Abstract
Forty strains of Salmonella enterica (S. enterica) subspecies salamae (II), arizonae (IIIa), diarizonae (IIIb), and houtenae (IV) were isolated from human or environmental samples and tested for bacteriophage production. Production of bacteriophages was observed in 15 S. enterica strains (37.5%) belonging to either the subspecies salamae (8 strains) or diarizonae (7 strains). Activity of phages was tested against 52 pathogenic S. enterica subsp. enterica isolates and showed that phages produced by subsp. salamae had broader activity against pathogenic salmonellae compared to phages from the subsp. diarizonae. All 15 phages were analyzed using PCR amplification of phage-specific regions and 9 different amplification profiles were identified. Five phages (SEN1, SEN4, SEN5, SEN22, and SEN34) were completely sequenced and classified as temperate phages. Phages SEN4 and SEN5 were genetically identical, thus representing a single phage type (i.e. SEN4/5). SEN1 and SEN4/5 fit into the group of P2-like phages, while the SEN22 phage showed sequence relatedness to P22-like phages. Interestingly, while phage SEN34 was genetically distantly related to Lambda-like phages (Siphoviridae), it had the morphology of the Myoviridae family. Based on sequence analysis and electron microscopy, phages SEN1 and SEN4/5 were members of the Myoviridae family and phage SEN22 belonged to the Podoviridae family.
Introduction
At present, the Salmonella genus contains two species: S. enterica and S. bongori (V). S. enterica is further subdivided into six subspecies: enterica (I), salamae (II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV), and indica (VI) [1–3]. Salmonelloses (infections transferred between humans and animals, mainly by contaminated food) represent an important, global, public health problem, with S. enterica subsp. enterica causing about 99% of the human cases [3–6]. Out of more than 2,600 S. enterica subsp. enterica serovars [7], relatively few serovars are important causative agents of salmonelloses [3]. While S. enterica subsp. enterica is typically found in warm-blooded animals, S. enterica subsp. arizonae and S. bongori are associated with cold-blooded animals. The other four subspecies of S. enterica can be isolated from both host types and can occasionally cause human infections [8–13].
Bacteriophages are the most abundant group of biological agents in our environment [14]. Tailed phages represent the dominant morphotype among characterized bacterial viruses as shown through electron microscopic examination of more than 5500 phages, in which 96% of phages were tailed [15]. Tailed phages also dominate with regard to the number of complete genome sequences; analysis of 607 complete phage genomes in the GenBank database revealed 506 (80%) tailed phages [16]. Tailed phages are in the Caudovirales order and are subdivided according to tail morphology into 3 families–Myoviridae (phages with long, contractile tails), Siphoviridae (phages with long, non-contractile tails), and Podoviridae (phages with short tails).
Based on a comparison of genome sequences of tailed enterobacterial phages [17], 77 Salmonella phages were found in 23 clusters including lytic phages (e.g. T4-like, T7-like, and SETP3-like) and temperate phages (e.g. Lambda-like, P2-like, and P22-like). In addition, 9371 prophages from 3298 Salmonella genomes were recently identified; out of them, 744 prophages belonged to the P22-like group and 4758 to the P2-like group [18].
Salmonella phages or prophages have been predominantly studied in strains of S. enterica subsp. enterica. In this communication, we focused on phage production by strains in the less common S. enterica subspecies salamae, arizonae, diarizonae, and houtenae. Genetic relatedness of identified phages and their activity spectra were determined. In addition, phages SEN1, SEN4/5, SEN22, and SEN34 were characterized by electron microscopy and whole genome sequencing.
Materials and Methods
Bacterial strains and culture media
From 1999–2005, 40 non-pathogenic strains of S. enterica subspecies salamae, arizonae, diarizonae, and houtenae (referred as Sen 1–40) were collected in the Czech Republic from human clinical samples and from environmental samples. All S. enterica strains were provided by The National Reference Laboratory for Salmonella, National Institute of Public Health (NIPH), Prague, Czech Republic. Salmonella strains were serotyped using the White-Kauffmann-Le Minor classification scheme [19] and their serotype characteristics are listed in S1 Table.
For cultivation of bacteria in liquid culture, tryptone yeast (TY) medium with the following composition was used: yeast extract (HiMedia, Mumbai, India) 5g/L, casein enzyme hydrolysate (HiMedia) 8g/L, and NaCl (Penta, Prague, Czech Republic) 5g/L. TY agar plates were supplemented with a 1.5% (w/v) of agar (Hi-Media).
Identification of lysogenic strains
Forty S. enterica strains were tested for bacteriophage production using a cross test method where each strain was tested as a possible producer as well as a possible indicator strain. In addition, four standard E. coli indicators (K12-Row, C6 (φ), B1, and P400), which were available in our laboratory strain collection [20–21], were used to detect phage producers. Briefly, each Salmonella strain was inoculated from a fresh TY broth culture into an agar base layer, 1.5% (w/v), using a sterile needle and then cultivated at 37°C for 48 hr. The resulting macrocolony was killed using chloroform vapors (30 min) and the plate was overlaid with a top layer of 0.7% (w/v) agar enriched with a suspension of 107 indicator bacteria. Phage production was evaluated after overnight cultivation at 37°C.
Determination of phage titer and inducibility of phages
Phage producers were inoculated into TY broth and incubated overnight. The fresh culture was diluted hundredfold with TY broth and cultivated for an additional 7 hr (37°C, 200 rpm). Subsequently, the culture was centrifuged at 4000 × g for 15 min to remove bacteria. The supernatant was transferred to a sterile tube and stored at 4°C with a few drops of chloroform to ensure sterility. To determine the concentration of phage particles, the sterile supernatant was serially diluted 10 times; each dilution was added to a suspension of phage-susceptible bacteria (107 bacteria in 3 ml of the melted top layer of 0.7% agar) and spread on agar plates. After overnight cultivation, plaques were counted and the resulting bacteriophage titer was expressed as plaque forming units (PFU)/ml.
