Abstract
Antibiotic resistance in pathogens requires new targets for developing novel antibacterials. The bacterial type III secretion system (T3SS) is an attractive target for developing antibacterials as it is essential in the pathogenesis of many Gram-negative bacteria. The T3SS consists of structural proteins, effectors and chaperones. Over 20 different structural proteins assemble into a complex nanoinjector that punctures a hole on the eukaryotic cell membrane to allow the delivery of effectors directly into the host cell cytoplasm. Defects in the assembly and function of the T3SS render bacteria non-infective. Two major classes of small molecules, salicylidene acylhydrazides and thiazolidinones, have been shown to inhibit multiple genera of bacteria through the T3SS. Many additional chemically and structurally diverse classes of small molecule inhibitors of the T3SS have been identified as well. While specific targets within the T3SS of a few inhibitors have been suggested, the vast majority of specific protein targets within the T3SS remain to be identified or characterized. Other T3SS inhibitors include polymers, proteins and polypeptides mimics. In addition, T3SS activity is regulated by its interaction with biologically relevant molecules, such as bile salts and sterols, which could serve as scaffolds for drug design.
Keywords: type III secretion system, antibiotics, antibiotic resistance, small molecule inhibitors, virulence, salicylidene acylhydrazides, thiazolidinones
Introduction
Pathogenic Gram-negative bacteria pose a significant global health impact with an estimated 2 million infections and 23,000 deaths per year in just the United States (1). Examples of these organisms include Pseudomonas, Shigella, Salmonella, Chlamydia, enteropathogenic E. coli (EPEC), and Yersinia. The appearance and rapid evolution of multidrug resistant strains has become of great concern for public health (2–4). Unfortunately, the development of new antibiotics presents a difficult challenge. Since 2009 three antibiotics targeting Gram-negative bacteria, though not exclusively, have been approved as the deadline approaches for the initiative of the Infectious Diseases Society of America’s for at least 10 new antibiotics by 2020 (5). The rate of entry of new antibiotics into the pipeline is extremely slow (6–9). This is largely due to several factors, namely, (i) the lack of novel antibiotic targets (10), (ii) high throughput screens often turn up known targets or novel targets that do not make it past early stages of drug development due to toxicity or off target effects (11), and (iii) the disinterest of big pharmaceutical companies to discover new antibiotics or conduct clinical trials due the problem of antibiotic resistance and poor investment return has only exacerbated the situation (11, 12). The rapid emergence of multidrug resistant strains coupled with the dearth of novel antibiotics suggests a need for identifying novel targets for development of antibiotics.
Traditional antibiotics often fall into two classes: bactericidal compounds that cause cell death and/or bacteriostatic compounds that inhibit cellular growth (11). In either case, these drugs often induce a selection pressure on bacteria to develop drug resistance, which is usually obtained via horizontal gene transfer between bacteria or by de novo mutations (11). Targeting virulence pathways of pathogenic bacteria has been suggested as an alternative strategy (13, 14). One current theory is that the use of antivirulence or anti-infective drugs, in contrast to antibiotics, will dampen the selection pressure for the emergence of resistant strains because these drugs do not directly harm the organism (15, 16). Notably, there have been documented cases of resistance to antivirulence drugs, though it has been argued that the existence of such mechanisms for resistance does not suggest it will become a problem in a clinical setting (15). Another advantage is that because virulence mechanisms are used by pathogenic bacteria, antivirulence drugs are hypothesized to have less of an influence on the host commensal flora when compared to traditional broad spectrum antibiotics (17).
The Type III Secretion System – Multiple Targeting Opportunities
Overview of the T3SS
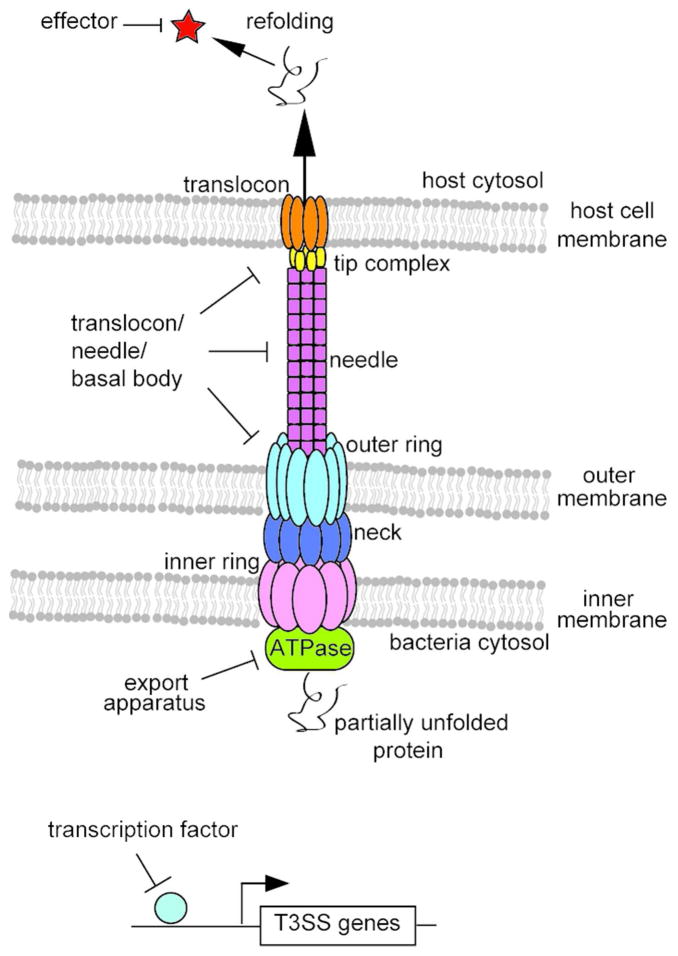
The type III secretion system functions as a conduit for delivery of virulence factors by translocating proteins from the bacterial cytoplasm into the eukaryotic host cell cytoplasm to facilitate infection (18). The structural component of the T3SS, the needle complex, was first visualized by Galan & coworkers in 1998 (19) and since then the structures and functions of many T3SS proteins have been elucidated [reviewed in (20–23)]. T3SS proteins are highly homologous in sequence, structure and function among different bacteria (20, 24, 25). Therefore, protein-protein interactions within the system among different bacteria and with host cells are thought to occur through similar mechanisms. This theory is supported by the similarities observed between the assembly of the Salmonella and Shigella T3SS needles (26) and conserved structural motifs within the basal structure (27). Importantly, disruption of many aspects of the T3SS often abolishes pathogenicity. For these reasons, the potential of using the T3SS as a pseudo-broad spectrum antivirulence target is of great interest (13). As the T3SS is complex, there are many different potential targeting strategies relating to various aspects of the system, which are outlined in Figure 1 and described below.
Figure 1.
Potential Targeting Strategies for T3SS Inhibitors. Cartoon showing potential targeting strategies of the T3SS, including the T3SS apparatus, effector proteins and transcription factors.
The T3SS needle apparatus
The T3SS needle apparatus is made up of around 25 different proteins that assemble together to regulate and facilitate the secretion of effector proteins into host cells (21). Together, the membrane-embedded export apparatus controls the secretion of proteins and anchors the apparatus into the bacterial membrane (28). An ATPase provides recognition for chaperone/effector complexes and is thought to provide energy for insertion and unfolding of effector proteins into the apparatus (29). The needle provides a ~25Å diameter extracellular channel for the secretion of partially unfolded effector proteins (26, 28). The tip complex regulates secretion and is a scaffold for translocon assembly (30). Finally, the translocon creates a pore in host cell membrane (31). Many subsections of the apparatus, such as the tip, needle, inner ring and outer ring, are assembled by oligomers of a single protein whose affinities are governed by the sum of weak protein-protein interactions (32). Furthermore, the structures of many T3SS proteins are similar between different organisms (20). Two possible modes of targeting the needle apparatus directly could be protein-protein interactions within each component parts such as the needle monomer interactions or interactions between subsections such as the tip-translocon interaction (Figure 1).
