Abstract
The acetogen Clostridium ljungdahlii is capable of syngas fermentation and microbial electrosynthesis. Biofilm formation could benefit both these applications, but was not yet reported for C. ljungdahlii. Biofilm formation does not occur under standard growth conditions, but attachment or aggregation could be induced by different stresses. The strongest biofilm formation was observed with the addition of sodium chloride. After 3 days of incubation, the biomass volume attached to a plastic surface was 20 times higher with than without the addition of 200 mM NaCl to the medium. The addition of NaCl also resulted in biofilm formation on glass, graphite and glassy carbon, the latter two being often used electrode materials for microbial electrosynthesis. Biofilms were composed of extracellular proteins, polysaccharides, as well as DNA, while pilus-like appendages were observed with, but not without, the addition of NaCl. A transcriptome analysis comparing planktonic (no NaCl) and biofilm (NaCl addition) cells showed that C. ljungdahlii coped with the salt stress by the upregulation of the general stress response, Na+ export and osmoprotectant accumulation. A potential role for poly-N-acetylglucosamines and D-alanine in biofilm formation was found. Flagellar motility was downregulated, while putative type IV pili biosynthesis genes were not expressed. Moreover, the gene expression analysis suggested the involvement of the transcriptional regulators LexA, Spo0A and CcpA in stress response and biofilm formation. This study showed that NaCl addition might be a valuable strategy to induce biofilm formation by C. ljungdahlii, which can improve the efficacy of syngas fermentation and microbial electrosynthesis applications.
Introduction
The acetogen Clostridium ljungdahlii is of high interest for industrial applications, because of its specific metabolic capacities. Firstly, C. ljungdahlii is capable of converting CO2/H2 and CO to acetate and ethanol [1]. Mixtures of these gases are produced during steel production and the gasification of biomass. The fermentation of this syngas by acetogenic bacteria, such as C. ljungdahlii, allows the production of renewable chemicals and biofuels [2–4]. Secondly, C. ljungdahlii is capable of microbial electrosynthesis, i.e. the reduction of CO2 to acetate with electrons derived from an electrode [5]. C. ljungdahlii potentially has a direct electron uptake mechanism [5], but it could also indirectly derive electrons from an electrode by consuming electrolytically generated H2 [6]. Independent of the mechanism, microbial electrosynthesis is seen as a promising strategy to convert electrical energy into biofuels and other organic commodities [7–9]. Furthermore, a genetic system has recently been developed for C. ljungdahlii, enabling the engineering of strains towards higher-value end-products and increasing the economic feasibility of both syngas fermentations and microbial electrosynthesis [10–13].
Most syngas fermentation reactors today rely on planktonic growth, but to improve the mass transfer from the gas to the liquid phase, also some setups with attached biomass have been suggested [14]. In addition, direct electron uptake from an electrode depends on the attachment of the bacteria to the electrode [15], while there is also evidence that hydrogen mediated electron uptake benefits from biofilm formation on the electrode [16, 17]. One study found that the multi-species biofilm that had developed in a syngas fermenting hollow-fiber membrane biofilm reactor was dominated by C. ljungdahlii [18]. In addition, attachment of C. ljungdahlii to the cathode of a microbial electrosynthesis reactor was observed, but the attachment was limited to thin patches which did not completely cover the electrode [5]. The further optimization of both syngas fermentations and microbial electrosynthesis will therefore rely on a good understanding and the improvement of attachment and biofilm formation by C. ljungdahlii. At this moment, however, a fundamental understanding of biofilm formation by C. ljungdahlii is still completely lacking. Biofilm formation by Clostridium species in general is not well understood and the limited available knowledge is restricted to the pathogens C. perfringens and C. difficile [19], which phylogenetically differ strongly from C. ljungdahlii [20, 21].
