upstream Activator Sequence
- Cl
cell lethal
- Bi
bifid
- DV
Dorso-ventral
- GFP
Green Fluorescent Protein
- FLP
Flippase
- ACF
after clone formation
- RNAi
RNA interference
- N
Notch
- Hh
Hedgehog
- Neu
Neuralized
- Mib
Mind Bomb
- Yki
yorkie
- Cul-3
Cullin-3
- RNF
Ring finger proteins
- NIG
National Institute of Genetics
- Twi
twist
- FRT
Flippase recombination targets
- AEL
after egg laying
- Dlg
disc large
- β
beta
- Elav
embryonic lethal, abnormal vision
- IgG
Immunoglobulin G
- FITC
Fluorescein isothiocyanate
- Cy3
Cyanines 3 dye
- Cy5
Cyanines 5 dye
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Baehrecke
Supplementary Material
References
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol 1976; 53: 217–240. [DOI] [PubMed] [Google Scholar]
- Ma C, Moses K. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development 1995; 121: 2279–2289. [DOI] [PubMed] [Google Scholar]
- Tare M, Puli OR, and, Singh AMolecular genetic mechanisms of axial patterning: mechanistic insights into generation of axes in the developing eye. In: Singh A, and Kango-Singh M (eds). Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Springer: NewYork, 2013, pp 37–75. [Google Scholar]
- Treisman JE, Rubin GM. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 1995; 121: 3519–3527. [DOI] [PubMed] [Google Scholar]
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 1997; 16: 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold HC, Yang YX, Chen L, Cadigan KM. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta Physiol (Oxf) 2011; 204: 74–109. [DOI] [PubMed] [Google Scholar]
- Swarup S, Verheyen EM. Drosophila homeodomain-interacting protein kinase inhibits the Skp1-Cul1-F-box E3 ligase complex to dually promote Wingless and Hedgehog signaling. Proc Natl Acad Sci USA 2011; 108: 9887–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J, Jassim O, Bao S, Cagan R. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech Dev 2004; 121: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development 2004; 131: 2409–2418. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 2004; 7: 491–501. [DOI] [PubMed] [Google Scholar]
- Singh A, Shi X, Choi KW. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development 2006; 133: 4771–4781. [DOI] [PubMed] [Google Scholar]
- Singh A, Lim J, Choi K-WDorso-ventral boundary is required for organizing growth and planar polarity in the Drosophila eye. In: Mlodzik M (ed). Planar Cell Polarization during Development: Advances in Developmental Biology and Biochemistry. Elsevier Science & Technology Books: New York, 2005, pp 59–91. [Google Scholar]
- Singh A, Tare M, Puli OR, Kango-Singh M. A glimpse into dorso-ventral patterning of the Drosophila eye. Dev Dyn 2012; 241: 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, White K. Cell death in development: signaling pathways and core mechanisms. Semin Cell Dev Biol 2015; 39: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. Grim, a novel cell death gene in Drosophila. Genes Dev 1996; 10: 1773–1782. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 1995; 9: 1694–1708. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science 1994; 264: 677–683. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol 2002; 4: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 2002; 4: 416–424. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 2000; 19: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T et al. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell 2004; 7: 897–907. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell 2004; 7: 909–915. [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development 2005; 132: 2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development 1994; 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada T, Fujimura-Kamada K, Nishida Y, Matsumoto K. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 1999; 400: 166–169. [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada T, O'Connor MB. Mechanisms for removal of developmentally abnormal cells: cell competition and morphogenetic apoptosis. J Biochem 2004; 136: 13–17. [DOI] [PubMed] [Google Scholar]
- Tare M, Modi RM, Nainaparampil JJ, Puli OR, Bedi S, Fernandez-Funez P et al. Activation of JNK signaling mediates amyloid-ss-dependent cell death. PloS One 2011; 6: e24361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis 2009; 14: 1021–1028. [DOI] [PubMed] [Google Scholar]
- Igaki T, Miura M. The Drosophila TNF ortholog Eiger: emerging physiological roles and evolution of the TNF system. Semin Immunol 2014; 26: 267–274. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Chan J, Chern JJ, Choi KW. Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics 2005; 171: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. Targeting protein ubiquitylation: DDB1 takes its RING off. Nat Cell Biol 2009; 11: 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Keaton M, Dutta A. Regulation of TGF-beta signaling, exit from the cell cycle, and cellular migration through cullin cross-regulation: SCF-FBXO11 turns off CRL4-Cdt2. Cell Cycle 2013; 12: 2175–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 2011; 144: 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Duronio RJ. Developmental regulation of replication-coupled protein destruction. Cell Cycle 2011; 10: 859–860. [DOI] [PubMed] [Google Scholar]