In parallel, induction of phage production was tested using the same protocol with a slight modification, which included the addition of mitomycin C (final concentration 0.1 μg/ml) to the culture of lysogenic strains during the exponential growth phase (i.e., after 4 hr of cultivation at 37°C, 200 rpm); cultivation continued for an additional 3 hr. Phage production was determined using a plaque counting technique. A comparison of phage titers counted from cultures with and without addition of mitomycin C indicated inducibility of phage production.
Activity of phages against clinically important Salmonella isolates
Using the identified phage producers, we examined the activity of phages against a set of 52 clinically important S. enterica subsp. enterica isolates using the cross-test method described above. Clinical isolates belonged to common pathogenic serovars, i.e. Enteritidis (22 isolates), Typhimurium (13 isolates), Choleraesuis (4 isolates), Indiana (3 isolates), Derby (2 isolates), Mikawashima (2 isolates), Agona (1 isolate), Chester (1 isolate), Infantis (1 isolate), Java (1 isolate), Ohio (1 isolate), and San Diego (1 isolate). All these pathogenic strains were collected by The National Reference Laboratory for Salmonella, National Institute of Public Health (NIPH), Prague, Czech Republic, from patients living in the Czech Republic. The characteristics of the pathogenic strains are shown in S1 Table.
Analysis of PCR amplification profiles of phages
DNA from all phages was isolated using a QIAGEN Lambda Midi Kit (Qiagen, Hilden, Germany) per the protocol recommended by the manufacturer. Total bacteriophage DNA was digested using EcoRI enzyme (New England Biolabs, Ipswich, USA), cloned into the pUC19 cloning vector (New England Biolabs); the resulting clones were sequenced. The resulting sequences were compared with the GenBank database using BLAST to identify similarities to other phages. Subsequently, 12 DNA regions representing different phage genes were selected for primer design. The DNA from all phages was used as a template for PCR amplification (Table 1) and up to 2 genetic regions originating from the same phage were used for further screening. A list of genetic loci and the corresponding primer sequences are shown in Table 1. Individual PCR reactions contained 0.4 μl of 10 mM dNTP mix, 1.8 μl of 10× ThermoPol Reaction buffer, 0.25 μl of each primer (100 pmol/μl), 0.05 μl of Taq polymerase (5000 U/ml), 1 μl of the tested phage DNA sample, and 15.2 μl of PCR grade water. PCR amplification was performed under the following cycling conditions: 94°C (2 min); 94°C (30 s), 60°C (30 s), 72°C (60 s), 30 cycles; 72°C (10 min). Resulting PCR products were sequenced with amplification primers. The resulting phage sequences were deposited in the GenBank under accession numbers KM202139, KM202140, KM202142 –KM202144, KM202151 –KM202154, KM202157, KM202158, KM202160, KM202164 –KM202167, KM202171 –KM202176, KM202179, KM202181 –KM202184, and KM202186 –KM202195.
Table 1. List of primers used for amplification and sequencing of several phage genome regions.
| Analyzed genome region | Phage template for primer design | Primer name | Primer sequence (5'-3') | Amplicon length (bp) | Target region encoding |
|---|---|---|---|---|---|
| GA1 | SEN22 | F4c5-F | CAACTTGCGACTGCTCTTTG | 474 | Terminase small subunit |
| F4c5-R | CGAAGGAAGCGTTAGACCTG | ||||
| GA2 | SEN2 | F3c7-F | GTTGGGTGGAAGAAGCTGAA | 413 | Terminase large subunit |
| F3c7-R | AGCATCTGGCCCTGTATCTG | ||||
| GA3 | SEN8 | F20c3-F | TTCAGTGGTTGGTTCCATGA | 471 | Portal protein |
| F20c3-R | GCGTTAAGAAGCCAGAAACG | ||||
| GA4 | SEN8 | F20c5-F | TTATTTGCTGCGCTGACATC | 494 | Scaffolding protein |
| F20c5-R | TCAGAAGAAGGCCTGGCTAA | ||||
| GA5 | SEN23 | F6c1-F | GGCCGATCAGTTGCTTAAAA | 232 | Integrase |
| F6c1-R | CTCATGCCCCAACATTTTCT | ||||
| GA6 | SEN4 | F9c2-F | GCCGTCTGATAACCCAGAAA | 456 | Methyltransferase |
| F9c2-R | CGTTTTCCAGATCAGCCTGT | ||||
| GA7 | SEN34 | F38c2-F | AAGTGGGGAACTGCTGAAGA | 487 | Replication protein O |
| F38c2-R | AACGCTGCCATAAGCTGACT | ||||
| GA8 | SEN1 | F1c3-F | AAAGGCCGGTTAGGTAGCTC | 456 | Replication protein |
| F1c3-R | ATCGCTCGCATGTTTAACG | ||||
| GA9 | SEN22 | F4c4-F | AATGGATGCAGTCAGGGAAG | 421 | Nin locus |
| F4c4-R | GTCAATCATCGCGTTTTCCT | ||||
| GA10 | SEN5 | F10c1-F | TTCGCAAATGAAATCGAGTG | 443 | Hypothetical protein |
| F10c1-R | ATCGGCAACTTACCGTCATC | ||||
| GA11 | SEN23 | F6c4-F | GTTGTTCAGGCCGTTGATTT | 284 | Hypothetical protein |
| F6c4-R | GTACATCGCCTGAAGGGAGA | ||||
| GA12 | SEN34 | F38c1-F | GTTTATGGCGCTGAAAAGGA | 457 | Hypothetical protein |
| F38c1-R | GTTGTTCAGGCCGTTGATTT |
Preparation of phage stock
Bacteriophage stock was prepared from a single selected plaque [22] that was resuspended in 100 μl of TY broth containing 0.01M CaCl2. This phage suspension was added to 0.7% TY agar containing 107 phage-susceptible bacteria (i.e., they did not produce their own bacteriophages) and spread on agar plate. After overnight cultivation, the bacteriophage-containing top layer of TY agar, with confluent lysis, was collected into a sterile tube and resuspended in 3 ml of SM buffer (100 mM NaCl, 8 mM MgSO4*7H2O, 50 mM Tris-Cl, 0.01% (w/v) gelatine) and 0.5 ml of chloroform. The tube was incubated at 37°C for 20 min and the suspension was centrifuged at 4000 × g for 15 min to remove bacteria and agar residue. The supernatant, containing the phage particles, was transferred to a sterile tube and few drops of chloroform were added.