For these reasons, the goal of this study was to examine biofilm formation by C. ljungdahlii. No biofilm formation could be observed when C. ljungdahlii was grown in standard growth conditions. Even in rich medium conditions, for which biofilm formation by C. perfringens and difficile was reported [22–24], C. ljungdahlii did not form a biofilm. Therefore, we tried to induce biofilm formation by applying different stress conditions, as biofilms are known to protect cells from a harsh environment [19, 25]. We found that the addition of NaCl to the medium strongly induced biofilm formation, while also other stress conditions led to attachment or aggregation. We further characterized the NaCl induced biofilms of C. ljungdahlii, assessed attachment to different materials and analyzed the biofilm matrix composition. In addition, we performed a RNA sequencing analysis comparing gene expression of biofilm and planktonic cells to obtain some mechanistic understanding of biofilm formation by C. ljungdahlii.
Materials & Methods
Strain and growth conditions
Clostridium ljungdahlii DSM13528 (ATCC55383) was taken from the laboratory’s culture collection. Working stocks were stored at -80°C in 10% dimethyl sulfoxide. These stocks were revived in PETC 1754 medium (ATCC), adapted as previously described [11]. The revived culture was transferred to the same medium once and the transfer was used in late log-phase to inoculate the experiments with 5% volume. Biofilm formation by C. ljungdahlii was tested in a rich medium, adapted from the previously described YTF medium [11]. This medium consisted of 17 g·L-1 Bacto tryptone (BD) and 10 g·L-1 yeast extract (Fisher Scientific) at pH 6.0, was bubbled with nitrogen and autoclaved, after which 5 g·L-1 fructose and 1 mM cysteine were added from anoxic and sterile stock solutions. Sodium chloride or other components were also added from anoxic and sterile stock solutions, depending on the experiment.
For biofilm quantification using the crystal violet assay and RNA extraction, C. ljungdahlii was grown in 6-well, polystyrene, non-treated, flat-bottom plates (CytoOne). Wells were filled with 5 mL medium inside of a glovebox with N2:CO2:H2 (83:10:7) atmosphere. The plates were incubated in an Anaeropack rectangular jar (Mitsubishi), which was kept inside the glovebox. For confocal microscopy, C. ljungdahlii was grown in 2-well chamber slides of Permanox plastic (Lab Tek). Wells were filled with 1.5 mL medium and the slides were incubated inside the glovebox. For electron microscopy and growth curves, C. ljungdahlii was grown in Balch tubes filled with 10 mL medium. To test biofilm formation on different materials, a piece of glass, graphite or glassy carbon (3 x 1 cm) was placed vertically in the tubes. All incubations were performed at 37°C. All experiments were repeated several times and representative experiments are depicted in the figures.
Crystal violet assay
Biofilm formation was quantified using the well-established crystal violet assay [22]. As a first step, the supernatant was removed from the wells and its optical density at 600 nm (OD 600 nm) was determined as a measure for the amount of planktonic cells. The remaining biofilm was washed twice with 5 mL PBS buffer (Cold Spring Harbor Protocols). All pipetting steps were carefully performed in order to not disturb the biofilms. In a next step, the biofilm was stained with 5 mL 0.2% crystal violet. After 30 min of incubation, the crystal violet solution was removed and the excess stain was rinsed away with two washes of PBS buffer. Pictures of the stained biofilms were taken with a Canon SX50 HS camera. Finally, crystal violet was extracted from the biofilm using 5 mL methanol and after 30 min of incubation the absorbance of the methanol solution was measured at 570 nm (A 570 nm). This absorbance is an often used measure for biofilm formation [22]. Highly colored methanol solutions were diluted 10 to 25 times before measurement. Optical densities and absorbances were measured with a Genesys 2 spectrophotometer (Spectronic Instruments). All biofilm quantification experiments used three biological replicates.