Salmonella contains genetic loci that encode three T3SS operons with distinct functions. The Salmonella pathogenicity island-1 (SPI-1) encoded T3SS is the most studied and functions primarily in the initial invasion of non-phagocytic cells (33, 34). The SPI-2 T3SS is involved in the maintenance of Salmonella containing vacuoles and bacterial replication within host cells, though it’s structure has not been as extensively characterized (35). In addition, there is a flagellar T3SS (24). Other T3SS families include the Ysc and Ysa T3SS of Yersinia and the Sct T3SS of Chlamydia (36). Since part of the T3SS needle apparatus is exposed to the extracellular environment prior to and during infection, disrupting its assembly is a potential target for developing inhibitors.
Chaperones and effectors
T3SS effectors have a wide range of functions within the host cell, but often involve manipulating host cell signaling, secretory trafficking, cytoskeletal dynamics, or the inflammatory response (23, 37). It has been shown that effectors work in concert for infection (38). Regardless, deletions of many effectors attenuate virulence so they are considered potential drug targets (39–41). While in the bacterial cytoplasm, effectors are often in complex with chaperone proteins that target them to the export apparatus and protect from aggregation (42, 43). Disrupting chaperone/effector interactions could prevent secretion of effector proteins. However, some effectors, such as YopE and SopE, have been shown to retain partial secretion even the absence of their chaperone binding domain (43, 44). Therefore, disruption of the chaperone/effector interactions may not necessarily result in a decrease in infectivity.
T3SS gene transcription
As stated above, the T3SS is organized into genetic loci whose gene expression is highly regulated by various transcription factors. Transcriptional regulators of both SPI-1 (45) and SPI-2 (46) loci have been identified in Salmonella. T3SS transcriptional regulators have also been characterized in other bacteria, such as Pseudomonas (47), Yersinia (48) and Shigella (49). Deletions of various T3SS transcription factors lead to the disappearance of the needle apparatuses on the bacterial surface due to essential constituents not being expressed (50). For this reason, transcription factors are potential drug targets as well.
Literature of T3SS inhibitors
Salicylidene acylhydrazides
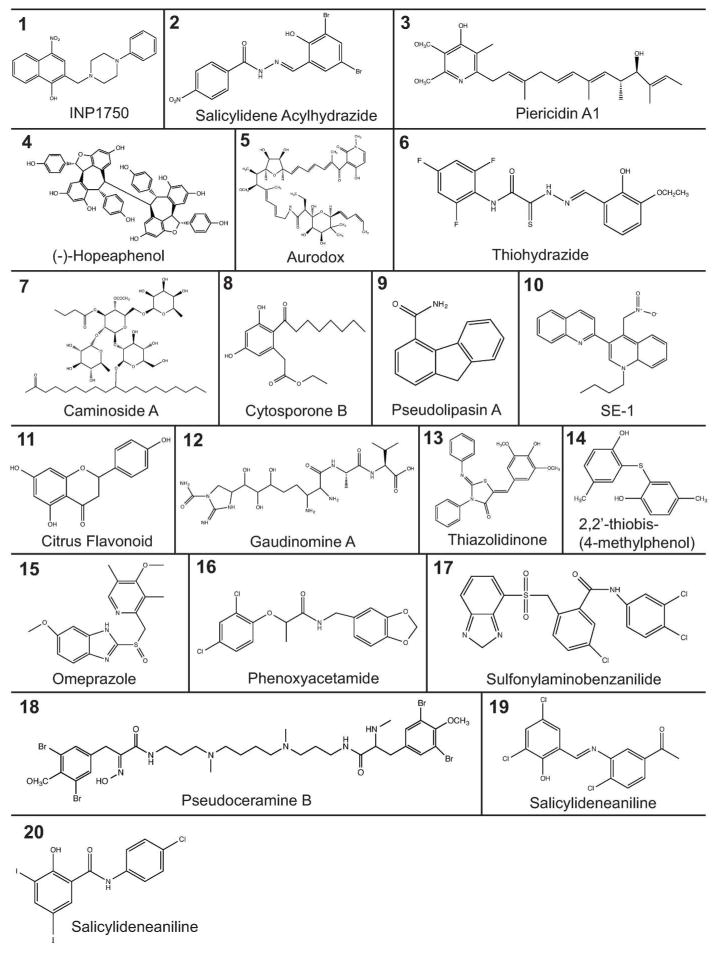
Many different classes of structurally diverse small molecule compounds have been identified as inhibitors of the T3SS (Figure 2 and Table 1). The most well studied class of T3SS inhibitors is the salicylidene acylhydrazides (SAHs). One of the first reports comes from Kauppi et al. (51, 52) where they performed a whole-cell assay screen against T3SS gene expression using a reporter of the effector protein YopE fused with luciferase in Yersinia. Out of 9,400 compounds tested, a few compounds were identified, including SAHs. A disadvantage of the assay was that it was based on coupling between Yop effector expression and secretion of the negative regulator LcrQ, therefore it was unclear whether the compounds acted directly against the T3SS (51). A follow up study showed that their SAH compounds directly blocked effector secretion of the T3SS in a dose-dependent manner and they were suggested to act at the level of the T3SS machinery (52, 53). SAHs have been shown to broadly inhibit the T3SS of many bacteria genera, including Chlamydia (54–57), Shigella (58), Salmonella (59–63) and pathogenic E. coli (64–66). Notably, most SAHs have been shown to have no negative affect on bacterial growth (51, 55, 61). Interestingly, some SAHs derivatives inhibited bacterial motility by acting on the flagella (51, 59, 60), while others did not affect bacterial motility (64).
Figure 2.
Structure of T3SS Inhibitors. Structure of the classes of T3SS inhibitors outlined in Table 1. Structures were made using ChemBioDraw 13.0.
Table 1.
Chemical inhibitors of the T3SS and their putative targets.
