Abstract
The type III secretion system (T3SS) is essential for the pathogenesis of many bacteria including Salmonella and Shigella, which together are responsible for millions of deaths worldwide each year. The structural component of the T3SS consists of the needle apparatus, which is assembled in part by the protein-protein interaction between the tip and the translocon. The atomic detail of the interaction between the tip and the translocon proteins is currently unknown. Here, we used NMR methods to identify that the N-terminal domain of the Salmonella SipB translocon protein interacts with the SipD tip protein at a surface at the distal region of the tip formed by the mixed α/β domain and a portion of its coiled-coil domain. Likewise, the Shigella IpaB translocon protein and the IpaD tip protein interact with each other using similar surfaces identified for the Salmonella homologs. Furthermore, removal of the extreme N-terminal residues of the translocon protein, previously thought to be important for the interaction, had little change on the binding surface. Finally, mutations at the binding surface of SipD reduced invasion of Salmonella into human intestinal epithelial cells. Together, these results reveal the binding surfaces involved in the tip-translocon protein-protein interaction and advance our understanding of the assembly of the T3SS needle apparatus.
Keywords: type III secretion system, tip, translocon, SipB, IpaB, SipD, IpaD, NMR spectroscopy
INTRODUCTION
Many Gram-negative bacteria assemble a protein nanoinjector called the type III secretion system (T3SS) to inject virulence factors into the cytoplasm of host cells and initiate infectious diseases. This includes the causative agents of food poisoning/typhoid (Salmonella Typhimurium/Typhi) and dysentery (Shigella flexneri). Together, these pathogens are responsible for millions of deaths worldwide each year, primarily in children under the age of six.1,2 The T3SS is essential for pathogenicity as deletions of its protein components render bacteria non-invasive.3 The structural component of the T3SS, the needle apparatus, is composed of a base anchored at the bacterial cell membranes, an extracellular needle, a tip complex and a translocon.4 The needle is formed by the oligomerization of PrgI in Salmonella and MxiH in Shigella and the atomic structure of the polymerized needle has been determined for both Salmonella and Shigella.5,6 At the needle tip sits a complex of an estimated pentameric copy of SipD in Salmonella 7 and IpaD in Shigella.8,9 The tip complex functions as a signal transducer for host cell contact, a platform for the assembly of the translocon, and a regulator of secretion of effector proteins.7,10,11 The crystal structures of SipD12,13 and IpaD14,15 reveal three domains: an N-terminal α-helical hairpin, a central coiled-coil and a mixed α/β region (Fig. 1). The N-terminal α-helical hairpin functions as a self-chaperone that prevents self-oligomerization and premature interactions with other T3SS proteins, such as the needle.15,16 A current hypothesis is that during assembly at the tip of the needle, the N-terminal α-helical hairpin of SipD/IpaD is displaced to allow interaction with the needle.15,16
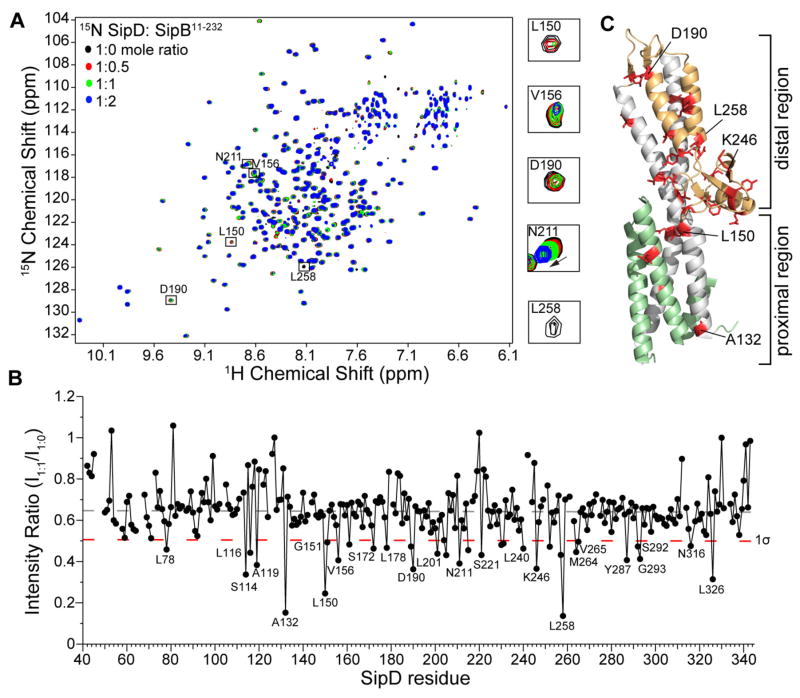
Figure 1.
Amide titrations of SipDC244S with SipB11-232. (A) Overlay of four 1H-15N TROSY spectra of 15N/ILV SipDC244S with increasing concentrations of unlabeled SipB11-232. (B) Plot of ratio of peak intensities of complex (I1:1) vs free (I1:0) for the SipDC244S:SipB11-232 complex at 1.1:1.0 molar ratio shown with the average peak intensity ratio (dashed gray line) and one standard deviation from the average (1σ, dashed red line). (C) Residues (colored red) with peak intensity ratio (I1:1/I1:0) lower than 1σ were mapped onto the crystal structure of SipD [colored green (N-terminal hairpin), gray (coiled-coil), and orange (mixed α/β domain)].
Upon host cell contact, bacteria assemble a ~3 nm wide translocon pore in the host membrane to allow the passage of effectors into the host cytoplasm.17–19 The translocon is assembled from two transmembrane proteins termed the major and the minor translocon protein based on their molecular weights. The major and minor translocon proteins in Salmonella are SipB and SipC, respectively; and their counterparts in Shigella are IpaB and IpaC, respectively.3,4,17 The translocon proteins also function as effectors.20–22 The topology of SipB and IpaB are predicted to contain an N-terminal ectodomain, a transmembrane helical region, and a C-terminal amphipathic helix.23–27 The atomic structure of the assembled translocon or the full length translocon proteins is currently unknown, however, crystal structures of the N-terminal ectodomains of SipB and IpaB revealed structurally similar coiled-coil motifs.28 Further, Nguyen et al.29 recently proposed a crystallography-derived model of the membrane-insertion and topology of the SipB/IpaB counterpart in Aeromonas hydrophila, AopB.