Fluorescent staining and confocal laser scanning microscopy
After the required incubation time, the supernatant was removed from chamber slides and the wells were washed twice with PBS buffer. Material pieces were transferred using a previously glued string to twice 5 mL PBS buffer for washing. All biofilms were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.3) for 30 min at 4°C. Next, the biofilms were incubated in staining mixture for 15 min in the dark. For live/dead staining, the mixture consisted of 1.5 μL·mL-1 of SYTO9 (green) and propidium iodide (red) (Live/Dead BacLight Bacterial Viability Kits, Life Technologies) in PBS buffer. SYPRO Ruby Red Biofilm Matrix Stain (Invitrogen) was used as provided by the manufacturer. Calcofluor white (Sigma) was used in a concentration of 5 μg·mL-1. In a next step, the biofilms were incubated in PBS buffer for 15 min in the dark to remove excess stain. Afterwards, excess liquid was let to evaporate for a few minutes and chambers were removed from the slides. Before the biofilms were completely dry, mounting medium, prepared with the Prolong Antifade Kit (Life Technologies) or using a simple recipe (9 mL glycerol, 1 mL 1M Tris HCl buffer at pH 8.3, 0.05 g n-propyl gallate, heated till all powder was dissolved) was applied on the biofilms. The biofilms grown in chamber slides and on graphite and glassy carbon pieces were covered with a cover slide, which was sealed with nail polish. A diamond pen was used to cut the cover slides to the right size. Graphite and glassy carbon pieces were glued onto a microscope slide for easy handling in the microscope. Glass pieces were placed on a microscope slide and then sealed with nail polish.
Biofilms were visualized using a Leica DMI6000 confocal laser scanning microscope. SYTO9 and propidium iodide were excited using a 488 nm Argon laser. The emission of SYTO9 and propidium iodide was measured between 490 and 520 nm and between 600 and 720 nm, respectively. Unstained biofilms were used to exclude autofluorescence of the samples and biofilms stained with only one stain were used to exclude cross-feeding. SYPRO Ruby Red was excited using a 458 nm laser and its emission was measured between 550 and 720 nm The biofilm stained with calcofluor white was analyzed using a Nikon A1R confocal laser scanning microscope. Calcofluor white was excited with a 405 nm laser and its emission was measured between 500 and 550 nm. Images were taken using LAS AF or NIS Elements and further analyzed using FIJI and COMSTAT (ImageJ).
Transmission electron microscopy
The presence of cell appendages was studied using transmission electron microscopy (TEM). After the required incubation time, the supernatant was removed from the tube and the biofilm was washed twice with PBS buffer. If not yet detached, cell aggregates were loosened from the tube wall by gently shaking in 1 mL of PBS buffer. The solution with the cell aggregates was then transferred to an Eppendorf tube and was washed twice with mQ water by centrifugation at 3000 rpm for 30 s. In case of aggregation instead of biofilm formation, the aggregates were allowed to sediment down in between the PBS buffer washing steps. In case of planktonic cells, centrifugation at 6000 rpm was used in between all washing steps.
Carbon films on a 400 square mesh copper grid (Electron Microscopy Sciences) were treated in a Harrick Plasma Cleaner, after which 10 μL of cell suspension was placed on the grids for 5 min. The grids were then negatively stained using 1% uranyl acetate and destained using mQ water for 30 s each. The samples were visualized using a JEOL JEM-2000FX at an acceleration voltage of 200 kV.