Isolation of bacteriophage DNA for whole genome sequencing
The bacterial stocks originated from single plaques multiplied in indicator strains (i.e., did not produce their own phages) were used for propagation of phages and isolation of phage DNA (see above). Phages were propagated per previously published protocols [22]. Briefly, 200 ml of TY broth was inoculated with 2 ml of fresh phage indicator culture and cultivated for 4.5 hr (37°C, 250 rpm). Thereafter, 200 μl of 0.02M CaCl2 and phage particles (≈ 108) from our phage stock were added and cultivation continued for an additional 4.5 hr (37°C, 250 rpm). Finally, the bacterial culture was lysed using chloroform (10 min, 37°C, 250 rpm). Bacterial nucleic acids in lysates were degraded using RNase A and DNase I (final conc. 1 μg/ml, 45 min, RT) and phage particles were precipitated using NaCl (1M, 1 hr, 4°C) and PEG 8000 (10%, 16 hr, 4°C). After centrifugation (11,000 × g, 11 min, 4°C), the pellet was resuspended in 5 ml of SM buffer and phage particles were purified using isopycnic ultracentrifugation in a CsCl gradient (87,000 × g, 2 hr, 4°C). The collected bacteriophage suspension was dialyzed with dialysis buffer (10 mM NaCl, 50 mM Tris-Cl, 10 mM MgCl2) for 2 ×1 hr. The dialyzed phage suspension was treated with EDTA (final conc. 20 mM), proteinase K (final conc. 50 μg/ml) and SDS (final conc. 0.5%) for 1 hr at 56°C. Phage DNA was isolated by phenol-chloroform, precipitated by isopropanol, and finally diluted in 50 μl of TE buffer.
Whole-genome sequencing and analysis of phage genomes
Genomic DNA of 5 phages was sequenced using Ion Torrent™ next-gen sequencing technology (Life Technologies, Carlsbad, USA) at SeqOmics Biotechnology Ltd (Mórahalom, Hungary). All genomes were assembled de novo using SeqMan NGen software from a Lasergene v.10 genomics package (DNASTAR, Madison, USA). Genomes were annotated using RAST (Rapid Annotation using Subsystem Technology), a fully-automatic annotation service [23], followed by manual correction based on the BLASTX algorithm [24]. Genomes were deposited in GenBank under the following numbers: SEN1 (KT630644); SEN4 (KT630645), SEN5 (KT 630646), SEN22 (KT 630648), and SEN34 (KT 630649). Genomes of Salmonella phages were analyzed using the Lasergene program package (DNASTAR). BLAST comparisons of phage genomes were produced with Easyfig [25]. Average nucleotide identity (ANI) of phages was calculated by the JSspeciesWeb Server running MUMmer 3.0 software [26, 27].
Electron microscopy
Phage samples for electron microscopy were prepared from phage stocks (originated from a single plaque) according to previously published protocols [22]. Phage suspensions were subsequently precipitated with polyethylene glycol (PEG) and purified using centrifugation in a CsCl gradient [22]. Bacteriophage particles were visualized using a negative staining method. Briefly, phage suspensions (5 μl) were placed on glow discharge activated carbon-coated grids [28]. After an adsorption period (30 s), the unabsorbed liquid material was removed using filter paper. The electron-microscopic grids were then stained with 10 μl of 1% phosphotungstic acid, 2% uranyl acetate, or 2% ammonium molybdate for 10–30 s; excess staining solution was removed using filter paper. Samples were viewed using a MORGAGNI 268D (FEI, Hillsboro, OR, USA) or a Philips CM 100 transmission electron microscope (FEI).
Results
Characterization of non-pathogenic Salmonella enterica strains and identification of lysogenic strains
All non-pathogenic S. enterica strains used in this study (n = 40) were classified into subspecies based on biochemical markers and serotyped using the White-Kaufmann-LeMinor scheme. A total of 16 strains belonged to S. enterica subsp. salamae, 5 strains to subsp. arizonae, 15 strains to subsp. diarizonae, and 4 strains to subsp. houtenae. Strains mostly came from human feces or other human material, and from waste water sludge. In 7 cases, the origin of the strain was unknown (S1 Table).
All forty S. enterica strains were tested against each other (i.e., in 1,600 individual tests), as potential phage producers and phage indicators. In addition to S. enterica strains, four standard E. coli phage-indicator strains (i.e., K12-Row, C6 (φ), B1, and P400) were used. The results are summarized in Table 2. Production of bacteriophages was observed in 15 non-pathogenic S. enterica strains (37.5%) (Sen1, Sen2, Sen4, Sen5, Sen6, Sen8, Sen14, Sen16, Sen22, Sen23, Sen24, Sen30, Sen31, Sen34, and Sen35) belonging to either subspecies salamae (8 strains) or diarizonae (7 strains). All phages formed small turbid plaques without a lytic halo. Plaque sizes varied from 0.3–1.6 mm in diameter. Bacteriophage titers obtained from non-induced cultures ranged from 1.3 × 103 to 4.0 × 106 PFU/ml. Moreover, mitomycin C induction of phages was effective in 8 out of 15 phage producers (Table 2), increasing the phage titer by two orders of magnitude.