How the translocon proteins interact with the tip protein is poorly understood. Dickenson et al.23 reported the interaction between the Shigella IpaD tip protein with the N-terminal ectodomain of IpaB (residues 11-226) by FRET and showed that the interaction required the presence of the bile salt deoxycholate and that IpaB residues 11–27 were essential for the interaction. Results showing that the proximal region of SipD (defined here as the bottom of the coiled-coil, Fig. 1, which is pointed towards the T3SS needle) is essential for its interaction with the needle protein PrgI16,30 led us to hypothesize that the distal region of SipD (defined here as the upper portion of the coiled-coil and mixed α/β domain, Fig. 1), is likely the site of interaction for the translocon. Here, we show results of NMR titrations between the tip proteins and the translocon proteins of Salmonella and Shigella. Our results show that the tip protein of both Salmonella and Shigella interacts with the N-terminal domain of their respective major translocon protein. The regions of the tip proteins involved in the interaction, the distal region, are conserved between the two bacteria. Additionally, SipD mutations at the distal region resulted in decreased invasion of Salmonella into human epithelial cells. Our results suggest a model of protein-protein interaction between the tip and the major translocon protein of the T3SS.
MATERIALS AND METHODS
Protein Expression and Purification
The expression and purification of SipDC244S (residues 39-343 C244S) has been described previously.13 IpaD (residues 38-332) and IpaDC322S (residues 38-332 C322S) were subcloned into the NdeI/SalI sites of a modified pET-21a expression vector containing an N-terminal His6 tag followed by a tobacco etch virus (TEV) protease cleavage site. Both tip proteins retained a GHM artifact after cleavage with TEV protease. We previously reported that the C244S point mutation does not alter the crystal structure of SipD and Salmonella harboring the SipDC244S mutation are fully functional in assembling the T3SS and invading eukaryotic cells.13 This suggests that Cys to Ser point mutations do not drastically alter the structure and function of T3SS tip proteins. SipB11-232, SipB82-312, IpaB9-226 and IpaB74-224 were subcloned into the NdeI/XhoI sites of pET-22b. The SipB and IpaB constructs retained a non-cleavable C-terminal LEH6 tag for protein purification. Unlabeled SipB and IpaB proteins were expressed in E. coli BL21(DE3) DNAY cells in 1 liter of Luria Broth media containing 30 μg/mL kanamycin and 100 μg/mL carbenicillin. Cells were grown at 37°C until an OD600 of ~0.8, induced with isopropyl-β-D-thiogalactopyrandoside (IPTG) (1 mM IPTG for SipB and 0.5 mM IPTG for IpaB) and grown overnight at 15°C to a final OD600 of ~2.6. ILV labeling where the isoleucine Cδ1, the leucine Cδ, and the valine Cγ methyl groups are 13C-labeled was used. Uniformly 15N/ILV-labeled tip proteins were expressed in 1 liter of 1× M9 minimal media at 37°C containing 1 g/L 15NH4Cl. At an OD600 of ~ 0.4 the growth medium was supplemented with 60 mg of 2-ketobutyric acid-4-13C (Sigma #571342; which labels the isoleucine 13Cδ1 methyl group) and 100 mg of 2-keto-3-(methyl-13C)-butyric-4-13C acid (Sigma 571334; which labels the leucine 13Cδ and the valine 13Cγ methyl groups). For assignment of ILV 13C methyl resonances, perdeuterated 15N/ILV-labeled SipDC244S was prepared by cell growth in M9 minimal media in 1 liter of D2O with 15N- and ILV-labels, and 2 g/L deuterated D-glucose-1,2,3,4,5,6,6-d7 (Cambridge Isotope Laboratories #CLM-2062). Protein expression was induced with 1 mM IPTG at an OD600 of 0.8 and cell growth was continued overnight at 15°C. Bacterial cells were harvested by centrifugation (4000 rpm, 10 min), resuspended with binding buffer (500 mM NaCl, 20 mM Tris-HCl, 5 mM imidazole, pH 8.0, 0.1 mM phenylmethanesulfonyl fluoride) and sonicated. The cell lysate was centrifuged (13,000 rpm, 10 min) to remove cellular debris and 700 μL of 5% polyethyleneimine was added to the supernatant to precipitate the nucleic acids and the mixture was centrifuged (13,000 rpm, 10 min). The supernatant was loaded into a 10 mL Ni2+-affinity column (Gold Biotechnology), and the column was washed with 150 mL binding buffer (500 mM NaCl, 20 mM Tris-HCl, 5 mM imidazole, pH 8.0), and eluted with 40 mL elution buffer (500 mM NaCl, 20 mM Tris-HCl, 250 mM imidazole, pH 8.0). For SipD and IpaD constructs, the eluted fractions were pooled and incubated in 0.02% by volume of 0.1 mM recombinant tobacco etch virus (TEV) protease31 in buffer (50 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 1 mM DTT, 20 mM NaCl) at room temperature overnight. The mixture was purified by Ni2+-affinity chromatography to separate the protein (which flowed through the Ni2+ column) from the affinity tag (which was retained in the Ni2+ column). Purified proteins were dialyzed into NMR buffer (100 mM NaCl, 20 mM sodium phosphate, pH 7.4 and 10% D2O) and concentrated using Amicon Ultra 3K (Millipore) filtration columns. Protein concentrations were measured by A280.