RNA sequencing
The gene expression of biofilm cells (NaCl addition) was compared with that of planktonic cells (no NaCl addition) by RNA sequencing. After the required incubation time, 0.5 mL RNAlater RNA Stabilization Reagent (Qiagen) was added inside of the glovebox to three replicate wells for each condition. After incubating for 10 min, the wells were scraped to detach attached cells and the culture medium was transferred to a centrifuge tube and spinned for 5 min at 4000 rpm. Cell pellets were frozen at -80°C until RNA extraction. Total RNA was extracted using the RiboPure Bacteria Kit (Ambion, Life Technologies) and rRNA was removed using the MICROBExpress Kit (Ambion, Life Technologies). The quality of the remaining mRNA was checked using the Experion Automated Electrophoresis System with the RNA StdSens Analysis kit (BioRad). cDNA libraries were prepared using the ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre, Illumina) and for each sample a different Epicentre ScriptSeq Index PCR primer (Illumina) was used. The indexed libraries were pooled and 100 bp single reads were obtained from an Illumina HiSeq2000 (Deep Sequencing Core Facility, Umass Medical School). The obtained reads were cleaned and rRNA sequences were removed using the Galaxy platform (https://usegalaxy.org). Rockhopper [26] was used to map the reads against the C. ljungdahlii DSM13528 genome (ASM14368v1) (Ensembl Bacteria), assemble the transcripts, normalize the reads for the different replicates and experiments and test for differential expression between the two conditions. Insight into the regulation of different pathways was obtained by comparing differentially expressed genes against the pathways described for C. ljungdahlii in KEGG (http://www.genome.jp/kegg/). Genes are considered as significantly differentially expressed if the q-value (false discovery rate) is < 0.001 and the log2 fold change (FC) of the gene expression in the biofilm versus the planktonic condition is <-1 (downregulated) or >1 (upregulated). The obtained sequences are deposited at the NCBI SRA under accession number SRP094966.
Results
Screening of the C. ljungdahlii biofilm forming capacity
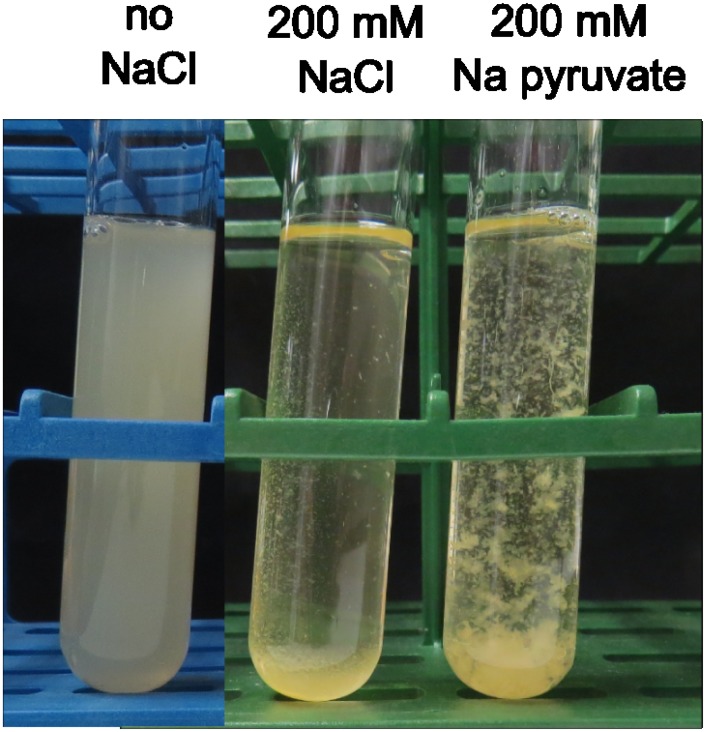
The investigation of biofilm formation by C. ljungdahlii was initiated by observing its growth in Balch tubes. Other Clostridium species are known to form biofilms when grown in rich media [22–24], but only planktonic growth could be observed for C. ljungdahlii when grown in the adapted YTF medium (Fig 1, left). For this reason, biofilm formation under different stress conditions was tested (Table 1). Salt stress induced by sodium chloride led to the formation of a very sticky cell pellet at the bottom of the tube, as well as clear attachment to the wall of the tube (Fig 1, middle). The addition of potassium chloride led to a similar phenotype as observed with NaCl, while no attachment and only attachment to the bottom of the tube was observed with respectively calcium and magnesium chloride (Table 1). Another salt that led to a phenotype clearly different from planktonic growth was sodium pyruvate, which was added as a substrate instead of fructose. With this salt, large and strong aggregates were formed, that could not be suspended by shaking (Fig 1, right; Table 1). However, no attachment to the tube wall could be observed with the addition of sodium pyruvate.