Table 2. Phage producers and indicator strains of identified phages.
| Phage producer (S. enterica subspecies) | Susceptible indicator strains | PFU/ml | Plaque diameter (mm) | |
|---|---|---|---|---|
| S. enterica | E. coli | |||
| Sen1 (salamae)* | Sen4, Sen5, Sen6, Sen9, Sen11 | P400, B1 | 2.8 × 104 | 1.5 ± 0.06 |
| Sen2 (salamae)* | Sen11, Sen17, Sen26 | - | 2.9 × 106 | 0.8 ± 0.06 |
| Sen4 (salamae) | Sen38 | P400, B1 | 9.2 × 103 | 0.5 ± 0.05 |
| Sen5 (salamae) | Sen38 | P400, B1 | 6.9 × 103 | 0.5 ± 0.06 |
| Sen6 (salamae)* | Sen9, Sen33 | - | 2.7 × 104 | 0.3 ± 0.06 |
| Sen8 (salamae) | Sen6, Sen9, Sen10, Sen36, Sen38, Sen40 | - | 1.5 × 105 | 0.7 ± 0.05 |
| Sen14 (salamae)* | Sen11 | P400, B1 | 6.1 × 104 | 0.5 ± 0.05 |
| Sen16 (salamae) | Sen38, Sen6, Sen9, Sen10, Sen11, Sen40, Sen36 | P400, B1 | 1.6 × 105 | 1.6 ± 0.05 |
| Sen22 (diarizonae)* | Sen26, Sen39, Sen34 | - | 4.0 × 106 | 1.2 ± 0.08 |
| Sen23 (diarizonae)* | Sen11, Sen22, Sen24, Sen39 | - | 3.4 × 104 | 0.4 ± 0.07 |
| Sen24 (diarizonae) | Sen26, Sen39 | - | 1.5 × 104 | 1.1 ± 0.07 |
| Sen30 (diarizonae)* | Sen9 | - | 7.2 × 103 | 1.1 ± 0.05 |
| Sen31 (diarizonae) | Sen9 | - | 1.3 × 103 | 1.1 ± 0.06 |
| Sen34 (diarizonae)* | Sen23, Sen24, Sen26, Sen39 | - | 5.2 × 104 | 1.3 ± 0.07 |
| Sen35 (diarizonae) | - | P400, B1 | 6.5 × 104 | 0.7 ± 0.05 |
*phage production was induced by mitomycin C; PFU–plaque forming unit
Indicator strains used for preparation of phage stocks are underlined.
Analysis of phage activity against pathogenic S. enterica clinical isolates
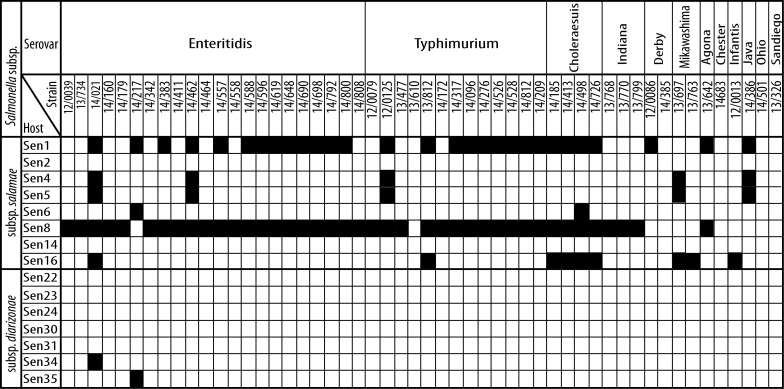
Activity of all 15 phages from identified phage producers was tested against 52 pathogenic S. enterica subsp. enterica isolates belonging to common pathogenic serovars. The activity spectra of phages are shown in Fig 1. Two strains, i.e. Sen1 and Sen8, produced phages, which lysed the majority of pathogenic isolates, while phages produced by Sen4, Sen5, Sen6, Sen16, Sen34, and Sen35 strains inhibited less than 20% of isolates. Moreover, all pathogenic Salmonella isolates were resistant to phages produced by strains Sen2, Sen14, Sen22, Sen23, Sen24, Sen30, and Sen31. In general, phages from subsp. salamae showed broader activity against pathogenic salmonellae compared to phages from subsp. diarizonae.
Fig 1. Activity of phages against pathogenic Salmonella enterica subsp. enterica isolates.
Susceptibility of clinical isolates to phages is shown in black. “Host” refers to identified phage producers. Note the difference in the spectra of phages produced by strains from subsp. salamae and diarizonae.
Analysis of phage PCR amplification profiles
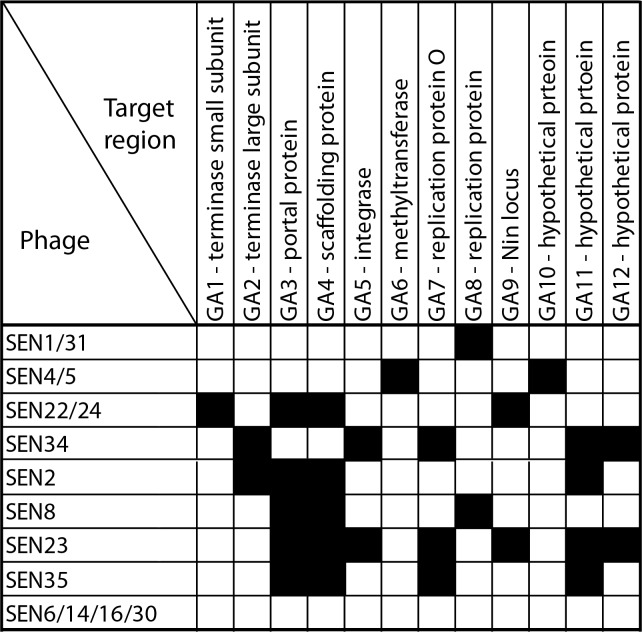
Variability of identified phages (denoted as “SEN” throughout the manuscript) was analyzed using PCR amplification of several loci obtained during construction of small libraries of phages. Altogether, 12 short fragments (200–300 bp) from different genes were tested in all 15 phages (Fig 2). Phages SEN6, SEN14, SEN16, and SEN30 were negative for all PCR detected DNA regions. Three pairs of phages (i.e., SEN1 and SEN31, SEN4 and SEN5, and SEN22 and SEN24) had identical amplification profiles. The five remaining phages showed a unique spectra of PCR positive reactions. Altogether, nine different amplification profiles were identified. Finally, phages SEN1, SEN4, SEN5, SEN22, and SEN34 were selected for further analysis based on: i) variability in PCR profiles, ii) differences in the activity against pathogenic salmonellae, iii) classification of phage producers into different subspecies, and iv) quality of isolated DNA.