NMR Spectroscopy
NMR data were collected using a Bruker Avance 800 MHz spectrometer equipped with a cryogenic triple resonance probe and were processed using NMRPipe32 and analyzed using NMRView.33 Two-dimensional 1H-15N TROSY spectra were acquired at 30°C using ~0.4 mM of 15N/ILV SipDC244S dissolved in NMR buffer with and without unlabeled SipB11-232 or SipB82-312. Two-dimensional 1H-15N-TROSY spectra were acquired at 30°C using ~0.2 mM 15N/ILV IpaD or 15N/ILV IpaDC322S dissolved in NMR buffer with and without unlabeled IpaB9-226 or IpaB74-224. Two IpaD constructs were used in the NMR titrations reported herein. Native IpaD has a cysteine residue at position 322, thus the NMR buffer also contained 5 mM dithiothreitol (DTT), whereas samples with IpaDC322S did not require DTT in the NMR buffer. The similarity of the 2D 1H-15N TROSY spectra of IpaD and IpaDC322S (Fig. S1) indicated that the C322S mutation did not drastically alter the tertiary structure of IpaD. IpaDC322S was a better-behaved sample in solution with sharper NMR peaks and the sample was more stable over time, hence, IpaDC322S was used in some of the titrations reported herein. The previously reported amide resonance assignments of SipD34 and IpaD35 were used in the NMR analysis. Residues perturbed during the titrations were mapped onto the crystal structure of SipD (PDB ID 3NZZ)13 or IpaD (PDB ID 2J0O).15
Two-dimensional 1H-13C HSQC spectra were acquired for SipDC244S ILV-labeled samples with and without SipB. The ILV 13C-methyl resonances of SipDC244S were assigned following the method of Xiao et al.36 Briefly, 500 μL of ~0.9 mM perdeuterated 15N/ILV SipDC244S in NMR buffer was lyophilized and resuspended in 100% D2O. A 3D 1H-13C-13C HMQC-NOESY-HMQC dataset was acquired using 8 scans with 2048 complex points (1H), 80 complex points (13C) and 100 complex points (NOE 13C) with a 300 ms mixing time and a recycle delay of 2 s. Sweep widths were 10.0 ppm for 1H centered at 4.69 ppm and 20.0 ppm for 13C centered at 19.0 ppm. To aid in the ILV assignment, twelve isoleucine to leucine point mutations were introduced into SipDC244S using Quikchange (Stratagene). The Ile-to-Leu SipDC244S mutants were expressed in M9 media and labeled at the isoleucine 13Cδ1 methyl group and 2D 1H-13C HSQC spectra were acquired to identify the 13Cδ1 methyl peak. The rest of the leucine and valine 13C-methyl peaks were assigned from nuclear Overhauser effects (NOEs) from the 3D dataset in combination with distance information from the crystal structure of SipD (PDB ID 3NZZ).13
Salmonella Invasion Assay
A Salmonella invasion assay was used to test the effect of mutations at the distal end of SipD on the ability of Salmonella typhimurium to invade a cultured human intestinal epithelial cell line (Henle-407 cells, American Type Culture Collection CCL-6) as described previously.13 Wild type (SL1344) and sipD− knockout strains of Salmonella typhimurium were used. Single, double, or triple point mutations were introduced by Quikchange kit (Stratagene) in the plasmid pRK2-SipD,13 which harbored full length sipD. Percent invasiveness was calculated relative to the sipD− knockout strain complemented with the wild type SipD in pRK2-SipD. Error bars were determined from triplicate experiments.
Immunoblotting
Salmonella typhimurium strains with the sipD− knockout and pRK2-SipD (with wild type and mutant SipD) were grown in 15 mL of LB broth containing 0.3 M NaCl and 1 mM IPTG. Cell growth was continued at 37°C at 80 rpm until the OD600 reached 0.9. Bacterial cells were harvested by centrifugation at 4000 rpm for 20 minutes then passed through a 0.45 μm pore-size filter. Proteins from the cultured supernatants were precipitated by incubating with 10% (v/v) trichloroacetic acid at 4°C for 1 hour and recovered by centrifugation at 4000 pm for 40 minutes. Pellets were resuspended in 2 mL 10% (v/v) SDS and 8 mL ice-cold acetone then incubated overnight at −20°C. After centrifugation at 4000 pm for 40 minutes, the pellets were dried and resuspended in 100 μL 8M Urea and 100 μL 2× SDS-loading dye. Proteins from whole-cell lysates (cell pellets) were resuspended in 1 mL 1× PBS and recovered by centrifugation at 13000 rpm for 15 minutes. The pellets were resuspended in 50 μL 1× PBS and 50 μL 2× SDS-loading dye. For immunoblotting, proteins were separated by SDS-PAGE and transferred to a PVDF membrane for 1.5 hours at 70 mA using an ECL Semi-Dry Blotter (Amersham Biosciences). After transfer, the membrane was incubated in 5% (v/v) non-fat dry milk with shaking for 1 hr at room temperature and then incubated with the primary antibody solution (1% non-fat dry milk, 0.1% Tween-20, 1× TBS, rat anti-SipD IgG antibody) with shaking at 4°C overnight. Following this incubation step, the membrane was rinsed three times with wash buffer (0.1% Tween-20, 1× TBS pH 7.4). Membrane was then incubated in the secondary antibody solution (anti-rat IgG antibody conjugated to Alexa Fluor-680 in wash buffer) for 1 hr at room temperature with shaking. The membrane was then rinsed three times with wash buffer. The blot was analyzed using an ODYSSEY Infrared Imaging System (LI-COR Biosciences).
RESULTS
The N-terminal domain of SipB interacts with the distal region of SipD
We used NMR methods to identify the surfaces involved in the tip-translocon interaction by titrating the Salmonella SipB11-232 translocon protein into 15N/ILV labeled SipDC244S and monitoring the titrations by acquiring 2D 1H-15N TROSY spectra. Results of NMR titrations showed a concentration dependent decrease in intensities of many SipDC244S peaks (Fig. 1A) indicating complex formation to be primarily in the intermediate exchange NMR time scale. In addition, some SipDC244S residues, such as N211 (Fig. 1A), G220 and K302, and showed changes in the chemical shift with increasing concentrations of SipB11-232 indicating these residues were in fast exchange NMR time scale. Importantly, there were also many peaks, such as A99, G176, S270, that did not undergo signal broadening, even at a SipDC244S:SipB11-232 ratio of 1:2, indicating that complex formation did not result in protein aggregation, which would have broadened out all the peaks of SipDC244S. To identify the SipDC244S residues that were perturbed the most by SipB11-232, the peak intensity ratio (I1:1/I1:0) was calculated for each non-overlapped SipDC244S peak at a SipDC244S:SipB11-232 molar ratio of 1:1 (Fig. 1B). Residues with peak intensity ratios lower than the average intensity minus one standard deviation were mapped onto the structure of SipD (Fig. 1C). SipDC244S residues that were affected the most by SipB11-232 were primarily hydrophobic (L78, L116, A132, L150, G151, V156, L178, L240, L248, and L326) and some polar (S114, N211, N316) residues. The affected residues clustered in the distal region of SipD (Fig. 1C), suggesting that this region of SipD is involved in the interaction with SipB.