| Compound | Structure # (Figure 2) | Organism | T3SS Target | Reference |
|---|---|---|---|---|
|
| ||||
| (−)-Hopeaphenol | 4 | Yersinia, Pseudomonas, Chlamydia | ? | (123) |
|
| ||||
| 1-butyl-4-nitromethyl-3-quinolin-2-yl-4H-quinoline [SE-1] | 10 | Shigella | VirF / transcription factor | (79) |
|
| ||||
| 2-imino-5-arylidene thiazolidinone | 13 | Yersinia, Salmonella also inhibits T2S of Pseudomonas and T4-Pilli Secretion of Francisella | Secretin? | (71, 72, 124) |
|
| ||||
| 2,2′-thiobis-(4-methylphenol) | 14 |
Yersinia Pseudomonas |
YopD / translocon | (73) |
|
| ||||
| 8-hydroxyquinolines [INP1750] | 1 | Yersinia, Chlamydia | ? | (119) |
|
| ||||
| Aurodox | 5 | EPEC, Citrobacter | ? | (84) |
|
| ||||
| Benzimidazole | Yersinia | LcrF / transcription factor | (76) | |
| Pseudomonas | ExsA / transcription factor | (77, 78) | ||
|
| ||||
| Caminoside A-D | 7 | EPEC | ? | (125, 126) |
|
| ||||
| Citrus Flavonoids | 11 | Vibrio | ? | (87) |
|
| ||||
| Cytosporone B | 8 | Salmonella | ? | (86) |
|
| ||||
| Gaudinomine A-D | 12 | EPEC | ? | (85) |
|
| ||||
| Omeprazole | 15 | Salmonella | ATPase inhibitor, effects through nitric oxide production | (62, 81) |
|
| ||||
| Phenoxyacetamide | 16 | Pseudomonas | PscF / needle protein | (74, 75) |
|
| ||||
| Piericidin A1 | 3 | Yersinia | ? | (127) |
|
| ||||
| Pseudoceramine | 18 | Yersinia | ? | (128) |
|
| ||||
| Pseudolipasin A | 9 | Pseudomonas | ExoU / effector | (10) |
|
| ||||
| Salicylanilide | 20 | Yersinia | ? | (51, 88, 129) |
|
| ||||
| Salicylidene Acylhydrazide |
2 | Chlamydia | HemG / heme metabolism | (54–57, 120, 130) |
| Yersinia | ? | (51, 52) | ||
| Shigella | needle assembly? | (58) | ||
| Salmonella | FlhA / flagellar inner membrane protein | (59–63) | ||
| EPEC | ?, possibly due to non-T3SS cell metabolism enzymes, such as WrbA/Tpx/FolX | (64–66) | ||
|
| ||||
| Salicylideneaniline | 19 | Yersinia, EPEC | ? | (64) |
|
| ||||
| Sulfonylaminobenzanilide | 17 | Yersinia | ? | (51, 53) |
|
| ||||
| Thiohydrazones of Thiohydrazide | 6 | Chlamydia | ? | (131) |
|
| ||||
| Various Compounds | not shown | Yersinia, EPEC | ? | (132, 133) |
|
| ||||
| Various Compounds | not shown | Yersinia | ? | (83) |
|
| ||||
| Various Compounds | not shown |
Yersinia Burkholderia |
YscN / ATPase BsaS / ATPase |
(80) |
|
| ||||
| Viscous Polymers [PEG8000 / Alginate / Mucin] | not shown | Pseudomonas | inhibits T3SS apparatus rotation | (90) |
|
| ||||
| Lactoferrin | not shown | Shigella, EPEC, Salmonella | degradation of tip and translocon proteins | (97) |
|
| ||||
| Polypeptide Mimics | not shown | Salmonella/Shigella | SipB, IpaB / Translocon | (134) |
| EPEC | CdsN ATPase | (98) | ||
| EspA / tip | (100) | |||
The specific targets within the T3SS itself of many SAHs remain unknown or ambiguous, though putative targets have been suggested. First, it has been suggested that these compounds target the formation or assembly of the SPI-1 needle apparatus directly (58). Veenendaal et al. (58) showed by electron microscopy that Shigella needles were reduced in number by ~40% and the distribution of observed needle lengths was altered by one compound. Martinez-Argudo et al. (59) isolated SAH resistant Salmonella strains with a mutation in FlhA, a flagellar inner membrane protein, suggesting the compounds target the conserved basal body. Second, it has been suggested that SAHs target transcription factors or induce changes in cellular metabolism that affect the T3SS (65, 66). Wang et al. used affinity chromatography of conjugated beads of SAHS against E. coli lysate and identified three specific binding partners involved in bacterial cellular metabolism, WrbA, Tpx and FolX, and speculated targeting these proteins result in changes in T3SS gene expression by altering cellular metabolism (66). Importantly, binding was observed between the SAHs and homologs of WrbA and Tpx from other T3SS containing bacterial pathogens such as Salmonella and Pseudomonas (66). Since then, the crystal structure of Tpx has been solved and models of its binding to SAHs have been described (67). Finally, it has been suggested SAHs interfere with the T3SS through indirect methods, such as altering iron availability due to chelation (61, 63). In Salmonella and Chlamydia, the addition of exogenous iron attenuates the inhibitory effect of SAHs (55, 63, 68). Furthermore, a mutation in hemG, an enzyme involved with heme synthesized, conferred resistance to SAHs in Chlamydia (69). In addition, changes in gene expression of iron metabolism related genes has been reported in Salmonella, but not observed in E. coli, which suggests other mechanisms are at work (63, 65). Recently, a gallium(III)–salicylidene acylhydrazide complex has been shown to disrupt secretion and expression of T3SS proteins in addition to inhibiting biofilm formation in Pseudomonas (70). The current data on SAHs suggest they target the T3SS through multiple mechanisms or that different SAH derivatives inhibit the system by different mechanisms.
Thiazolidinones
Another well studied class of T3SS inhibitors is the thiazolidinones. Felise et al. (71) identified thiazolidinone from a whole-cell screen assay in Salmonella against protein secretion using a reporter construct of the effector SipA fused to the Yersinia phospholipase YplA. Cleavage of a supplied substrate, PED6, by the reporter construct resulted in measurable fluorescence, which was proportional to the amount of the secreted SipA (71). Out of 92,000 screened compounds, a 2-imino-5-arylidene thiazolidinone was identified as a promising candidate, as it did not affect bacterial growth or T3SS transcription, and therefore was suggested to target formation or assembly of the needle apparatus (71). The compound was additionally shown to inhibit the T3SS of Yersinia, as well as the type II secretion system in Pseudomonas and the type IV pili secretion system of Francisella, though it did not target the flagellar-specific T3SS (71). Because of the broad range of action, it was hypothesized that the thiazolidinones target the conserved outer membrane Secretin protein. A follow up study showed that thiazolidinone dimers inhibited the T3SS more potently and it was suggested that the compounds act along a large oligomeric protein-protein interaction surface (72).
Other classes of T3SS inhibitors
Many other chemically and structurally diverse classes of chemical inhibitors that affect the T3SS have been identified (Figure 2). Although the specific targets within the T3SS for most of these inhibitors have not been elucidated, the putative targets of a few compounds have been hypothesized. Yop secretion was inhibited by 2,2′-thiobis-(4-methylphenol) through an interaction with the minor translocon protein YopD of Yersinia (73). Unfortunately, this compound was shown to be toxic to eukaryotic cells and requires structural modification to be considered a suitable drug candidate (73). Mutations in the needle protein PscF of Pseudomonas conferred resistance against phenoxyacetamide inhibitors, suggesting PscF as their molecular target (74, 75). However, no biochemical binding assays showing a direct interaction between the two have been reported. Benzimidazoles have been shown to target the T3SS through inhibition of DNA binding by transcription factors such as LcrF in Yersinia (76, 77) and the Pseudomonas homolog ExsA (78). Similarly, 1-butyl-4-nitromethyl-3-quinolin-2-yl-4H-quinoline inhibited the DNA binding of the Shigella transcription factor VirF, which controls transcription of Shigella T3SS genes (79). A variety of compounds have been shown to inhibit ATPases, such as YscN in Yersinia and its homolog BsaS in Burkholderia (80). Further, omeprazole inhibited the ATPase of the SPI-2 T3SS of Salmonella, possibly through regulation of nitric oxide production (81). Pseudolipasin A inhibited phospholipase A2 activity of the Pseudomonas effector ExoU (82).
There are many small molecule inhibitors without known specific targets within the T3SS as shown in Table 1. A screen in Yersinia by Harmon et al. (83) identified various chemically diverse hydrophobic compounds that inhibited translocation of effectors into eukayrotic cells, but not in vitro secretion or expression, suggesting they disrupted the formation of a functional translocon or that they inhibited interaction with the host cell. Various compounds showing inhibition of T3SS-mediated hemolysis, such as aurodox and the gaudinomines, have also been identified (84, 85). Other compounds, such as salicylanilides, salicylideneanilines, sulfonylaminobenzanilides, cytosporone B, and citrus flavonoids are hypothesized to broadly inhibit T3SS gene transcription through unknown mechanisms (53, 64, 86–88).