Fig 2. Phage PCR amplification profiles.
Based on sequencing of cloned phage libraries, 12 phage DNA regions were selected for PCR amplification from all fifteen phages. Positive amplification results are shown in black.
Morphology of phage particles
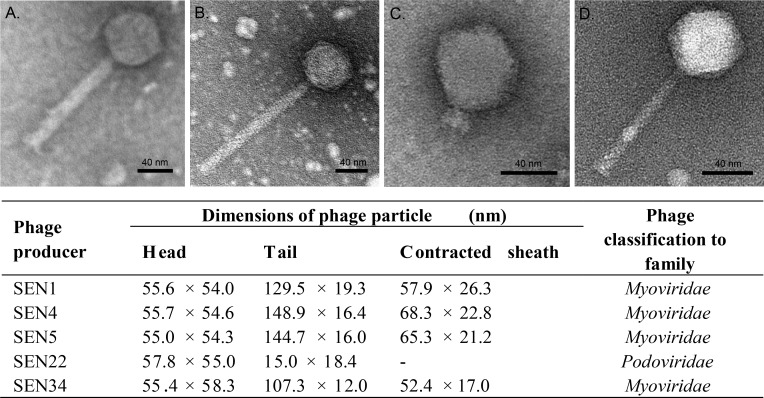
Phages SEN1, SEN4, and SEN5 from producers belonging to subsp. salamae, and phages SEN22 and SEN34 from producers belonging to subsp. diarizonae were characterized using electron microscopy. A phage suspension obtained from a single plaque was negatively stained and examined with a transmission electron microscope to determine phage morphology. The morphology of phage particles is summarized in Fig 3. Phages SEN1, SEN4, SEN5, and SEN34, shared the same morphology, i.e. a head with icosahedral symmetry and a long contractile tail. Diameters of icosahedral capsids ranged from 54.0–58.3 nm and the length of the tail, including the contractile sheath, ranged from 107.3–148.9 nm. In contrast, phage SEN22 had an icosahedral head (57.8 nm in width; 55 nm in length) with very short noncontractile tail (18.4 nm in width; 15.0 nm in length). Based on previous classifications [29], the first morphological group corresponds to the Myoviridae family, and the second morphological group corresponds to the Podoviridae family.
Fig 3. Morphology of Salmonella phages.
Electron microscopy of negatively stained phage particles isolated from the producer strain Sen1 (A), Sen4 and Sen5 (phage SEN4/5) (B), Sen22 (C), and Sen34 (D). While phage SEN22 belongs to family Podoviridae, other phages showed morphology of family Myoviridae. Bar scale represents 40 nm. Dimensions of phage virions are shown in table.
Whole genome analysis
Whole genome sequencing of phages SEN1, SEN4, SEN5, SEN22, and SEN34 was performed. The main characteristics of phage genomes are shown in Table 3. Genome length of the sequenced phages ranged from 29.7–41.3 kb with G+C content ranging from 47.8–53.4%. Genome lengths were correlated with genome sizes, which were estimated from PFGE (data not shown), indicating that complete genome sequences had been obtained. While the cos site, identical to predicted cohesive ends of phage Psp3 (ggcgtggcggggaaagcat), was found for SEN1, sticky ends of phages SEN4 and SEN5 (ggcgaggcgggggaacgag) were more similar to the cos site of phage P2. In addition, a pac sequence (gaagatttatctgaagtcgtta) identical to that of P22 phage was identified for phage SEN22. Due to unusual SEN34 genome (see below), the packaging-responsible sequences were not identified in the SEN34 genome. All sequenced phages encoded integrase, a characteristic of temperate phages.
Table 3. Genome characteristics of newly sequenced Salmonella phages.
| Phage | Host (S. enterica subsp.) | Genome size (bp) | No. of reads (average coverage) | G+C content (%) | No. of predicted genes | The most similar phage (e-value/ANI %)* | Genbank Acc. No. |
|---|---|---|---|---|---|---|---|
| SEN1 | salamae | 29733 | 9163 (67×) | 53.01 | 43 | Enterobacteria phage PsP3 (0.0/97.11%) | KT630644 |
| SEN4 | salamae | 33509 | 14313 (94×) | 53.36 | 47 | no similarity# | KT630645 |
| SEN5 | salamae | 33509 | 14619 (95×) | 53.36 | 47 | no similarity | KT630646 |
| SEN22 | diarizonae | 41338 | 15702 (72×) | 47.83 | 55 | Salmonella phage epsilon34 (0.0/94.11%) | KT630648 |
| SEN34 | diarizonae | 40740 | 19506 (108×) | 49.91 | 63 | no similarity | KT630649 |
*BLASTN analysis of whole genome phage sequences was performed against a database of viruses (taxid:10239) and results were ranked based on the total score. Average nucleotide identity (ANI) of the most similar phages was calculated by the JSspeciesWeb Server.