The extreme N-terminal 11-27 residues of IpaB were previously reported to be essential for the interaction with IpaD.23 Residues 11-27 of IpaB are homologous to residues 13–34 of SipB, and this region is present in the SipB11-232 construct used in the NMR titration with SipD shown above (Fig. 1). To test if residues 11–34 of SipB are essential in the interaction with SipD, we used another construct, SipB82-312, that lacked the N-terminal 13–34 residues for titrations with SipDC244S and additionally contained residues 233–312, which are predicted to contain an additional α-helix. Titration of 15N/ILV SipDC244S with unlabeled SipB82-312 resulted in decreased peak intensities of specific SipDC244S peaks (Fig. S2A) indicating that SipB82-312 interacted with SipDC244S despite lacking the N-terminal 13–34 residues. Plot of the peak intensity ratios of (I1:1/I1:0) of the SipDC244S:SipB82-312 complex at 1:1 molar ratio (Fig. S2B) and mapping the results on the structure of SipD showed that the affected residues of SipB82-312 were located at the distal end of SipD (Fig. S2C). Again, many of the residues that were affected were hydrophobic or polar residues, such as L150, L194, V225, Y252 and Q297. There were some differences in the residues affected between the two different SipB constructs, however some residues were affected in both experiments, such as L150, D190 and N211 (Fig. 1B & Fig. S2B). These data suggested that SipDC244S interacted with the N-terminal domain of SipB and that removal of the extreme N-terminal residues (11 to 81) of SipB did not significantly alter the affected SipD surface. Notably, we additionally attempted a titration between 15N/ILV SipDC244S and SipB11-312, but the NMR data was not acquired because sample precipitation was observed even at a SipDC244S: SipB11-312 molar ratio of 1:1.
Assignment of ILV 13C methyl resonances of SipD
Side-chain specific isotope-labeling techniques, such as the 13C methyl groups of isoleucine, leucine and valine (or ILV labeling), are utilized for NMR spectroscopy of high molecular weight proteins. Although ILV probes are useful in studying protein structures and dynamics because of their sharp NMR resonances and high sensitivity,37 they have been sparsely used in studies of the T3SS tip proteins because of the current lack of ILV assignments for this protein family. The 1H-13C HSQC spectra of 15N/ILV SipDC244S showed 114 of 114 predicted methyl ILV resonances (Fig. S3). We report here that all the ILV 13C methyl resonances of SipDC244S were assigned (Fig. S3) through a combination of site-directed mutagenesis of isoleucine to leucine (Fig. S4) and through-space 1H-1H nuclear Overhauser effects (NOEs) observed using 3D HMQC-NOESY of perdeuterated ILV SipDC244S and distance information from the previously reported crystal structure of SipD (Fig. S5).13 These are currently the only available ILV assignments for any T3SS tip protein and should facilitate further studies of molecular interactions of SipD.
Titrations of ILV labeled SipD with SipB11-232
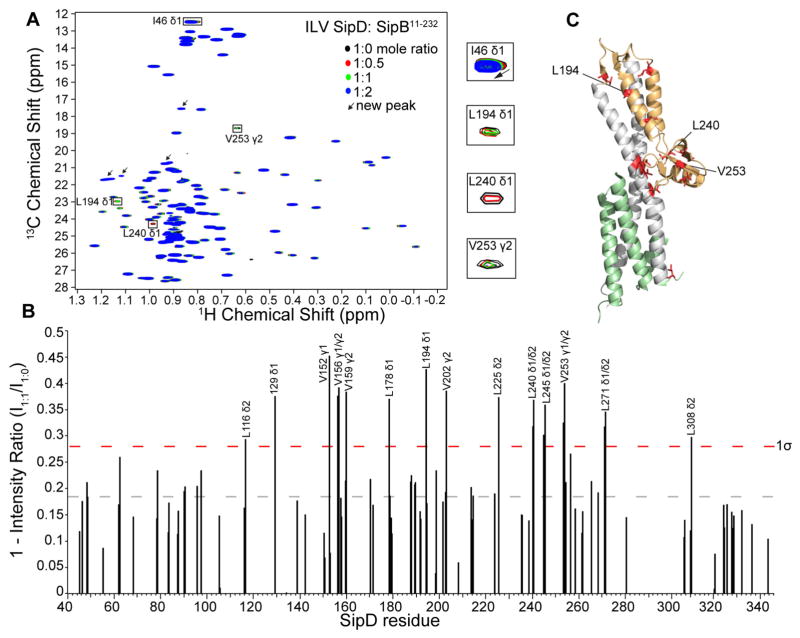
The simultaneous labeling of SipDC244S with 15N and ILV allowed the titration with SipB11-232 to be monitored firstly by 2D 1H-15N HSQC to query the effect of the interaction on the 15N amides of SipDC244S, the results of which are shown above (Fig. 1), and secondly by 2D 1H-13C HSQC to query the effect of SipB11-232 on the ILV methyl resonances of SipDC244S. Like the results of the amide titrations (Fig. 1), most of the affected ILV resonances showed decreased peak intensities with a few residues showed chemical shift deviations, such as I46 (Fig. 2A) and I129. To identify which SipDC244S ILV peaks were perturbed upon complex formation with SipB11-232, the (I1:1/I1:0) peak intensity ratio was plotted as a difference from peak intensity ratio of 1, which signified no change in the peak intensity upon complex formation (Fig. 2B). In agreement with the results of the amide titrations (Fig. 1C), the affected ILV methyl residues clustered at the distal end of SipD (Fig. 2C). Importantly, there were residues that were affected in ILV methyl titrations that were also affected in the amide titrations, such as V156, L178, V225, L240 (Fig. 1B & Fig. 2B). Unlike the 15N titrations, we observed the appearance of new slow-exchange methyl ILV peaks in the SipDC244S 1H-13C HSQC spectra upon titration with SipB11-232 (Fig. 2A, arrows). The new peaks could be due to a conformational change in the side-chain methyls that was not observed in the 15N titrations. Together, the results of ILV methyl and amide titrations are in agreement that the distal region of SipD is the primary surface involved in the interaction with the N-terminal ectodomain of SipB.