Fig 1. Visual effect of different growth conditions.
C. ljungdahlii was grown in tubes without (left) or with (middle) the addition of 200 mM NaCl to the medium. In addition, C. ljungdahlii was grown with 200 mM sodium pyruvate, while fructose and was omitted from the medium (right). The pictures were taken after 3 days of incubation.
Table 1. Overview of the C. ljungdahlii phenotypes obtained in different stress conditions.
The lower limit of the given concentration range reflects the lowest concentration at which the phenotype was observed, while at concentrations above the given upper limit, the induced stress was so strong that no growth was observed after ten days of incubation.
| Stress factor | Phenotype observed in tubes | Concentration |
|---|---|---|
| NaCl | sticky cell pellet at the bottom of the tube, attachment to the tube wall | 200–280 mM |
| KCl | sticky cell pellet at the bottom of the tube, attachment to the tube wall | 200–280 mM |
| CaCl2 | slower growth, all planktonic, no attachment or aggregation | 125–175 mM |
| MgCl2 | sticky cell pellet at the bottom of the tube, but no attachment to the tube wall | 125–175 mM |
| Na pyruvate | large aggregates, no attachment | 200–280 mM |
| Na2S | small aggregates, no attachment | 4–8 mM |
| thiamphenicol | small aggregates, no attachment | 1.5–3 μg·mL-1 |
| Lower temperature | slower growth, all planktonic, no attachment or aggregation | 25°C |
Biofilm formation was further tested in other types of stress conditions, including antibiotic stress from thiamphenicol, chemical stress from sodium sulfide and incubation at a suboptimal temperature. All these conditions led to a slower growth of C. ljungdahlii and sometimes to the formation of small aggregates (Table 1). However, for none of these conditions, attachment to the tube wall could be observed. Since the most pronounced attachment was obtained with the addition of NaCl to the medium, we further focused this study on NaCl induced biofilm formation by C. ljungdahlii.
The effect of sodium chloride on C. ljungdahlii biofilm formation
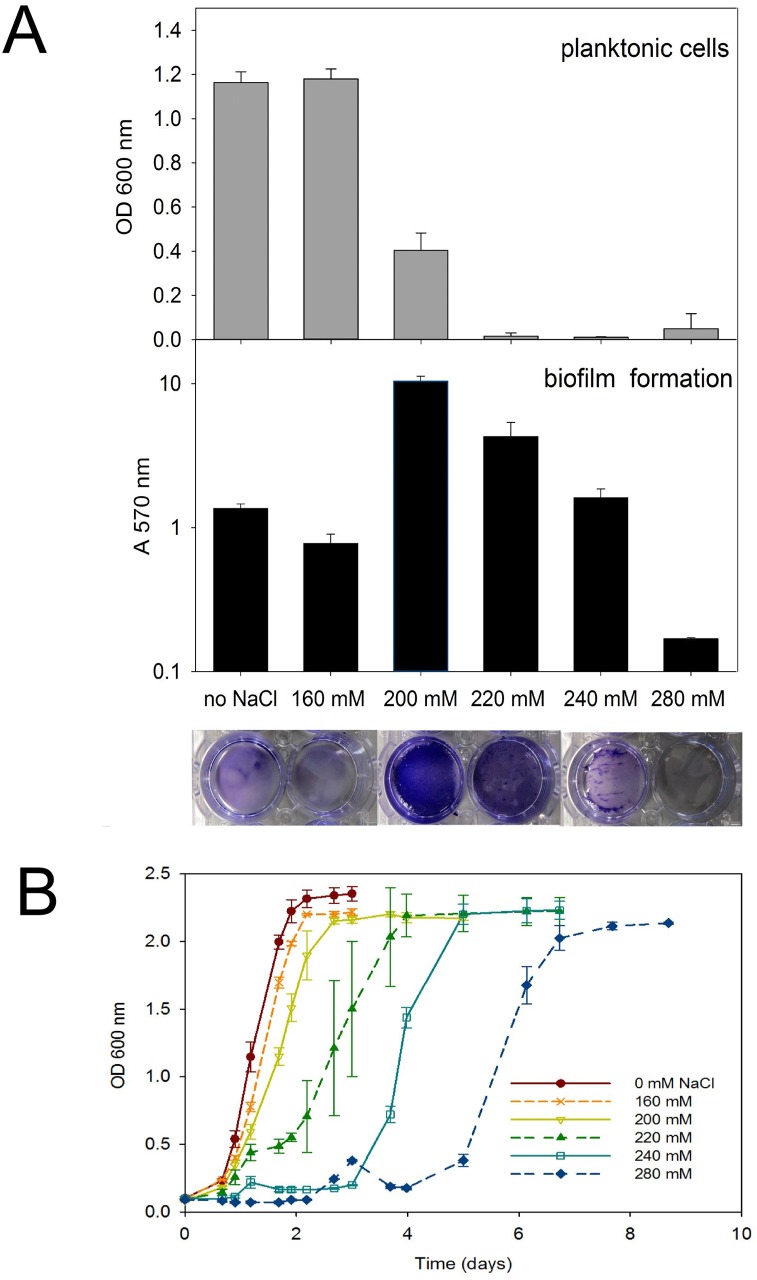
NaCl induced biofilms of C. ljungdahlii were further studied in well plates and the crystal violet assay was used to quantify biofilm formation. Firstly, the effect of the NaCl concentration (ranging from 0 to 280 mM) was investigated (Fig 2A). After 3 days of incubation, there was a high number of planktonic cells (high OD 600 nm values) and limited attachment (low A 570 nm values), without the addition of NaCl to the