#phage genome sequences showing similarity in less than 10% of genome length were marked as “no similarity”
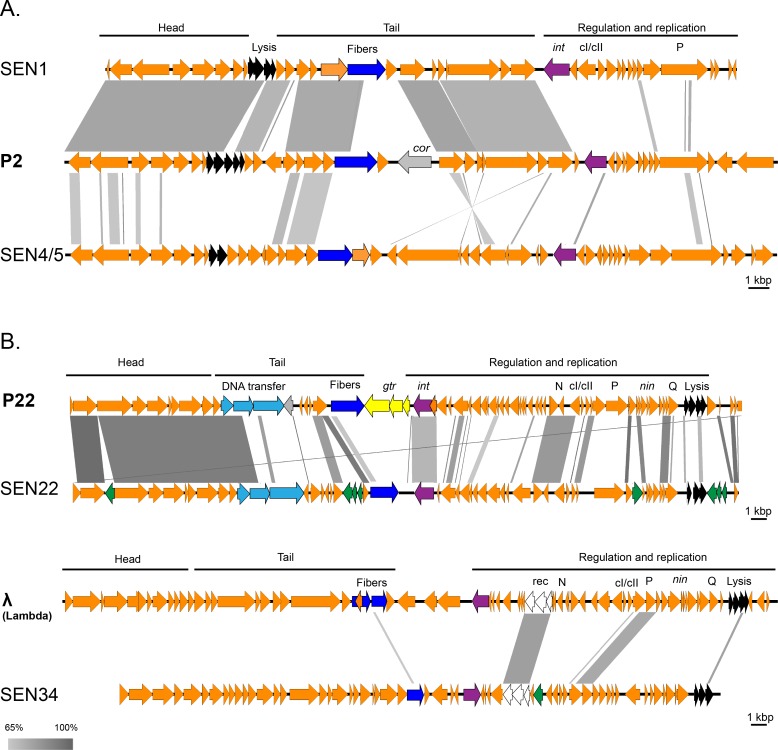
A BLASTN analysis of whole genomes (Table 3) revealed substantial similarity (ANI: 97.11%) between phage SEN1 and phage PsP3 (a phage belonging to the P2-like group) (GenBank No. AY135486). The genome of phage SEN1 was also very similar (ANI: 81.69%) to the prototype phage P2 (GenBank No. KC618326), especially in the morphogenesis region. Besides the genetic variability found in the region responsible for replication, the lysogenic conversion locus was not present in the SEN1 genome and the region encoding tail fibers differed between SEN1 and P2 phages (Fig 4A). In fact, the tail fiber gene of SEN1 (gp19) was related to another P2-like phage, Salmonella phage RE-2010 (GenBank No. HM770079).
Fig 4. Genome analyses of sequenced phages SEN1, SEN4/5, SEN22, and SEN34.
Genome comparison of phages isolated from subsp. salamae (A) and from subsp. diarizonae (B). Schematic organization of genes in phage genomes (head-tail-regulation-replication) is shown with bold black lines. The most variable genes (i.e., those encoding DNA transfer proteins and tail fibers) are shown in blue. Genes for integration (int) are violet, genes encoding lysis are black, the locus responsible for O antigen modification (gtr) is yellow, the locus for lysogenic conversion (cor) is grey, and the locus for recombination (rec) is white. Mobile elements (transposons, HNH endonucleases) are green and the remaining genes are orange. Comparisons of phage genomes were prepared using Easyfig software [25]; similarity between phages (> 65%) is shown using a gray scale (bottom left).
Whole genome sequencing of phages SEN4 and SEN5 revealed that these phages were identical (Table 3), thus representing a single phage type, denoted as phage SEN4/5. Whole genome analysis of SEN4/5, based on BLASTN, found similarities to the prophage sequence within the genome of Enterobacter cloacae complex (GenBank No. CP012162), but not to any known phages (Table 3); however, BLASTP analysis of predicted SEN4/5 proteins showed a distant similarity to several P2-like phages (Fig 4A and S2 Table). Nonetheless, the average nucleotide identity with the P2 phage was calculated to be 0.00%. Thus, SEN4/5 appears to belong to the P2-like phages, but it is clearly different from previously known members.
Both SEN22 and SEN34 phages, isolated from subsp. diarizonae, showed larger genomes compared to subsp. salamae phages (Table 3). The genome of phage SEN22 was found to be related to several P22-like phages having the greatest similarity (ANI: 94.11%) to Salmonella phage ɛ34 (GenBank No. NC_011976). The SEN22 genome region encoding the head of the virion was more similar to prototype phage P22 than the region encoding the virion tail and the region responsible for regulation and replication (Fig 4B). In addition, four insertions of mobile elements were identified in the SEN22 genome, including a unique insertion of a homing endonuclease gene between the terminase gene and the portal protein gene.
BLASTN analysis of the SEN34 genome sequence revealed a low degree of similarity to the prophage sequence within the genome of S. enterica subsp. enterica serovar Newport str. CVM 21550 (GenBank No. CP010283), but not to any known phages (Table 3), while the BLASTP analysis of predicted proteins showed that there was a distant similarity between the SEN34 virion assembly genes and two uncharacterized phages including the Burkholderia phage Bups phi1 and Acinetobacter phage Ab105-1phi (S2 Table). Additionally, the region responsible for replication and regulation of SEN34 was similar to several Lambda-like phages (Fig 4B and S2 Table). When all these observations are taken together, SEN34 appears to be a new phage type (ANI with Lambda phage: 0.00%).
Discussion
To date, almost 200 phage types have been identified in the genus Salmonella [30] and if prophages are included, the number is greater than 9,000 [18]. Prophages appear to be very common in Salmonella, as shown by production of 136 functional phages from 173 S. enterica subsp. enterica (serovar Typhimurium) isolates [31]. It has been calculated that there are, on average, 2.8 known prophages present per Salmonella genome [18]. While phages and prophages of subsp. enterica have been intensively studied (reviewed in [18, 30, 32]), information on phages in other Salmonella subspecies is scarce. In fact, putative prophage sequences were only recently identified in the whole genome sequences of two strains belonging to non-enterica subspecies, i.e. in subsp. arizonae (GenBank: CP000880.1; prophage Sari1; [33]), and subsp. salamae (GenBank: ATFA01000000; [34]). This study focused on phage production within strains S. enterica subspecies salamae, arizonae, diarizonae, and houtenae. While fifteen phage producers were identified in subspecies salamae and diarizonae, no producers were identified in subspecies arizonae and houtenae; however, it is unclear if this difference is due to the relatively low number of investigated strains in the second group (9 out of 40) or due to other factors.