Figure 2.
ILV titrations of SipDC244S with SipB11-232. (A) Overlay of four 1H-13C HSQC spectra of 15N/ILV SipDC244S with increasing concentrations of unlabeled SipB11-232. (B) Plot of peak intensities subtracted from one (1 − I1:1/I1:0) of the SipDC244S:SipB11-232 complex at molar ratio of 1.1:1.0 shown with the average value (dashed gray line) and one standard deviation from the average (1σ, dashed red line). (C) Residues (colored red) with (1 − I1:1/I1:0) value higher than 1σ were mapped onto the crystal structure of SipD [colored green (N-terminal hairpin), gray (coiled-coil), and orange (mixed α/β domain)].
The N-terminal domain of IpaB interacts with the distal end of IpaD
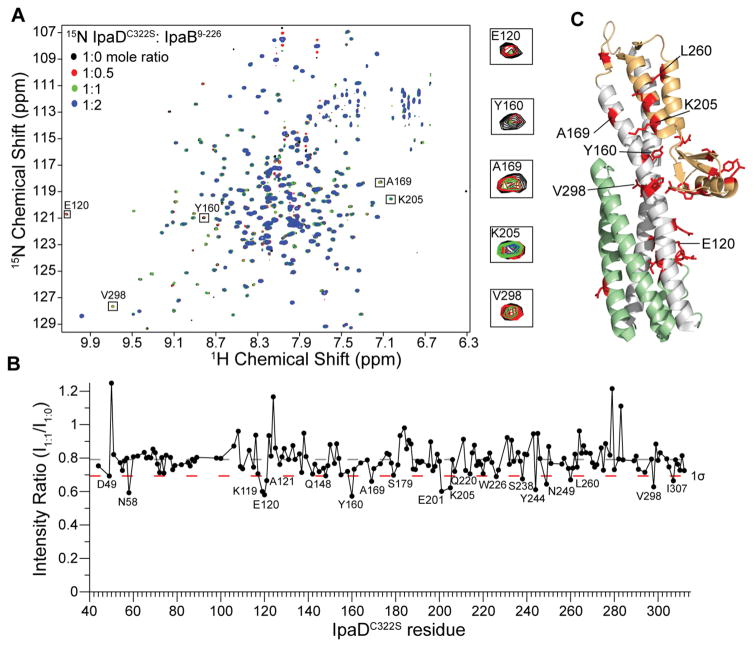
To compare the above results obtained for Salmonella on the interaction between the tip protein and the major translocon protein of Shigella, 15N/ILV labeled IpaD was titrated with unlabeled N-terminal domain of IpaB (residues 9-226). Results of NMR titrations monitored by 2D 1H-15N TROSY showed that, similar to the Salmonella SipD and SipB interaction, stepwise addition of unlabeled IpaB9-226 to IpaDC322S resulted in an IpaB9-226 concentration dependent decrease in peak intensities of specific IpaDC322S amide peaks (Fig. 3A). Importantly, there were residues that did not undergo signal broadening, even at an IpaDC322S:IpaB9-226 ratio of 1:2 indicating complex formation did not result in protein aggregation. The intensity ratio (I1:1/I1:0) for each non-overlapped IpaDC322S residue at an IpaDC322S:IpaB9-226 ratio of 1:1 was plotted to identify the tip residues that were perturbed the most upon addition of IpaB9-226 (Fig. 3B) and mapped onto the crystal structure of IpaD (Fig. 3C). Similar to the results observed for the Salmonella tip-translocon interaction, the affected residues clustered in the distal region of IpaD. IpaDC322S residues that were perturbed the most by IpaB9-226 were primarily hydrophobic, such as Y160, V298 and I307, or polar residues, such as N58, E120 and K205 (Fig. 3B).
Figure 3.
Amide titrations of IpaDC322S with IpaB9-226. (A) Overlay of four 1H-15N TROSY spectra of 15N/ILV IpaDC322S with increasing concentrations of unlabeled IpaB9-226. (B) Plot of ratio of peak intensities of complex (I1:1) vs free (I1:0) for the IpaDC322S:IpaB9-226 complex at 1.1:1.0 molar ratio shown with the average peak intensity ratio (dashed gray line) and one standard deviation from the average (1σ, dashed red line). (C) Residues (colored red) with peak intensity ratio (I1:1/I1:0) lower than 1σ were mapped onto the crystal structure of IpaD [colored green (N-terminal hairpin), gray (coiled-coil), and orange (mixed α/β domain)].
To test if the N-terminal region of IpaB (residues 11 to 27) was essential for the interaction between IpaD and IpaB as reported previously,23 we titrated a shorter IpaB construct lacking the N-terminal region, IpaB74-224, into IpaD. Titration of 15N/ILV IpaD with unlabeled IpaB74-224 resulted in a concentration dependent decrease in peak intensity of many IpaD NMR peaks (Fig. S6A), indicating that IpaB interacted with IpaD despite lacking the N-terminal residues 11–27. Analysis of the peak intensity ratio (I1:1/I1:0) of IpaD:IpaB74-224 complex at 1:1 molar ratio (Fig. S6B) showed that complex formation with IpaB74-224 affected a cluster of IpaD residues at the distal region (Fig. S6C). Many of the IpaD residues that were affected were hydrophobic or charged polar residues, including R55, H131, I145, L195, Q239, M250 and L297 (Fig. S6B). There were some differences in the residues affected between the two different IpaB constructs, however many residues were affected in both experiments, such as E120, K205, S238, L260 and V298 (Fig. 3B & Fig. S6B). Together, these data suggest that, like the Salmonella homologs, IpaD interacts with the N-terminal domain of IpaB and that removal of the extreme N-terminal residues of IpaB did not alter its interaction with IpaD.