Non-small molecule inhibitors of T3SS
Non-small molecule inhibitors of T3SS have been reported. These include polymers, proteins and polypeptide mimics. Ohgita et al. (89) reported that the proton-motive force dependent rotation of the Pseudomonas T3SS was inhibited by the addition of the viscous polymer polyethylene glycol (PEG) 8000 and this was hypothesized to occur by the resistance of physical rotation due to solution viscosity. This hypothesis was further supported by a follow up study showing that other viscous polymers, such as alginate and mucin, inhibited T3SS rotation while low viscosity polymers such as PEG200 do not (90). PEG derivatives are commonly used excipients in drug formulations (91) and because these experiments showed a direct correlation between the rotation of the T3SS needle apparatus and secretion of effectors, the addition of viscous polymers such as PEG as excipients to future antivirulence drug formulations is of interest (90, 91).
Proteins and polypeptides have also been reported as inhibitors of T3SS. For example, the glycoprotein Lactoferrin has been shown to decrease virulence of Salmonella, Shigella and E. coli by targeting the T3SS (92–95). In Shigella, Lactoferrin induced the loss and degradation of the translocon proteins IpaB and IpaC at the bacterial surface (94). Similarly, Lactoferrin caused the loss and degradation of the E. coli tip and translocon proteins (EspA, EspB and EspD) at the bacterial surface (92, 93, 96). Lactoferrin-mediated inhibition of the T3SS is thought to occur through two mechanisms. First, its ability to bind lipopolysaccharide on the bacterial surface is thought to cause instability of virulence proteins at the bacterial surface by disrupting essential protein-protein interactions (97). Second, the degradation of T3SS proteins occurs via the intrinsic serine protease activity of Lactoferrins (97). Additionally, Lactoferrin inhibits infection and has a bacteriostatic effect due to its ability to sequester iron, which is an essential micronutrient needed by many bacterial pathogens (97).
Polypeptide mimics targeting T3SS components have also been described. In Chlamydia, a 28 amino acid polypeptide mimic targeting the CdsN ATPase via its interaction with the putative inner membrane-tethering protein CdsL inhibited bacterial invasion of eukaryotic host cells (98). In Salmonella, Hayward et al. (99) showed that a polypeptide derived from the C-terminus of the translocon protein SipB was shown to be a potent inhibitor of the membrane fusion activity of both wild-type SipB and the Shigella homolog IpaB in vitro. This polypeptide mimic also blocked the entry of Salmonella and Shigella into cultured eukaryotic cells (99). Finally, coiled-coiled peptides designed against the EPEC tip protein EspA inhibited T3SS-mediated hemolysis, EspA polymerization and secretion of effector proteins (100).
Small molecules that bind T3SS proteins
Biologically relevant small molecules interact with structural and effector proteins of the T3SS and could potentially be used as scaffolds for drug design. These small molecules include sterols and lipids. Bile salts in the intestines of hosts are thought to act as environmental sensors for infection by Salmonella and Shigella (101). In Shigella, bile salts increase invasiveness to epithelial cells (102), while in Salmonella bile salts decrease invasiveness (103). Because the tip protein is assembled on the needle at the bacterial surface prior to host cell contact, it has been suggested to function as a sensor for the host environment by interacting with bile salts (101). NMR titrations suggested different interaction sites for bile salts such as deoxycholate and chenodeoxycholate on the Salmonella SipD and the Shigella IpaD tip proteins (104, 105). This is further supported by differences in the binding of deoxycholate in co-crystal structures with SipD (106) or IpaD (107). Interestingly, binding of bile salts was reported to induce a conformation change in IpaD, but not SipD (107), which possibly explains the difference in the responses observed between Salmonella and Shigella in the presence of bile salts.
The translocon proteins interact with sterols, for example, the Shigella IpaB translocon protein interacts with cholesterol and sphingolipids on host cell lipid rafts to mediate infection (108, 109). Binding of IpaB to sterol was proposed to cause the disorganization of the Golgi and other recycling networks by altering the distribution of cholesterol on the cell membrane and the sorting of the eukaryotic cell surface that promoted bacterial uptake (110). Interaction of cholesterol with homologous translocon proteins in other bacteria such as the Salmonella SipB and the Pseudomonas PopB/PopD proteins is required for infection (111, 112). Data also suggests interaction of translocon proteins with cholesterol is essential after initial infection. For example, the T3SS effectors IcsB from Shigella and BopA from Burkholderia bound to cholesterol leading to evasion of autophagy in host cells (113). In Salmonella, the SPI-2 effector SseJ caused esterification of cholesterol to maintain the Salmonella containing vacuole (114). Currently, a detailed mechanism of the molecular interactions of T3SS proteins with cholesterol or sphingolipids and their role in pathogenesis remain to be worked out.
Future Directions
Identification of T3SS targets
Drug discovery and development of small molecule inhibitors is lengthy and costly, with a potential drug candidate taking an average of over 10 years and costing millions of dollars to reach the market as an approved drug. The process often begins with a high throughput screen to identify potential hits. This is followed by target identification and validation, often in parallel with structure activity relationships to identify the most potent lead compound. Next, in vivo animal models are tested. If successful, pharmacokinetics and dynamics are analyzed and product is formulated as needed. Finally, clinical trials are performed.
The most pressing concern in the field is the identification and characterization of specific targets within the many protein components of the T3SS. Even with the most well studied classes of inhibitors, SAHs and thiazolidinones, the specific T3SS targets remain unknown or ambiguous. This is likely due to the fact that most high throughout screens utilized assays that broadly monitored the secretion of effectors or gene transcription rather than using a targeted approach, such as a specific protein-protein interaction involved in the assembly of the T3SS needle apparatus. In addition, derivatives within a class of T3SS inhibitors have unique chemical structure that could potentially interact with non-T3SS targets in other organisms leading to differences in potency or mechanisms of action. This could explain the complex nature of the results observed in the literature of SAHs. A targeted and more specific approach for future high throughout screens could help solve this problem. Many binding assays have been developed to monitor protein-protein interactions of the structural proteins of the T3SS needle apparatus that could be adapted for drug screening. For example, fluorescence polarization and FRET assays show binding of the tip and translocon protein in Shigella (115) and NMR spectroscopy showed binding of the tip and needle protein in Salmonella (116).
SAR design
Structure-activity relationships (SAR) provide a direct link between the chemical structure of a molecule and its observed activity to create a potent lead compound. The identification of a target facilitates SAR studies especially when the mechanism of interaction is known, although they can be performed in the absence of a target (117). SAR studies have been initiated for a few T3SS inhibitor classes and extensively with SAHs. The data from SAR analysis of SAHs is complex and difficult to interpret. The salicylic phenol group is necessary for inhibiting activity, but substitutions in other positions are generally tolerable (52, 118). SAR based optimization of future SAHs compounds could be complicated if they target T3SS through multiple mechanisms as suggested in the literature (52, 118). SAR data on other classes of small molecule inhibitors of T3SS are available as well. Thiazolidinone analogs show sensitivity against substitutions at the imino N-2, amido N-3, and 5-arylidene groups and SAR analysis led to the identification of a more soluble derivative (71). Hydrophobicity and lipophilicity were shown to be important factors for inhibition of the T3SS by sulfonylaminobenzanilides (53). Cytosporone derivatives with extensive carbon chains containing a phenyl acetic acetate ester group were most potent (86). Derivatives of 8-hydroxyquinoline required a fused pyridine ring and an aromatic hydroxyl group for inhibition of T3SS function (119). Benzimidazoles are sensitive to substitutions in the linker ringer and the middle phenyl ring (76). Finally, phenoxyacetamides were sensitive to changes on the A ring (where 2,4-dichlorophenyl is preferred) and their stereocenter was critical for T3SS inhibition and SAR analysis led to the identification of an 8-fold more potent compound than found in initial screens (74, 75).