medium, confirming the observation in tubes (Fig 1). Also with the addition of 160 mM NaCl to the medium, cells were mainly planktonic and attachment was limited (Fig 2A). In contrast, with the addition of 200 mM NaCl to the medium, a thick biofilm was formed, as the attachment was ten times higher and the optical density of the solution was three times lower than without the addition of NaCl (Fig 2A). With a further increase of the NaCl concentration, the attachment decreased, while the amount of planktonic cells remained low. The thinner biofilms at the higher NaCl concentrations can be explained by slower growth at those concentrations. This was confirmed by recording growth curves for C. ljungdahlii at different NaCl concentrations. Increasing NaCl concentrations were found to cause increasing lag phases (Fig 2B). For instance, with the addition of 160 mM NaCl, growth started only slightly later than without the addition of NaCl and was completed within two days, while with the highest NaCl concentration (280 mM), there was a lag phase of 5 days before C. ljungdahlii growth started, reflecting the increased stress. Nevertheless, final cell densities were similar for all tested NaCl concentrations.
Fig 2. Effect of the NaCl concentration on biofilm formation and growth.
A) C. ljungdahlii was grown in well plates and NaCl was added to the medium in concentrations ranging from 0 to 280 mM (n = 3). The crystal violet assay (described in text) was performed after 3 days of incubation. The pictures underneath the data bars show the corresponding, stained biofilms, before extraction with methanol. Remark that OD 600 nm values are given on a linear scale, while the A 570 nm values are on a logarithmic scale. B) C. ljungdahlii was grown in tubes and NaCl was added to the medium in concentrations ranging from 0 to 280 mM (n = 3). Tubes were vortexed at a low speed to break up the formed aggregates, before measurement of the OD 600 nm.
It should be noted that the minimum NaCl concentration and incubation time required for biofilm formation slightly varied between experiments, but mostly ranged between 200 to 220 mM and 2 to 3 days, respectively. For this reason, a combination of these concentrations and incubation times was used to induce biofilm formation in all further experiments. After prolonged incubation times, partial to complete degradation of the biofilm was often observed (results not shown). This biofilm degradation could be due to physical disintegration or to active dispersal of the cells out of the biofilm.