The bile salt deoxycholate was reported to be essential for the interaction between IpaD and IpaB.23 To test this, we titrated 15N labeled IpaDC322S with unlabeled and IpaB9-226 in the presence of 0.4 mM deoxycholate. Like the titration in the absence of deoxycholate (Fig. 3), we observed a concentration dependent decrease in the peak intensity of IpaDC322S NMR peaks (Fig. S7A). Analysis of the peak intensity ratio of IpaDC322S:IpaB9-226 complex in the presence of deoxycholate complex at a 1:1:2 molar ratio (Fig. S7B) showed that the affected IpaDC322S residues were primarily clustered at the distal region (Fig. S7C), similar to the results of the IpaDC322S:IpaB9-226 complex in the absence of deoxycholate (Fig. 3C). IpaD residues that were affected in both the absence and presence of deoxycholate included Q148, Y160, Q220, W226 and I307 (Fig. 3B & Fig. S7B). Together, this suggests that deoxycholate did not largely alter the affected IpaD and IpaB interaction surface or modulate the protein affinity as observed by NMR.
Mutations in the distal end of SipD reduce invasion of Salmonella into human epithelial cells
To correlate the biological relevance of our NMR titration results, we tested the effect of mutations along the distal region of SipD on the ability of Salmonella to invade human intestinal epithelial Henle-407 cells using a previously reported Salmonella invasion assay.13 Deletion of the sipD gene rendered Salmonella completely non-invasive (Fig. 4A). However, invasiveness could be rescued by complementing the sipD− null Salmonella strain with a plasmid expressing exogenous wild-type sipD (Fig. 4B). To test the significance of the distal end of SipD in invasion, point mutations were introduced into the mixed α/β region of the SipD rescue plasmid (Fig. 4B) and incorporated into the sipD− strain. Point mutations either moderately reduced invasion (N196D and V191D) or had no effect (L271D and L280D) (Fig. 4A). Because protein-protein interactions often occur across large surfaces,38 double or triple mutations in SipD were constructed (Fig. 4B). The double and triple mutations dramatically reduced the invasiveness of Salmonella (Fig. 4A). All mutant recombinant SipD showed similar protein fold to wild-type SipD based on circular dichroism (data not shown). Immunoblotting using anti-SipD antibodies of whole-cell lysates and supernatant showed that all the twelve SipD contructs used in the invasion assay were expressed in Salmonella, except for one triple mutant, N196D+V265D+L271D (Fig. S8). Thus, the decreased invasiveness of the SipD mutants (Fig. 4) could be ascribed to loss of function, except for one triple mutant, N196D+V265D+L271D, which was due to lost of protein expression (Fig. S8). These results suggest that the distal region of SipD is required for invasion and mutations that alter the surface of this region render Salmonella non-invasive.
Figure 4.
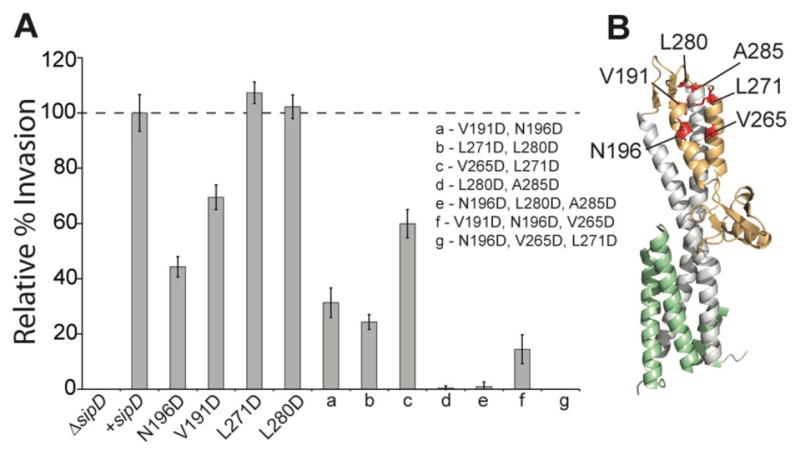
Mutations in the distal region of SipD decrease invasion of Salmonella. (A) Deletion of sipD (ΔsipD) renders Salmonella non-invasive to Henle-407 human intestinal epithelial cells. Invasion was rescued by complementing with a plasmid containing wild-type sipD. Point mutations in the mixed α/β domain of SipD reduced invasion (N196D and V191D) or had no effect (L271D and L280D). Double and triple mutations decreased invasion even further. (B) The residues chosen for mutation are shown in red on the structure of SipD.
DISCUSSION
The interaction of the tip protein and the major translocon protein is essential for the assembly of the T3SS needle apparatus. Dickenson et al.23 showed by FRET and fluorescence polarization that the Shigella IpaD tip protein interacts with the N-terminal ectodomain of the IpaB major translocon protein. Here, we used NMR methods to identify that the surfaces of the tip protein at the distal region, which includes the upper portion of the coiled-coil and the mixed α/β domain (Fig. 5C), are the sites of protein-protein interaction with the major translocon protein. The surfaces involved in the interaction are similar between SipD and SipB in Salmonella (Fig. 1 & Fig. 2) and IpaD and IpaB in Shigella (Fig. 3). Results of both amide (Fig. 1 & Fig. 3) and ILV titrations (Fig. 2) showed similar surfaces are involved in the interactions. Furthermore, in Salmonella the SipD residues affected were primarily hydrophobic and some uncharged polar residues (Fig. 1C), while in Shigella the IpaD residues affected were primarily charged polar and some hydrophobic residues (Fig. 3C). Changes in the NMR peaks brought about by protein-protein interaction were primarily in the intermediate exchange time scale, suggesting dissociation constant near the μM range (Fig. 1 & Fig. 3), which is in agreement with the reported Kd in the low μM range obtained by fluorescence spectroscopy.23 Our results are in agreement with the proposed interaction surface for the tip and the major translocon protein identified by paramagnetic relaxation enhancement.39 Additional data presented here show the role of deoxycholate and of the extreme N-terminal residues of the major translocon proteins in the interaction with the tip proteins; provides ILV methyl assignments of SipD; and provides biological invasion assay to test the proposed model of the tip-translocon interaction.