Animal models, pharmacokinetics and formulation
Preliminary studies using animal models have shown the effectiveness of different T3SS inhibitors against infection in vivo and validated the approach of targeting T3SS (61, 77, 84, 120). However, evaluation of pharmacokinetic parameters and formulation of potential drug molecule for delivery into host organisms is an essential step of drug discovery. Challenges with small molecule drug development include efflux, metabolism and membrane permeability. Notably, most of the literature describes relatively small and hydrophobic compounds that were identified through whole-cell screening, and therefore these compounds are likely to be able to pass through cellular membranes. A study examined the pharmacokinetics of SAHs in a mouse model (120). Many compounds were shown to have a short half-life, suggesting rapid metabolism or clearance, as well as problems with compound stability and solubility (120). Attempts at formulation with the non-ionic surfactant Poloxamer 407 and the polysaccharide (2-hydroxypropyl)-β-cyclodextrin (120, 121) were unsuccessful in improving efficacy. The ability of academia and the pharmaceutical industry to work together will speed the entry of new drugs into the pipeline by allowing for more extensive SAR optimization, pharmacokinetic analysis, formulation and in vivo testing of small molecule compounds (122).
Conclusions
The clinical application of small molecule inhibitors of bacterial virulence as anti-infectives remains to be exploited. Small molecule T3SS inhibitors that have been identified could be used to treat bacterial infection on their own because most of them are non-toxic to eukaryotic cells while still preventing secretion of effector proteins. Anti-infectives will not inhibit bacterial proliferation, thus bacteria must be cleared by other means such as the host immune system. These drugs may lead to increased immune cell memory. It is also possible that antivirulence drugs will need to be used in combination with other antibiotics for clearance. The discovery and validity of many classes of small molecule inhibitors targeting different aspects of the type III secretion systems of Gram-negative bacteria suggest that it is a promising approach that will applicable to clinical settings in the future.
Acknowledgments
Supported by NIH grants T32-GM008359 (A.C.M.) and AI074856 (R.N.D.).
Abbreviations
- SPI
Salmonella Pathogenicity Island
- T3SS
Type III Secretion System
- T3S
Type III Secretion
- SAR
Structure Activity Relationship
- EPEC
Enteropathogenic Escherichia coli
- SAHs
Salicylidene Acylhydrazides
References
- 1.CDC. Antibiotic Resistance Threats in the United States. 2013. [Google Scholar]
- 2.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–9. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 3.Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infectious Diseases. 2013;13:155–65. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and Evolution of Antibiotic Resistance. Microbiology and Molecular Biology Reviews. 2010;74:417–+. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert DN, Guidos RJ, Boucher HW, Talbot GH, Spellberg B, Edwards JE, et al. The 10 x ’20 Initiative: Pursuing a Global Commitment to Develop 10 New Antibacterial Drugs by 2020. Clinical Infectious Diseases. 2010;50:1081–3. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 6.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 7.Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 2013;66:571–91. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- 8.Schaberle TF, Hack IM. Overcoming the current deadlock in antibiotic research. Trends Microbiol. 2014;22:165–7. doi: 10.1016/j.tim.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Bassetti M, Merelli M, Temperoni C, Astilean A. New antibiotics for bad bugs: where are we? Annals of Clinical Microbiology and Antimicrobials. 2013;12 doi: 10.1186/1476-0711-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Jeong SH, Cha SS, Lee SH. A lack of drugs for antibiotic-resistant gram-negative bacteria. Nature Reviews Drug Discovery. 2007;6:938–9. [Google Scholar]
- 11.Coates ARM, Halls G, Hu YM. Novel classes of antibiotics or more of the same? British Journal of Pharmacology. 2011;163:184–94. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Current Opinion in Microbiology. 2003;6:427–30. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Baron C. Antivirulence drugs to target bacterial secretion systems. Current Opinion in Microbiology. 2010;13:100–5. doi: 10.1016/j.mib.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Keyser P, Elofsson M, Rosell S, Wolf-Watz H. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. Journal of Internal Medicine. 2008;264:17–29. doi: 10.1111/j.1365-2796.2008.01941.x. [DOI] [PubMed] [Google Scholar]
- 15.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nature Reviews Microbiology. 2014;12:300–8. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 16.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nature Chemical Biology. 2007;3:541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 17.Duncan MC, Linington RG, Auerbuch V. Chemical Inhibitors of the Type Three Secretion System: Disarming Bacterial Pathogens. Antimicrobial Agents and Chemotherapy. 2012;56:5433–41. doi: 10.1128/AAC.00975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis GR. The type III secretion injectisome. Nature Reviews Microbiology. 2006;4:811–25. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 19.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee S, Chaudhury S, McShan AC, Kaur K, De Guzman RN. Structure and Biophysics of Type III Secretion in Bacteria. Biochemistry. 2013;52:2508–17. doi: 10.1021/bi400160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izore T, Job V, Dessen A. Biogenesis, Regulation, and Targeting of the Type III Secretion System. Structure. 2011;19:603–12. doi: 10.1016/j.str.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Abrusci P, McDowell MA, Lea SM, Johnson S. Building a secreting nanomachine: a structural overview of the T3SS. Curr Opin Struct Biol. 2014;25C:111–7. doi: 10.1016/j.sbi.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. Fems Microbiology Reviews. 2011;35:1100–25. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 24.Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 1995;14:4187–95. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermant D, Menard R, Arricau N, Parsot C, Popoff MY. Functional Conservation of the Salmonella and Shigella Effectors of Entry into Epithelial-Cells. Molecular Microbiology. 1995;17:781–9. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 26.Demers JP, Sgourakis NG, Gupta R, Loquet A, Giller K, Riedel D, et al. The Common Structural Architecture of Shigella flexneri and Salmonella typhimurium Type Three Secretion Needles. Plos Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spreter T, Yip CK, Sanowar S, Andre I, Kimbrough TG, Vuckovic M, et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nature Structural & Molecular Biology. 2009;16:468–76. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarivach R, Vuckovic M, Deng WY, Finlay BB, Strynadka NCJ. Structural analysis of a prototypical ATPase from the type III secretion system. Nature Structural & Molecular Biology. 2007;14:131–7. doi: 10.1038/nsmb1196. [DOI] [PubMed] [Google Scholar]
- 30.Veenendaal AKJ, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Molecular Microbiology. 2007;63:1719–30. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 31.Mattei PJ, Faudry E, Job V, Izore T, Attree I, Dessen A. Membrane targeting and pore formation by the type III secretion system translocon. Febs Journal. 2011;278:414–26. doi: 10.1111/j.1742-4658.2010.07974.x. [DOI] [PubMed] [Google Scholar]