Out of 15 identified phages, SEN1, SEN4/5, SEN22, and SEN34 were further characterized. Bacteriophages SEN1, SEN4/5, and SEN34 belonged to the Myoviridae family, while SEN22 showed morphology typical of the Podoviridae family. These findings are in accordance with the fact that the majority of known phages belongs to the Myoviridae family [35].
Analysis of phage genomes revealed that these phages represent novel types. All sequenced phages encoded integrase, a characteristic of temperate phages. While phages SEN1 and SEN22 showed close sequence relatedness to prototype phages P2 and P22, respectively, phages SEN4/5 and SEN34 were quite different from known phages. Our findings are in accordance with a study by Casjens and Grose [18], where SEN4/5 genomic sequences formed a new, well defined, subcluster within the P2-like cluster. Phage SEN34 was similar to several disparate Lambda-like phages, but did not belong to any of the 17 known lambda clusters described by Grose and Casjens [17]. Thus, phage SEN34 is a representative of a new lambda cluster. This is in accordance with a recent comprehensive study of enterobacterial prophages [18], where the authors analyzed the available phage genomes from GenBank database and defined a new lambda phage cluster (“temperate 26”) with prototype phage SEN34. Although phage SEN34 is genetically related to Lambda-like phages, morphologically it is a member of the Myoviridae family (in contrast to Lambda phages, which belong to the Siphoviridae family). A similar discrepancy between genomic relatedness and morphology classification has also been shown for the LP65 phage, which had a Myoviridae morphology, however, it had a genome organization similar to the Lambda phage [36].
The additional 10 unsequenced phages identified in this study, likely represent distinct phage types as revealed by (i) differences in amplification profiles of various genomic regions, (ii) analyses of the sources of Salmonella strains, (iii) the spectra of indicator strains, and (iv) inducibility with mitomycin C.
Genomes of temperate phages sequenced in our study showed genome mosaicism, where several parts of the phage genomes were related to several different phages. This is in agreement with previous comparative analyses showing that the morphogenesis region is relatively conserved, while genome mosaicism is prevalent in the early regions of phage genomes [17, 37–40].
Besides common mosaicism in the early region, genes encoding proteins interacting with the host showed increased genetic diversity. The DNA encoding receptor-binding tailspike protein domain is among the most highly exchanged parts of the tailed phages. It is evolutionarily useful for phages to acquire new receptor specificities by swapping this domain through horizontal gene transfer [33]. While sequenced phages produced by the subsp. salamae showed a similarity to the region encoding tail fibers in the P2-like Salmonella phage RE-2010, no sequence similarity was found for the SEN22 and SEN34 phages.
Phages produced by strains belonging to subsp. salamae showed a broader activity spectra against pathogenic isolates of subsp. enterica compared to phages from subsp. diarizonae. This is in accordance with results from a phylogenetic study of Salmonella subspecies [41], which showed a close relationship between subsp. salamae and subsp. enterica. The observed broad activity of phages SEN1 and SEN8 against pathogenic salmonellae opens up the possibility of finding therapeutic applications for these phages, however, the presence of lysogenic cycles in these phages makes such therapeutic applications rather potential.
Conclusions
In this study, we determined phage production in S. enterica subspecies salamae, arizonae, diarizonae, and houtenae. Out of 15 identified phage producers, five complete phage genomes were determined and four different temperate phages were identified. Phages SEN1 and SEN4/5 clustered with P2-like phages, while phage SEN22 showed sequence relatedness to P22-like phages. Phage SEN34 was distantly related to Lambda-like phages (Siphoviridae), but had a morphology that was characteristic of Myoviridae.
Supporting Information
(DOCX)
(DOCX)
Acknowledgments
We thank Thomas Secrest (Secrest Editing, Ltd.) for his assistance with the English revision of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files. Genome sequence files are available from the GenBank database (accession numbers KT630644, KT630645, KT 630646,KT 630648, and KT 630649).