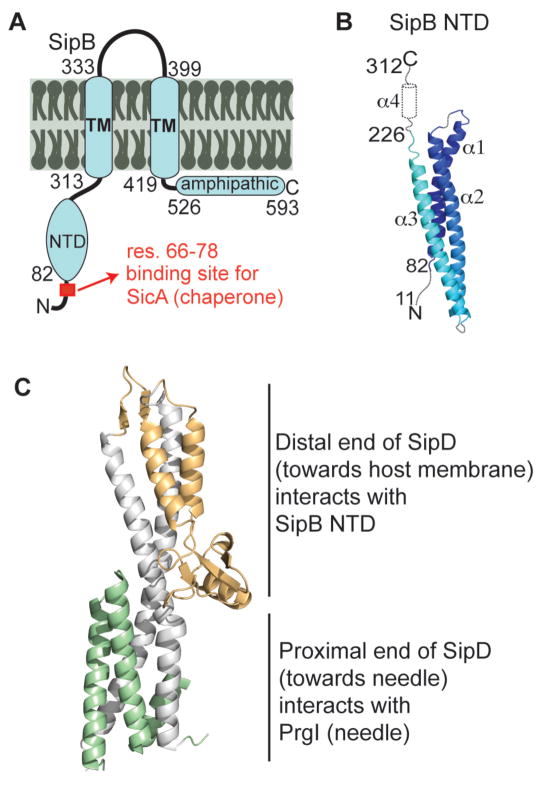
Figure 5.
Proposed model of the needle, tip and translocon interface. (A) Membrane topology of SipB showing the N-terminal domain (NTD). (B) Structure of the SipB N-terminal domain (82–226) with predicted secondary structure of regions of unknown structure present in our SipB constructs. (C) Model for the interaction hubs on the tip protein SipD, where the N-terminal domain of the translocon SipB interacts with the mixed α/β domain and the needle PrgI in interacts with the bottom of the coiled-coiled. The model of the Shigella IpaD and IpaB is expected to be homologous.
In contrast to the previous report that the bile salt deoxycholate is required for the IpaD-IpaB1-226 interaction,23 we observed IpaD-IpaB interaction in the absence of deoxycholate for the NMR titrations of IpaD-IpaB9-226 (Fig. 3) and IpaD-IpaB74-224 (Fig. S6). Bile salts such as deoxycholate are secreted in the intestinal tract as part of the digestive system and they affect the type III secretion of the enteric bacteria Shigella40,41 and Salmonella.42,43 Deoxycholate binds to IpaD35,41 and SipD41,44, and is hypothesized to function as an environmental sensor for the T3SS by inducing conformational change in IpaD that leads to latter steps in the assembly of the translocon.35 It has been proposed that the interaction of deoxycholate with IpaD induces a conformational change at the distal region of IpaD which then promotes binding to IpaB.23 We suspect that the difference in the results of fluorescence and NMR methods in observing IpaD-IpaB interaction with respect to the absence of deoxycholate lie in the concentrations of proteins used in the titrations. NMR requires 2–3 orders of magnitude more concentrated protein samples compared to fluorescence spectroscopy. Fluorescence polarization used 80 nM of fluorescein-labeled IpaB titrated with up to 10 μM of IpaD,23 whereas our NMR experiments used 0.2 mM IpaD titrated with up to 0.4 mM IpaB. Notably, we observed that the IpaD surface affected upon titration with IpaB was similar in the absence (Fig. 3C) and in the presence of 0.4 mM deoxycholate , or a 1:1:2 ratio of IpaD to IpaB to deoxycholate (Fig. S7C). Increasing the concentration of deoxycholate up to 1 mM, the concentration used by Dickenson et al.,23 resulted in sample precipitation.
Our results combined with previous data showing that the bottom of the coiled-coil of the tip protein is the site of protein-protein interactions with the needle protein16 leads to a model of the needle-tip-translocon interface. We propose a model of the needle-tip-translocon interface (Fig. 5) where the distal region of SipD interacts with the N-terminal ectodomain of SipB and the proximal end of SipD interacts with the needle PrgI. The in vivo stoichiometry of the tip and translocon complex is poorly understood. However, one current hypothesis is that the tip complex either forms a homopentameric complex with five IpaD or heteropentameric complex consisting of one copy of IpaB and four copies of IpaD.9 Similarly, the Salmonella system is hypothesized to be composed to five SipD copies interacting with five PrgI needle subunits.16 Our NMR results that the major translocon protein interacts with the distal region of the tip protein agree with the recent model of the Shigella tip-translocon complex derived by electron microscopy and cross-linking of IpaD and IpaB.9 Our results are also in agreement with a recently published topology of the assembled major translocon protein AopB (a homolog of SipB and IpaB) from Aeromonas hydrophila.29 Nguyen et al.29 show that both the N and C-termini of AopB are extracellularly exposed when the translocon is assembled and therefore the N-terminal ectodomain of the major translocon protein is available for interaction with the tip protein.29
CONCLUSIONS
We report here that the major translocon protein interacts with the distal surface of the tip protein in both Salmonella and Shigella. Mutations along the distal surface of the tip protein reduced the ability of Salmonella to invade intestinal epithelial cells, confirming the biological relevance of the distal surface of the tip protein. We also report the ILV methyl assignments of SipD and utilized the assignments in our interaction study to complement amide experiments (Fig. 2). Further structural studies are needed to determine which surface of the major translocon protein is affected upon interaction with the tip protein, whether the minor translocon protein is involved in the interaction with the tip and to build an atomic detail model of the assembled tip-translocon interface.