- 32.Deeds EJ, Bachman JA, Fontana W. Optimizing ring assembly reveals the strength of weak interactions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2348–53. doi: 10.1073/pnas.1113095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galan JE, Curtiss R. Cloning and Molecular Characterization of Genes Whose Products Allow Salmonella-Typhimurium to Penetrate Tissue-Culture Cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6383–7. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egan F, Barret M, O’Gara F. The SPI-1-like Type III secretion system: more roles than you think. Frontiers in Plant Science. 2014;5 doi: 10.3389/fpls.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology-Sgm. 2012;158:1147–61. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 36.Barret M, Egan F, O’Gara F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environmental Microbiology Reports. 2013;5:117–26. doi: 10.1111/j.1758-2229.2012.00394.x. [DOI] [PubMed] [Google Scholar]
- 37.Galan JE. SnapShot: Effector proteins of type III secretion systems. Cell. 2007;130:192-U5. doi: 10.1016/j.cell.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 38.Zhou DG, Galan J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes and Infection. 2001;3:1293–8. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 39.Figueira R, Watson KG, Holden DW, Helaine S. Identification of Salmonella Pathogenicity Island-2 Type III Secretion System Effectors Involved in Intramacrophage Replication of S. enterica Serovar Typhimurium: Implications for Rational Vaccine Design. Mbio. 2013;4 doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kidwai AS, Mushamiri I, Niemann GS, Brown RN, Adkins JN, Heffron F. Diverse Secreted Effectors Are Required for Salmonella Persistence in a Mouse Infection Model. Plos One. 2013;8 doi: 10.1371/journal.pone.0070753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clinical Microbiology Reviews. 2007;20:535–+. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–5. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 43.Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Molecular Microbiology. 2004;51:483–95. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 44.Boyd AP, Lambermont I, Cornelis GR. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: Role of the SycE chaperone binding domain of YopE. Journal of Bacteriology. 2000;182:4811–21. doi: 10.1128/jb.182.17.4811-4821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Current Opinion in Microbiology. 2007;10:24–9. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Osborne SE, Coombes BK. Transcriptional Priming of Salmonella Pathogenicity Island-2 Precedes Cellular Invasion. Plos One. 2011;6 doi: 10.1371/journal.pone.0021648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Molecular Microbiology. 2006;62:631–40. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 48.Li LM, Yan H, Feng LP, Li YL, Lu P, Hu YB, et al. LcrQ Blocks the Role of LcrF in Regulating the Ysc-Yop Type III Secretion Genes in Yersinia pseudotuberculosis. Plos One. 2014;9 doi: 10.1371/journal.pone.0092243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorman CJ, Porter ME. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Molecular Microbiology. 1998;29:677–84. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 50.Sukhan A, Kubori T, Wilson J, Galan JE. Genetic analysis of assembly of the Salmonella enterica serovar typhimurium type III secretion-associated needle complex. Journal of Bacteriology. 2001;183:1159–67. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem Biol. 2003;10:241–9. doi: 10.1016/s1074-5521(03)00046-2. [DOI] [PubMed] [Google Scholar]
- 52.Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. Small-molecule inhibitors specifically targeting type III secretion. Infect Immun. 2005;73:3104–14. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauppi AM, Andersson CD, Norberg HA, Sundin C, Linusson A, Elofsson M. Inhibitors of type III secretion in Yersinia: design, synthesis and multivariate QSAR of 2-arylsulfonylamino-benzanilides. Bioorg Med Chem. 2007;15:6994–7011. doi: 10.1016/j.bmc.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 54.Muschiol S, Normark S, Henriques-Normark B, Subtil A. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. Bmc Microbiology. 2009;9 doi: 10.1186/1471-2180-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slepenkin A, Chu H, Elofsson M, Keyser P, Peterson EM. Protection of Mice From a Chlamydia trachomatis Vaginal Infection Using a Salicylidene Acylhydrazide, a Potential Microbicide. Journal of Infectious Diseases. 2011;204:1313–20. doi: 10.1093/infdis/jir552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey L, Gylfe A, Sundin C, Muschiol S, Elofsson M, Nordstrom P, et al. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. Febs Letters. 2007;581:587–95. doi: 10.1016/j.febslet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 57.Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, et al. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2006;103:14566–71. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veenendaal AKJ, Sundin C, Blocker AJ. Small-Molecule Type III Secretion System Inhibitors Block Assembly of the Shigella Type III Secreton. Journal of Bacteriology. 2009;191:563–70. doi: 10.1128/JB.01004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez-Argudo I, Veenendaal AK, Liu X, Roehrich AD, Ronessen MC, Franzoni G, et al. Isolation of Salmonella Mutants Resistant to the Inhibitory Effect of Salicylidene acylhydrazides on Flagella-Mediated Motility. PLoS One. 2013;8:e52179. doi: 10.1371/journal.pone.0052179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrea A, Bjur E, Ygberg SE, Elofsson M, Wolf-Watz H, Rhen M. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrobial Agents and Chemotherapy. 2007;51:2867–76. doi: 10.1128/AAC.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hudson DL, Layton AN, Field TR, Bowen AJ, Wolf-Watz H, Elofsson M, et al. Inhibition of type III secretion in Salmonella enterica serovar typhimurium by small-molecule inhibitors. Antimicrobial Agents and Chemotherapy. 2007;51:2631–5. doi: 10.1128/AAC.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puiac S, Sem XH, Negrea A, Rhen M. Small-molecular virulence inhibitors show divergent and immunomodulatory effects in infection models of Salmonella enterica serovar Typhimurium. International Journal of Antimicrobial Agents. 2011;38:409–16. doi: 10.1016/j.ijantimicag.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Layton AN, Hudson DL, Thompson A, Hinton JC, Stevens JM, Galyov EE, et al. Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol Lett. 2010;302:114–22. doi: 10.1111/j.1574-6968.2009.01847.x. [DOI] [PubMed] [Google Scholar]
- 64.Gauthier A, Robertson ML, Lowden M, Ibarra JA, Puente JL, Finlay BB. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrobial Agents and Chemotherapy. 2005;49:4101–9. doi: 10.1128/AAC.49.10.4101-4109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tree JJ, Wang D, McInally C, Mahajan A, Layton A, Houghton I, et al. Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect Immun. 2009;77:4209–20. doi: 10.1128/IAI.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D, Zetterstrom CE, Gabrielsen M, Beckham KSH, Tree JJ, Macdonald SE, et al. Identification of Bacterial Target Proteins for the Salicylidene Acylhydrazide Class of Virulence-blocking Compounds. Journal of Biological Chemistry. 2011;286:29922–31. doi: 10.1074/jbc.M111.233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gabrielsen M, Beckham KS, Feher VA, Zetterstrom CE, Wang D, Muller S, et al. Structural characterisation of Tpx from Yersinia pseudotuberculosis reveals insights into the binding of salicylidene acylhydrazide compounds. PLoS One. 2012;7:e32217. doi: 10.1371/journal.pone.0032217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slepenkin A, Enquist PA, Hagglund U, de la Maza LM, Elofsson M, Peterson EM. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect Immun. 2007;75:3478–89. doi: 10.1128/IAI.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engstrom P, Nguyen BD, Normark J, Nilsson I, Bastidas RJ, Gylfe A, et al. Mutations in hemG Mediate Resistance to Salicylidene Acylhydrazides, Demonstrating a Novel Link between Protoporphyrinogen Oxidase (HemG) and Chlamydia trachomatis Infectivity. Journal of Bacteriology. 