Funding Statement
The author (O.B.) gratefully acknowledge the support by the project LO1509 of the Ministry of Education, Youth and Sports of the Czech Republic and by Operational Program Prague Competitiveness project (CZ.2.16/3.1.00/24023) supported by EU. This work was partly supported also by MUNI11/InGA04/2014 (to D.S.) and by GA16-21649S (to D.S.). This work was also supported by funds from the Faculty of Medicine, Masaryk University (ROZV/20/LF/2015), to junior researcher Lenka Mikalová-Paštěková. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Judicial Commission of the International Committee on Systematics of Prokaryotes. The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80. Int J Syst Evol Microbiol. 2005;55:519–20. 10.1099/ijs.0.63579-0 [DOI] [PubMed] [Google Scholar]
- 2.Tindall BJ, Grimont PAD, Garrity GM, Euzéby JP. Nomenclature and taxonomy of the genus Salmonella. Int J Syst Evol Microbiol. 2005;55:521–24. 10.1099/ijs.0.63580-0 [DOI] [PubMed] [Google Scholar]
- 3.Lan R, Reeves PR, Octavia S. Population structure, origins and evolution of major Salmonella enterica clones. Infect Genet Evol. 2009;9:996–1005. 10.1016/j.meegid.2009.04.011 [DOI] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J 2015;13:4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC). Surveillance for Foodborne Disease Outbreaks, United States, 2014, Annual Report. Atlanta, Georgia: US Department of Health and Human Services, CDC, 2016. [WWW document] URL www.cdc.gov/foodsafety/pdfs/foodborne-outbreaks-annual-report-2014-508.pdf [Google Scholar]
- 7.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161:26–9. 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 8.Snehalatha S, Mathai E, Jayasheela M, Chandy M, Lalitha MK, John TJ. Salmonella choleraesuis subsp. indica serovar bornheim causing urinary tract infection. J Clin Microbiol. 1992;30:2504–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giammanco GM, Pignato S, Mammina C, Grimont F, Grimont PAD, Nastasi A, et al. Persistent endemicity of Salmonella bongori 48:z(35):—in Southern Italy: molecular characterization of human, animal, and environmental isolates. J Clin Microbiol. 2002;40:3502–5. 10.1128/JCM.40.9.3502-3505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schröter M, Roggentin P, Hofmann J, Speicher A, Laufs R, Mac D. Pet snakes as a reservoir for Salmonella enterica subsp. diarizonae (Serogroup IIIb): a prospective study. Appl Environ Microbiol. 2004;70:613–5. 10.1128/AEM.70.1.613-615.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabarani CM, Bennett NJ, Kiska DL, Riddell SW, Botash AS, Domachowske JB. Empyema of preexisting subdural hemorrhage caused by a rare Salmonella species after exposure to bearded dragons in a foster home. J Pediatr. 2010;156:322–3. 10.1016/j.jpeds.2009.07.050 [DOI] [PubMed] [Google Scholar]
- 12.Abbott SL, Ni FCY, Janda JM Increase in extraintestinal infections caused by Salmonella enterica subspecies II-IV. Emerg Infect Dis. 2012;18:637–9. 10.3201/eid1804.111386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolker S, Itsekzon T, Yinnon AM, Lachish T. Osteomyelitis due to Salmonella enterica subsp. arizonae: the price of exotic pets. Clin Microbiol Infect. 2012;18:167–70. 10.1111/j.1469-0691.2011.03533.x [DOI] [PubMed] [Google Scholar]
- 14.Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–8. 10.1016/j.mib.2005.06.014 [DOI] [PubMed] [Google Scholar]
- 15.Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–43. 10.1007/s00705-006-0849-1 [DOI] [PubMed] [Google Scholar]
- 16.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75:610–35. 10.1128/MMBR.00011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468–470:421–43. 10.1016/j.virol.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casjens SR, Grose JH. Contributions of P2- and P22-like prophages to understanding the enormous diversity and abundance of tailed bacteriophages.Virology. 2016;496:255–76. 10.1016/j.virol.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France: [WWW document] URL www.scacm.org/free/Antigenic Formulae of the Salmonella Serovars 2007 9th edition.pdf [Google Scholar]
- 20.Šmarda J, Šmajs D, Horynová S. Incidence of lysogenic, colicinogenic and siderophore-producing strains among human non-pathogenic Escherichia coli. Folia Microbiol. 2006;51:387–91. [DOI] [PubMed] [Google Scholar]
- 21.Šmajs D, Micenková L, Šmarda J, Vrba M, Ševčíková, Vališová Z, et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol. 2010;10:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3rd ed, vol 1 Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 25.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benada O, Pokorny V. Modification of the polaron sputter-coater unit for glow-discharge activation of carbon support films. J Electron Microsc Tech. 1990;16: 235–9. 10.1002/jemt.1060160304 [DOI] [PubMed] [Google Scholar]
- 29.Ackermann HW, Eisenstark A. The present state of phage taxonomy. Intervirology. 1974;3:201–19. [DOI] [PubMed] [Google Scholar]
- 30.Kropinski AM, Sulakvelidze A, Konczy P, Poppe C. Salmonella phages and prophages—genomics and practical aspects. Methods Mol Biol. 2007;394:133–75. 10.1007/978-1-59745-512-1_9 [DOI] [PubMed] [Google Scholar]
- 31.Schicklmaier P, Moser E, Wieland T, Rabsch W, Schmieger H. A comparative study on the frequency of prophages among natural isolates of Salmonella and Escherichia coli with emphasis on generalized transducers. Antonie Van Leeuwenhoek. 1998;73 49–54. [DOI] [PubMed] [Google Scholar]
- 32.Switt AI, Sulakvelidze A, Wiedmann M, Kropinski AM, Wishart DS, Poppe C, et al. Salmonella phages and prophages: Genomics, taxonomy, and applied aspects. Methods Mol Biol. 2015;1225:237–87. 10.1007/978-1-4939-1625-2_15 [DOI] [PubMed] [Google Scholar]
- 33.Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. 10.1016/j.virol.2010.12.046 [DOI] [PubMed] [Google Scholar]
- 34.Sathyabama S, Kaur G, Arora A, Verma S, Mubin N, Mayilraj S, et al. Genome sequencing, annotation and analysis of Salmonella enterica subspecies salamae strain DMA-1. Gut Pathog. 2014;6:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann HW. Bacteriophage observations and evolution. Res Microbiol. 2003;154:245–51. 10.1016/S0923-2508(03)00067-6 [DOI] [PubMed] [Google Scholar]
- 36.Chibani-Chennoufi S, Dillmann ML, Marvin-Guy L, Rami-Shojaei S, Brüssow H. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J Bacteriol. 2004;186:7069–7083. 10.1128/JB.186.21.7069-7083.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. 10.1006/jmbi.2000.3729 [DOI] [PubMed] [Google Scholar]
- 38.Clark AJ, Inwood W, Cloutier T, Dhillon TS. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J Mol Biol. 2001;311:657–79. 10.1006/jmbi.2001.4868 [DOI] [PubMed] [Google Scholar]
- 39.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–82. [DOI] [PubMed] [Google Scholar]
- 40.Proux C, van Sinderen D, Suarez J, Garcia P, Ladero V, Fitzgerald GF, et al. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J Bacteriol. 2002;184:6026–36. 10.1128/JB.184.21.6026-6036.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Bhonagiri-Palsikar V, et al. Evolutionary genomics of Salmonella enterica subspecies. MBio. 2013;4(2):e00198–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Genome sequence files are available from the GenBank database (accession numbers KT630644, KT630645, KT 630646,KT 630648, and KT 630649).