Data Deposition
NMR assignment of SipDC244S ILV methyl resonances have been deposited into the BioMagResBank with the accession number 26739.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI074856 (R.N.D.), P30GM110761 (KU Bio NMR Facility), T32-GM008359 (A.C.M.) and NSF REU 1156856 (K.M.K.). We are grateful to Asokan Anbanandam for assistance with NMR spectroscopy.
References
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM International Collaboration on Enteric Disease 'Burden of Illness S. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 3.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee S, Chaudhury S, McShan AC, Kaur K, De Guzman RN. Structure and biophysics of type III secretion in bacteria. Biochemistry. 2013;52:2508–2517. doi: 10.1021/bi400160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demers JP, Habenstein B, Loquet A, Kumar Vasa S, Giller K, Becker S, Baker D, Lange A, Sgourakis NG. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nature communications. 2014;5:4976. doi: 10.1038/ncomms5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung M, Shen DK, Makino F, Kato T, Roehrich AD, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ. Three-dimensional electron microscopy reconstruction and cysteine-mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol. 2015;95:31–50. doi: 10.1111/mmi.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epler CR, Dickenson NE, Bullitt E, Picking WL. Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol. 2012;420:29–39. doi: 10.1016/j.jmb.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roehrich AD, Guillossou E, Blocker AJ, Martinez-Argudo I. Shigella IpaD has a dual role: signal transduction from the type III secretion system needle tip and intracellular secretion regulation. Mol Microbiol. 2013;87:690–706. doi: 10.1111/mmi.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell SW, De Guzman RN. The crystal structure of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barta ML, Guragain M, Adam P, Dickenson NE, Patil M, Geisbrecht BV, Picking WL, Picking WD. Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins. 2012;80:935–945. doi: 10.1002/prot.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathinavelan T, Lara-Tejero M, Lefebre M, Chatterjee S, McShan AC, Guo DC, Tang C, Galan JE, De Guzman RN. NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J Mol Biol. 2014;426:2958–2969. doi: 10.1016/j.jmb.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myeni SK, Wang L, Zhou D. SipB-SipC complex is essential for translocon formation. PLoS One. 2013;8:e60499. doi: 10.1371/journal.pone.0060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller CA, Broz P, Cornelis GR. The type III secretion system tip complex and translocon. Mol Microbiol. 2008;68:1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- 19.Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 20.Obregon C, Dreher D, Kok M, Cochand L, Kiama GS, Nicod LP. Human alveolar macrophages infected by virulent bacteria expressing SipB are a major source of active interleukin-18. Infect Immun. 2003;71:4382–4388. doi: 10.1128/IAI.71.8.4382-4388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickenson NE, Arizmendi O, Patil MK, Toth RTt, Middaugh CR, Picking WD, Picking WL. N-terminus of IpaB provides a potential anchor to the Shigella type III secretion system tip complex protein IpaD. Biochemistry. 2013;52:8790–8799. doi: 10.1021/bi400755f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim BH, Kim HG, Kim JS, Jang JI, Park YK. Analysis of functional domains present in the N-terminus of the SipB protein. Microbiology. 2007;153:2998–3008. doi: 10.1099/mic.0.2007/007872-0. [DOI] [PubMed] [Google Scholar]
- 25.McGhie EJ, Hume PJ, Hayward RD, Torres J, Koronakis V. Topology of the Salmonella invasion protein SipB in a model bilayer. Mol Microbiol. 2002;44:1309–1321. doi: 10.1046/j.1365-2958.2002.02958.x. [DOI] [PubMed] [Google Scholar]
- 26.Hume PJ, McGhie EJ, Hayward RD, Koronakis V. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol Microbiol. 2003;49:425–439. doi: 10.1046/j.1365-2958.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 27.Hayward RD, McGhie EJ, Koronakis V. Membrane fusion activity of purified SipB, a Salmonella surface protein essential for mammalian cell invasion. Mol Microbiol. 2000;37:727–739. doi: 10.1046/j.1365-2958.2000.02027.x. [DOI] [PubMed] [Google Scholar]
- 28.Barta ML, Dickenson NE, Patil M, Keightley A, Wyckoff GJ, Picking WD, Picking WL, Geisbrecht BV. The structures of coiled-coil domains from type III secretion system translocators reveal homology to pore-forming toxins. J Mol Biol. 2012;417:395–405. doi: 10.1016/j.jmb.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen VS, Jobichen C, Tan KW, Tan YW, Chan SL, Ramesh K, Yuan Y, Hong Y, Seetharaman J, Leung KY, Sivaraman J, Mok YK. Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly. Structure. 2015;23:2022–2031. doi: 10.1016/j.str.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Rathinavelan T, Tang C, De Guzman RN. Characterization of the interaction between the Salmonella type III secretion system tip protein SipD and the needle protein PrgI by paramagnetic relaxation enhancement. J Biol Chem. 2011;286:4922–4930. doi: 10.1074/jbc.M110.159434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Alminaite A, Vaheri A, Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 2010;151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. Conformational Changes in IpaD from Shigella flexneri upon Binding Bile Salts Provide Insight into the Second Step of Type III Secretion. Biochemistry. 2011;50:172–180. doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Y, Lee T, Latham MP, Warner LR, Tanimoto A, Pardi A, Ahn NG. Phosphorylation releases constraints to domain motion in ERK2. Proc Natl Acad Sci U S A. 2014;111:2506–2511. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto NK, Kay LE. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr Opin Struct Biol. 2000;10:585–592. doi: 10.1016/s0959-440x(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 38.Chen JM, Sawyer N, Regan L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur K, Chatterjee S, De Guzman RN. Characterization of the Shigella and Salmonella type III secretion system tip-translocon protein-protein interaction by paramagnetic relaxation enhancement. Chembiochem : a European journal of chemical biology. 2016 doi: 10.1002/cbic.201500556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 2008;283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect Immun. 2000;68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol Med Microbiol. 2004;41:177–185. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Nordhues BA, Zhong D, De Guzman RN. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry. 2010;49:4220–4226. doi: 10.1021/bi100335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.