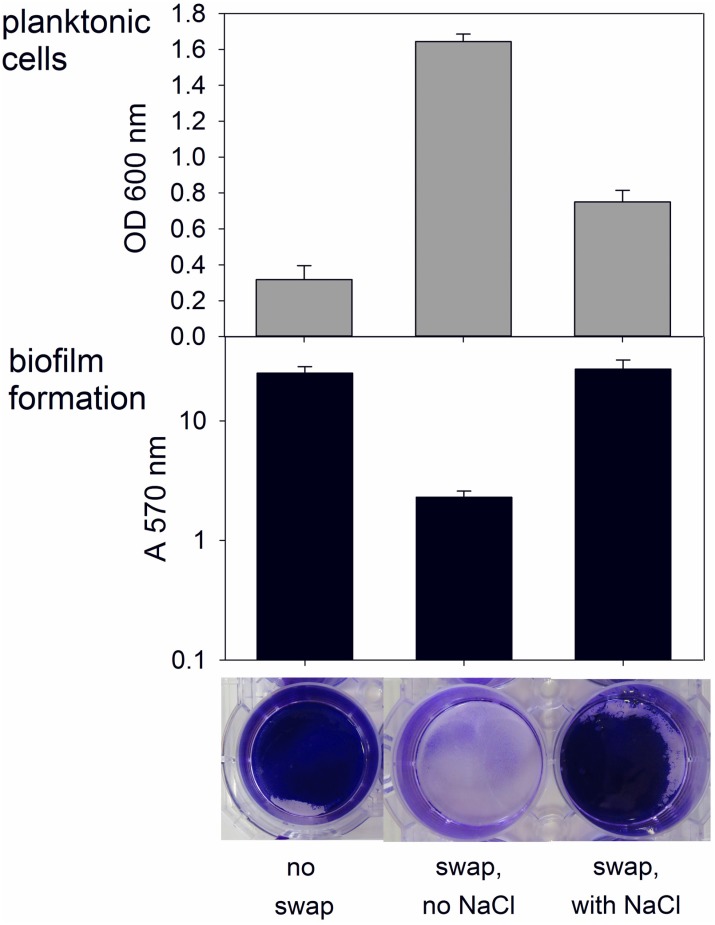
In addition, the crystal violet assay was used to investigate whether or not the addition of NaCl to the medium is required to maintain established biofilms. Hereto, biofilms were pregrown using NaCl and at the end of the incubation time, the supernatant was removed and fresh medium with or without NaCl was placed on the biofilms. One day after this medium swap, the wells with NaCl added to the fresh medium had similar biofilms as the control wells, for which the medium was not swapped (Fig 3). However, in the wells without NaCl addition to the fresh medium, the biofilms were completely degraded and only limited attachment remained. This was reflected by a ten times lower attachment and three times higher optical density of the supernatant than with the addition of NaCl to the fresh medium, demonstrating the need for NaCl in the medium to maintain established biofilms. It should be noted, however, that even with NaCl in the medium some biofilm degradation can be expected after prolonged incubation times.
Fig 3. Effect of a medium swap on established biofilms.
C. ljungdahlii biofilms were grown in well plates by adding 200 mM NaCl to the medium. The medium was not swapped in control wells (left), while, after 3 days of incubation, the supernatant of all other wells was carefully removed to not disturb the biofilms. Fresh medium without NaCl addition (middle) or with 200 mM NaCl (right) was added to the wells (n = 3). One day after the swap of the medium (four days after the start of the experiment), the crystal violet assay (described in text) was performed on all wells. The pictures underneath the data bars show the corresponding, stained biofilms, before extraction with methanol. Remark that OD 600 nm values are given on a linear scale, while the A 570 nm values are on a logarithmic scale.
