Abstract
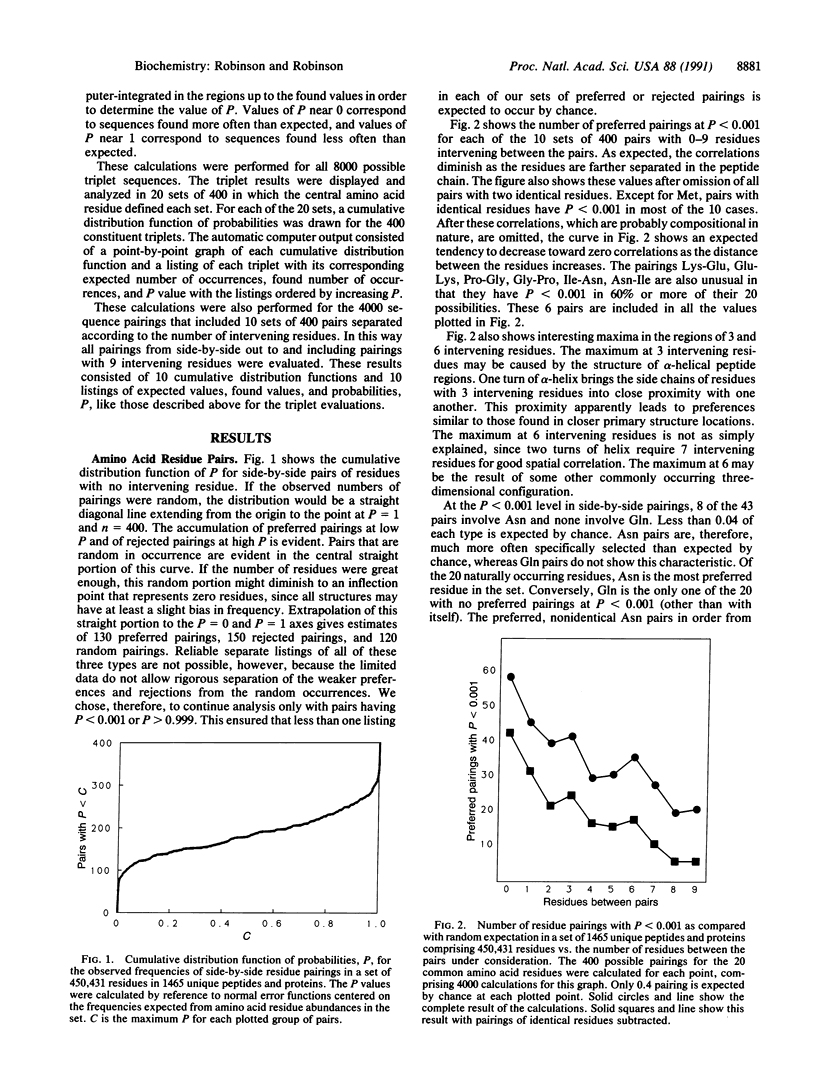
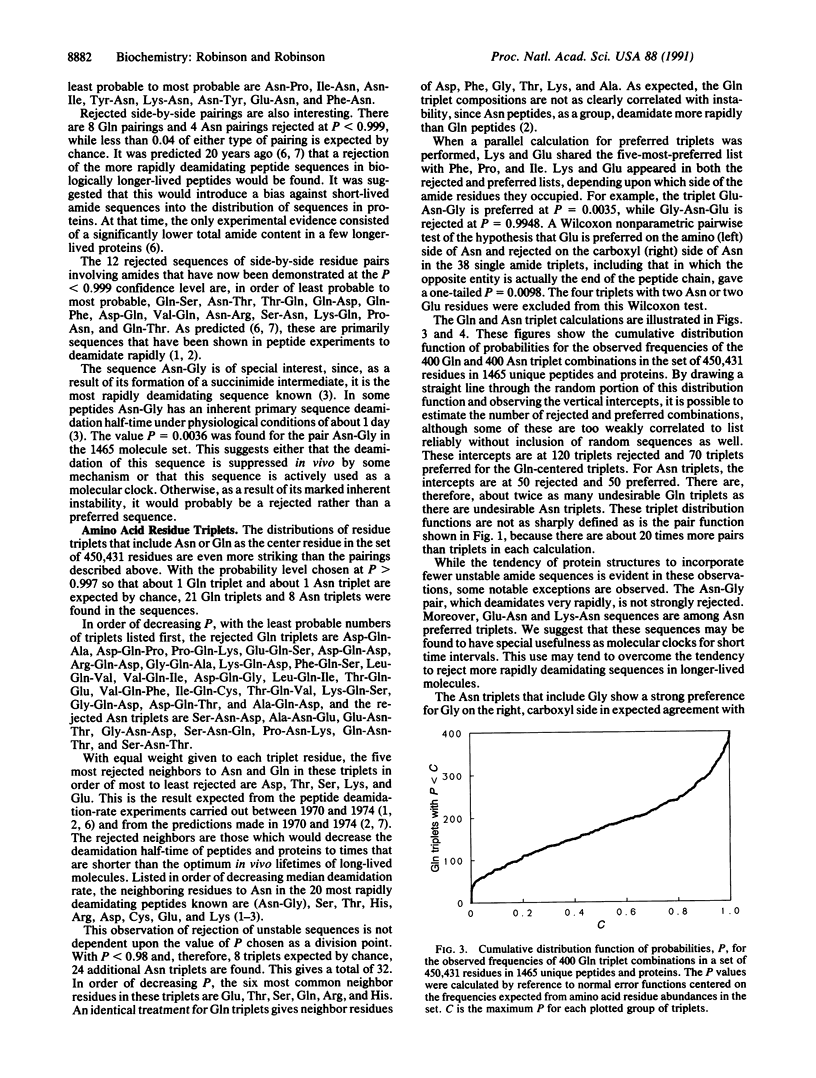
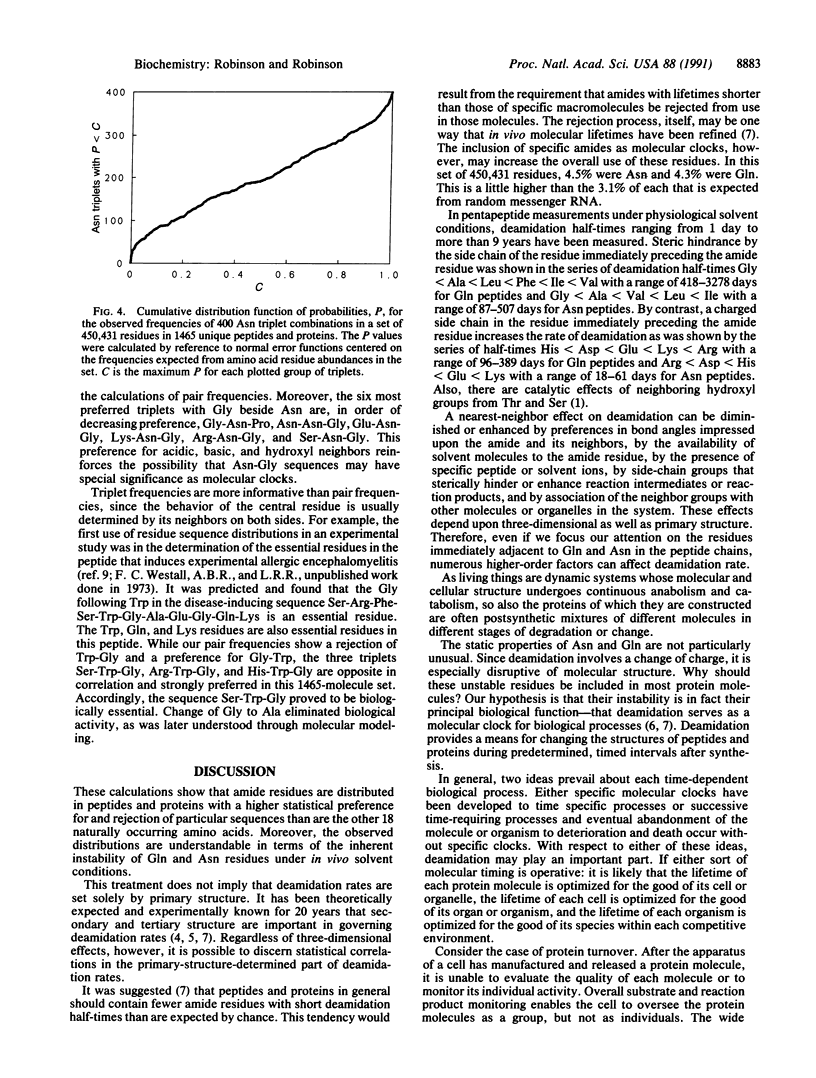
In a statistical study of neighboring residues in 1465 peptides and proteins comprising 450,431 residues, it was found that the preferences for residues neighboring to glutamine and asparagine residues are consistent with the hypothesis that the rates of deamidation of these residues are of biological significance. Some dipeptide and tripeptide structures have special usefulness and some are especially undesirable. More such structures exist for amide residues than for other residues, and their specific types are those most relevant to the deamidation of amide residues under biological conditions.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Flatmark T., Sletten K. Multiple forms of cytochrome c in the rat. Precursor-product relationship between the main component Cy I and the minor components Cy II and Cy 3 in vivo. J Biol Chem. 1968 Apr 10;243(7):1623–1629. [PubMed] [Google Scholar]
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987 Jan 15;262(2):785–794. [PubMed] [Google Scholar]
- Robinson A. B. Evolution and the distribution of glutaminyl and asparaginyl residues in proteins. Proc Natl Acad Sci U S A. 1974 Mar;71(3):885–888. doi: 10.1073/pnas.71.