2013;195:4221–30. doi: 10.1128/JB.00506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rzhepishevska O, Hakobyan S, Ekstrand-Hammarstrom B, Nygren Y, Karlsson T, Bucht A, et al. The gallium(III)-salicylidene acylhydrazide complex shows synergistic anti-biofilm effect and inhibits toxin production by Pseudomonas aeruginosa. J Inorg Biochem. 2014;138C:1–8. doi: 10.1016/j.jinorgbio.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 71.Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, et al. An Inhibitor of Gram-Negative Bacterial Virulence Protein Secretion. Cell Host & Microbe. 2008;4:325–36. doi: 10.1016/j.chom.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kline T, Barry KC, Jackson SR, Felise HB, Nguyen HV, Miller SI. Tethered thiazolidinone dimers as inhibitors of the bacterial type III secretion system. Bioorganic & Medicinal Chemistry Letters. 2009;19:1340–3. doi: 10.1016/j.bmcl.2009.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jessen DL, Bradley DS, Nilles ML. A Type III Secretion System Inhibitor Targets YopD while Revealing Differential Regulation of Secretion in Calcium-Blind Mutants of Yersinia pestis. Antimicrobial Agents and Chemotherapy. 2014;58:839–50. doi: 10.1128/AAC.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aiello D, Williams JD, Majgier-Baranowska H, Patel I, Peet NP, Huang J, et al. Discovery and Characterization of Inhibitors of Pseudomonas aeruginosa Type III Secretion. Antimicrobial Agents and Chemotherapy. 2010;54:1988–99. doi: 10.1128/AAC.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bowlin NO, Williams JD, Knoten CA, Torhan MC, Tashjian TF, Li B, et al. Mutations in the Pseudomonas aeruginosa needle protein gene pscF confer resistance to phenoxyacetamide inhibitors of the type III secretion system. Antimicrob Agents Chemother. 2014;58:2211–20. doi: 10.1128/AAC.02795-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim OK, Garrity-Ryan LK, Bartlett VJ, Grier MC, Verma AK, Medjanis G, et al. N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J Med Chem. 2009;52:5626–34. doi: 10.1021/jm9006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrity-Ryan LK, Kim OK, Balada-Llasat JM, Bartlett VJ, Verma AK, Fisher ML, et al. Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect Immun. 2010;78:4683–90. doi: 10.1128/IAI.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grier MC, Garrity-Ryan LK, Bartlett VJ, Klausner KA, Donovan PJ, Dudley C, et al. N-Hydroxybenzimidazole inhibitors of ExsA MAR transcription factor in Pseudomonas aeruginosa: In vitro anti-virulence activity and metabolic stability. Bioorganic & Medicinal Chemistry Letters. 2010;20:3380–3. doi: 10.1016/j.bmcl.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 79.Koppolu V, Osaka I, Skredenske JM, Kettle B, Hefty PS, Li JQ, et al. Small-Molecule Inhibitor of the Shigella flexneri Master Virulence Regulator VirF. Infection and Immunity. 2013;81:4220–31. doi: 10.1128/IAI.00919-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Swietnicki W, Carmany D, Retford M, Guelta M, Dorsey R, Bozue J, et al. Identification of Small-Molecule Inhibitors of Yersinia pestis Type III Secretion System YscN ATPase. Plos One. 2011;6 doi: 10.1371/journal.pone.0019716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Puiac S, Negrea A, Richter-Dahlfors A, Plant L, Rhen M. Omeprazole antagonizes virulence and inflammation in Salmonella enterica-infected RAW264.7 cells. Antimicrob Agents Chemother. 2009;53:2402–9. doi: 10.1128/AAC.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee VT, Pukatzki S, Sato H, Kikawada E, Kazimirova AA, Huang J, et al. Pseudolipasin A is a specific inhibitor for phospholipase A(2) activity of Pseudomonas aeruginosa cytotoxin ExoU. Infection and Immunity. 2007;75:1089–98. doi: 10.1128/IAI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harmon DE, Davis AJ, Castillo C, Mecsas J. Identification and Characterization of Small-Molecule Inhibitors of Yop Translocation in Yersinia pseudotuberculosis. Antimicrobial Agents and Chemotherapy. 2010;54:3241–54. doi: 10.1128/AAC.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kimura K, Iwatsuki M, Nagai T, Matsumoto A, Takahashi Y, Shiomi K, et al. A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J Antibiot (Tokyo) 2011;64:197–203. doi: 10.1038/ja.2010.155. [DOI] [PubMed] [Google Scholar]
- 85.Iwatsuki M, Uchida R, Yoshijima H, Ui H, Shiomi K, Kim YP, et al. Guadinomines, Type III secretion system inhibitors, produced by streptomyces sp K01–0509. Journal of Antibiotics. 2008;61:230–6. doi: 10.1038/ja.2008.33. [DOI] [PubMed] [Google Scholar]
- 86.Li JF, Lv C, Sun WY, Li ZY, Han XW, Li YY, et al. Cytosporone B, an Inhibitor of the Type III Secretion System of Salmonella enterica Serovar Typhimurium. Antimicrobial Agents and Chemotherapy. 2013;57:2191–8. doi: 10.1128/AAC.02421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vikram A, Jayaprakasha GK, Jesudhasan PR, Pillai SD, Patil BS. Suppression of bacterial cell-cell signalling, biofilm formation and type III secretion system by citrus flavonoids. Journal of Applied Microbiology. 2010;109:515–27. doi: 10.1111/j.1365-2672.2010.04677.x. [DOI] [PubMed] [Google Scholar]
- 88.Kauppi AM, Nordfelth R, Hagglund U, Wolf-Watz H, Elofsson M. Salicylanilides are potent inhibitors of type III secretion in Yersinia. Genus Yersinia: Entering the Functional Genomic Era. 2003;529:97–100. doi: 10.1007/0-306-48416-1_17. [DOI] [PubMed] [Google Scholar]
- 89.Ohgita T, Hayashi N, Hama S, Tsuchiya H, Gotoh N, Kogure K. A novel effector secretion mechanism based on proton-motive force-dependent type III secretion apparatus rotation. FASEB J. 2013;27:2862–72. doi: 10.1096/fj.13-229054. [DOI] [PubMed] [Google Scholar]
- 90.Ohgita T, Hayashi N, Gotoh N, Kogure K. Suppression of type III effector secretion by polymers. Open Biol. 2013;3:130133. doi: 10.1098/rsob.130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strickley RG. Solubilizing excipients in oral and injectable formulations. Pharm Res. 2004;21:201–30. doi: 10.1023/b:pham.0000016235.32639.23. [DOI] [PubMed] [Google Scholar]
- 92.Ochoa TJ, Clearly TG. Lactoferrin disruption of bacterial type III secretion systems. Biometals. 2004;17:257–60. doi: 10.1023/b:biom.0000027701.12965.d4. [DOI] [PubMed] [Google Scholar]
- 93.Ochoa TJ, Noguera-Obenza M, Ebel F, Guzman CA, Gomez HF, Cleary TG. Lactoferrin impairs type III secretory system function in enteropathogenic Escherichia coli. Infect Immun. 2003;71:5149–55. doi: 10.1128/IAI.71.9.5149-5155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gomez HF, Ochoa TJ, Carlin LG, Cleary TG. Human lactoferrin impairs virulence of Shigella flexneri. J Infect Dis. 2003;187:87–95. doi: 10.1086/345875. [DOI] [PubMed] [Google Scholar]
- 95.Mosquito S, Ochoa TJ, Cok J, Cleary TG. Effect of bovine lactoferrin in Salmonella ser. Typhimurium infection in mice. Biometals. 2010;23:515–21. doi: 10.1007/s10534-010-9325-1. [DOI] [PubMed] [Google Scholar]
- 96.Yekta MA, Verdonck F, Van Den Broeck W, Goddeeris BM, Cox E, Vanrompay D. Lactoferrin inhibits E. coli O157:H7 growth and attachment to intestinal epithelial cells. Veterinarni Medicina. 2010;55:359–68. [Google Scholar]
- 97.Ochoa TJ, Cleary TG. Effect of lactoferrin on enteric pathogens. Biochimie. 2009;91:30–4. doi: 10.1016/j.biochi.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stone CB, Bulir DC, Emdin CA, Pirie RM, Porfilio EA, Slootstra JW, et al. Chlamydia Pneumoniae CdsL Regulates CdsN ATPase Activity, and Disruption with a Peptide Mimetic Prevents Bacterial Invasion. Front Microbiol. 2011;2:21. doi: 10.3389/fmicb.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hayward RD, Hume PJ, McGhie EJ, Koronakis V. A Salmonella SipB-derived polypeptide blocks the ‘trigger’ mechanism of bacterial entry into eukaryotic cells. Mol Microbiol. 2002;45:1715–27. doi: 10.1046/j.1365-2958.2002.03124.x. [DOI] [PubMed] [Google Scholar]