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S et al. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 2011; 480: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Choi KW. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development 2003; 130: 6351–6360. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 2000; 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Lin HC, Wu JT, Tan BC, Chien CT. Cul4 and DDB1 regulate Orc2 localization, BrdU incorporation and Dup stability during gene amplification in Drosophila follicle cells. J Cell Sci 2009; 122: 2393–2401. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 1996; 381: 387–393. [DOI] [PubMed] [Google Scholar]
- Tare M, Puli OR, Moran MT, Kango-Singh M, Singh A. Domain specific genetic mosaic system in the Drosophila eye. Genesis 2013; 51: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott MK, Lengyel JA. Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics 1991; 129: 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 1997; 88: 789–799. [DOI] [PubMed] [Google Scholar]
- Heslip TR, Theisen H, Walker H, Marsh JL. Shaggy and dishevelled exert opposite effects on Wingless and Decapentaplegic expression and on positional identity in imaginal discs. Development 1997; 124: 1069–1078. [DOI] [PubMed] [Google Scholar]
- Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2007; 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev 2008; 22: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kipreos ET. Control of the Cdc6 replication licensing factor in metazoa: the role of nuclear export and the CUL4 ubiquitin ligase. Cell cycle 2008; 7: 146–150. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ 3rd, Reis T, Edgar BA et al. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 2008; 15: 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 1998; 391: 493–496. [DOI] [PubMed] [Google Scholar]
- Cho B, Fischer JA. Ral GTPase promotes asymmetric Notch activation in the Drosophila eye in response to Frizzled/PCP signaling by repressing ligand-independent receptor activation. Development 2011; 138: 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 2007; 134: 825–831. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 1999; 98: 453–463. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005; 122: 421–434. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Mille F, Thibert C. Morphogens and cell survival during development. J Neurobiol 2005; 64: 357–366. [DOI] [PubMed] [Google Scholar]
- Jumpertz S, Hennes T, Asare Y, Vervoorts J, Bernhagen J, Schutz AK. The beta-catenin E3 ubiquitin ligase SIAH-1 is regulated by CSN5/JAB1 in CRC cells. Cell Signal 2014; 26: 2051–2059. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Palmer L, Alexandre C, Kakugawa S, Beckett K, Gaugue I et al. Godzilla-dependent transcytosis promotes Wingless signalling in Drosophila wing imaginal discs. Nat Cell Biol 2016; 18: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem 2012; 81: 167–176. [DOI] [PubMed] [Google Scholar]
- Dennissen FJ, Kholod N, van Leeuwen FW. The ubiquitin proteasome system in neurodegenerative diseases: culprit, accomplice or victim? Prog Neurobiol 2012; 96: 190–207. [DOI] [PubMed] [Google Scholar]
- Singh A, Irvine KD. Drosophila as a model for understanding development and disease. Dev Dyn 2012; 241: 1–2. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu M, Su Y, Du J, Zhu AJ. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 (Bethesda) 2012; 2: 1563–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development 2002; 129: 4271–4280. [DOI] [PubMed] [Google Scholar]
- Kassis JA, Noll E, VanSickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. Proc Natl Acad Sci USA 1992; 89: 1919–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev 1998; 12: 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 1998; 125: 3741–3751. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell 1996; 87: 833–844. [DOI] [PubMed] [Google Scholar]
- Treier M, Bohmann D, Mlodzik M. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell 1995; 83: 753–760. [DOI] [PubMed] [Google Scholar]
- Blair SS. Genetic mosaic techniques for studying Drosophila development. Development 2003; 130: 5065–5072. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 1993; 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Steffensmeier AM, Tare M, Puli OR, Modi R, Nainaparampil J, Kango-Singh M et al. Novel neuroprotective function of apical-basal polarity gene crumbs in amyloid beta 42 (abeta42) mediated neurodegeneration. PloS One 2013; 8: e78717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Multiple Mechanisms Modulate Distinct Cellular Susceptibilities toward Apoptosis in the Developing Drosophila Eye. Dev Cell 2014; 30: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.