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. B., McKerrow J. H., Cary P. Controlled deamidation of peptides and proteins: an experimental hazard and a possible biological timer. Proc Natl Acad Sci U S A. 1970 Jul;66(3):753–757. doi: 10.1073/pnas.66.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. B., McKerrow J. H., Legaz M. Sequence dependent deamidation rates for model peptides of cytochrome C. Int J Pept Protein Res. 1974;6(1):31–35. doi: 10.1111/j.1399-3011.1974.tb02355.x. [DOI] [PubMed] [Google Scholar]
- Robinson A. B. Molecular clocks, molecular profiles, and optimum diets: three approaches to the problem of aging. Mech Ageing Dev. 1979 Feb;9(3-4):225–236. doi: 10.1016/0047-6374(79)90101-5. [DOI] [PubMed] [Google Scholar]
- Robinson A. B., Rudd C. J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr Top Cell Regul. 1974;8(0):247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- Robinson A. B., Scotchler J. W., McKerrow J. H. Rates of nonenzymatic deamidation of glutaminyl and asparaginyl residues in pentapeptides. J Am Chem Soc. 1973 Nov 28;95(24):8156–8159. doi: 10.1021/ja00805a032. [DOI] [PubMed] [Google Scholar]
- Westall F. C., Robinson A. B., Caccam J., Jackson J., Ylar E. H. Essential chemical requirements for induction of allergic encephalomyelitis. Nature. 1971 Jan 1;229(5279):22–24. doi: 10.1038/229022a0. [DOI] [PubMed] [Google Scholar]