- 100.Larzabal M, Mercado EC, Vilte DA, Salazar-Gonzalez H, Cataldi A, Navarro-Garcia F. Designed coiled-coil peptides inhibit the type three secretion system of enteropathogenic Escherichia coli. PLoS One. 2010;5:e9046. doi: 10.1371/journal.pone.0009046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneli surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infection and Immunity. 2007;75:2626–9. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pope LM, Reed KE, Payne SM. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun. 1995;63:3642–8. doi: 10.1128/iai.63.9.3642-3648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infection and Immunity. 2000;68:6763–9. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Nordhues BA, Zhong D, De Guzman RN. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry. 2010;49:4220–6. doi: 10.1021/bi100335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry. 2011;50:172–80. doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barta ML, Guragain M, Adam P, Dickenson NE, Patil M, Geisbrecht BV, et al. Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins. 2012;80:935–45. doi: 10.1002/prot.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lafont F, Tran Van Nhieu G, Hanada K, Sansonetti P, van der Goot FG. Initial steps of Shigella infection depend on the cholesterol/sphingolipid raft-mediated CD44-IpaB interaction. EMBO J. 2002;21:4449–57. doi: 10.1093/emboj/cdf457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD. Liposomes Recruit IpaC to the Shigella flexneri Type III Secretion Apparatus Needle as a Final Step in Secretion Induction. Infection and Immunity. 2009;77:2754–61. doi: 10.1128/IAI.00190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mounier J, Boncompain G, Senerovic L, Lagache T, Chretien F, Perez F, et al. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 2012;12:381–9. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 111.Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Molecular Microbiology. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- 112.Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. Embo Journal. 2003;22:4957–67. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kayath CA, Hussey S, El Hajjami N, Nagra K, Philpott D, Allaoui A. Escape of intracellular Shigella from autophagy requires binding to cholesterol through the type III effector, IcsB. Microbes and Infection. 2010;12:956–66. doi: 10.1016/j.micinf.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 114.Nawabi P, Catron DM, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol Microbiol. 2008;68:173–85. doi: 10.1111/j.1365-2958.2008.06142.x. [DOI] [PubMed] [Google Scholar]
- 115.Dickenson NE, Arizmendi O, Patil MK, Toth RTt, Middaugh CR, Picking WD, et al. N-Terminus of IpaB Provides a Potential Anchor to the Shigella Type III Secretion System Tip Complex Protein IpaD. Biochemistry. 2013 doi: 10.1021/bi400755f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rathinavelan T, Tang C, De Guzman RN. Characterization of the Interaction between the Salmonella Type III Secretion System Tip Protein SipD and the Needle Protein PrgI by Paramagnetic Relaxation Enhancement. Journal of Biological Chemistry. 2011;286:4922–30. doi: 10.1074/jbc.M110.159434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andricopulo AD, Montanari CA. Structure-activity relationships for the design of small-molecule inhibitors. Mini-Reviews in Medicinal Chemistry. 2005;5:585–93. doi: 10.2174/1389557054023224. [DOI] [PubMed] [Google Scholar]
- 118.Dahlgren MK, Zetterstrom CE, Gylfe S, Linusson A, Elofsson M. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg Med Chem. 2010;18:2686–703. doi: 10.1016/j.bmc.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 119.Enquist PA, Gylfe A, Hagglund U, Lindstom P, Norberg-Scherman H, Sundin C, et al. Derivatives of 8-hydroxyquinoline-antibacterial agents that target intra- and extracellular Gram-negative pathogens. Bioorganic & Medicinal Chemistry Letters. 2012;22:3550–3. doi: 10.1016/j.bmcl.2012.03.096. [DOI] [PubMed] [Google Scholar]
- 120.Ur-Rehman T, Slepenkin A, Chu H, Blomgren A, Dahlgren MK, Zetterstrom CE, et al. Pre-clinical pharmacokinetics and anti-chlamydial activity of salicylidene acylhydrazide inhibitors of bacterial type III secretion. J Antibiot (Tokyo) 2012;65:397–404. doi: 10.1038/ja.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ur-Rehman T, Nordfelth R, Blomgren A, Zetterstrom CE, Elofsson M, Gylfe A. Preliminary Pharmacokinetics of the Bacterial Virulence Inhibitor N′-(3,5-Dibromo-2-HydroxyBenzylidenene)-Nicotinic Acid Hydrazide. Advances in Yersinia Research. 2012;954:349–56. doi: 10.1007/978-1-4614-3561-7_42. [DOI] [PubMed] [Google Scholar]
- 122.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. British Journal of Pharmacology. 2011;162:1239–49. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zetterstrom CE, Hasselgren J, Salin O, Davis RA, Quinn RJ, Sundin C, et al. The Resveratrol Tetramer (−)-Hopeaphenol Inhibits Type III Secretion in the Gram-Negative Pathogens Yersinia pseudotuberculosis and Pseudomonas aeruginosa. PLoS One. 2013;8:e81969. doi: 10.1371/journal.pone.0081969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kline T, Felise HB, Barry KC, Jackson SR, Nguyen HV, Miller SI. Substituted 2-Imino-5-arylidenethiazolidin-4-one Inhibitors of Bacterial Type III Secretion. Journal of Medicinal Chemistry. 2008;51:7065–74. doi: 10.1021/jm8004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Linington RG, Robertson M, Gauthier A, Finlay BB, van Soes R, Andersen RJ. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Organic Letters. 2002;4:4089–92. doi: 10.1021/ol0268337. [DOI] [PubMed] [Google Scholar]
- 126.Linington RG, Robertson M, Gauthier A, Finlay BB, MacMillan JB, Molinski TF, et al. Caminosides B-D, antimicrobial glycolipids isolated from the marine sponge Caminus sphaeroconia. J Nat Prod. 2006;69:173–7. doi: 10.1021/np050192h. [DOI] [PubMed] [Google Scholar]
- 127.Duncan MC, Wong WR, Dupzyk AJ, Bray WM, Linington RG, Auerbuch V. An NF-kappa B-Based High-Throughput Screen Identifies Piericidins as Inhibitors of the Yersinia pseudotuberculosis Type III Secretion System. Antimicrobial Agents and Chemotherapy. 2014;58:1118–26. doi: 10.1128/AAC.02025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yin S, Davis RA, Shelper T, Sykes ML, Avery VM, Elofsson M, et al. Pseudoceramines A-D, new antibacterial bromotyrosine alkaloids from the marine sponge Pseudoceratina sp. Org Biomol Chem. 2011;9:6755–60. doi: 10.1039/c1ob05581j. [DOI] [PubMed] [Google Scholar]
- 129.Dahlgren MK, Kauppi AM, Olsson IM, Linusson A, Elofsson M. Design, synthesis, and multivariate quantitative structure-activity relationship of salicylanilides--potent inhibitors of type III secretion in Yersinia. J Med Chem. 2007;50:6177–88. doi: 10.1021/jm070741b. [DOI] [PubMed] [Google Scholar]
- 130.Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Molecular Microbiology. 2006;61:1543–55. doi: 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zigangirova NA, Zayakin ES, Kapotina LN, Kost EA, Didenko LV, Davydova DY, et al. Development of Chlamydial Type III Secretion System Inhibitors for Suppression of Acute and Chronic Forms of Chlamydial Infection. Acta Naturae. 2012;4:87–97. [PMC free article] [PubMed] [Google Scholar]
- 132.Pan N, Lee C, Goguen J. High throughput screening for small-molecule inhibitors of type III secretion in Yersinia pestis. Genus Yersinia: From Genomics to Function. 2007;603:367–75. doi: 10.1007/978-0-387-72124-8_34. [DOI] [PubMed] [Google Scholar]
- 133.Pan NJ, Brady MJ, Leong JM, Goguen JD. Targeting Type III Secretion in Yersinia pestis. Antimicrobial Agents and Chemotherapy. 2009;53:385–92. doi: 10.1128/AAC.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hayward RD, Hume PJ, McGhie EJ, Koronakis V. A Salmonella SipB-derived polypeptide blocks the ‘trigger’ mechanism of bacterial entry into eukaryotic cells. Molecular Microbiology. 2002;45:1715–27. doi: 10.1046/j.1365-2958.2002.03124.x. [DOI] [PubMed] [Google Scholar]