C. ljungdahlii biofilms on different materials
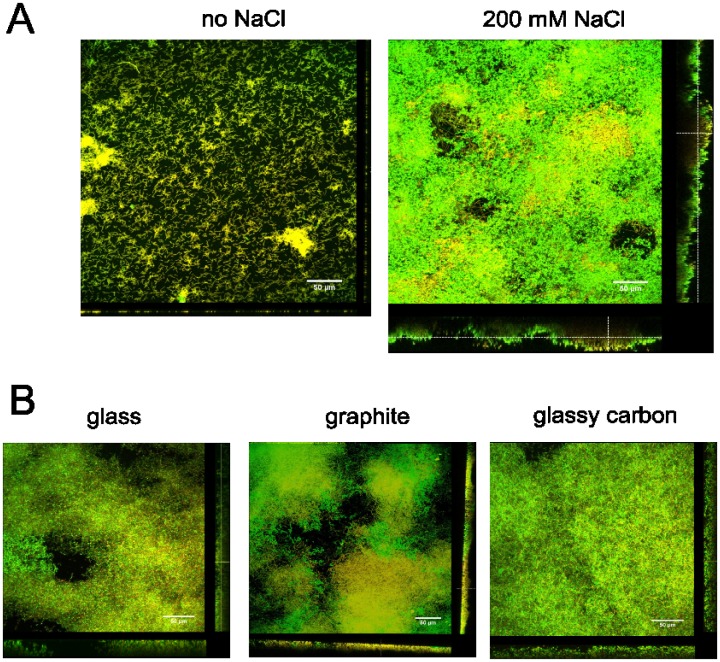
The C. ljungdahlii biofilms were further characterized and their thicknesses measured using live/dead staining and confocal laser scanning microscopy. The attachment with and without NaCl was compared for C. ljungdahlii grown in chamber slides (horizontal orientation). Without the addition of NaCl, individual cells and some cell aggregates of less than 7 μm thick were observed, covering only one third of the substrate (Fig 4A, left; Table 2). In contrast, with the addition of NaCl to the medium, structured biofilms with a thickness up to 48 μm and almost completely covering the substrate were visualized (Fig 4A, right; Table 2). These biofilms were mainly green with some yellowish or orangey patches, showing that a large amount of cells in the biofilm, but not all, had an intact membrane and were assumingly viable. Calculation of the biomass volume showed that with the addition of NaCl to the medium, the biofilm biomass was 20 times larger than without the addition of NaCl (Table 2).
Fig 4. Confocal laser scanning microscopy images of C. ljungdahlii biofilms.
A) Cells were grown in chamber slides without (left) or with (right) the addition of 200 mM NaCl to the medium. B) Cells were grown in tubes, in which a piece of glass (left), graphite (middle) or glassy carbon (right) was placed vertically and to which 200 mM NaCl was added. After 2 days of incubation, the biofilms were stained with live/dead staining as described in the text. The scale bars are 50 μm.
Table 2. Overview of the properties of the biofilms obtained with and without NaCl addition and on different materials.
The maximum biofilm thickness was obtained using COMSTAT, while the surface coverage was determined using FIJI. Averages and standard deviations were calculated from values derived from at least three different images taken at different areas of the biofilm. The biomass was calculated from the maximum biofilm thickness and the surface coverage and is therefore also a maximum value.
| 200 mM NaCl addition | no | yes | yes | yes | yes |
|---|---|---|---|---|---|
| Material | Permanox plastic | Permanox plastic | glass | graphite | glassy carbon |
| Orientation | horizontal | horizontal | vertical | vertical | vertical |
| Maximum biofilm thickness (μm) | 6.5 ± 1.7 | 48.4 ± 4.6 | 29.0 ± 4.7 | 31.5 ± 5.1 | 39.3 ±13.5 |
| Surface coverage (%)a | 31.4 ± 3.6 | 95.9 ± 2.1 | 92.9 ± 1.4 | 91.6 ± 2.0 | 99.0 ± 0.7 |
| Biomass (μm3·μm-2) | 2.0 | 46.4 | 26.9 | 28.9 | 39.1 |
In addition, the salt-induced attachment of C. ljungdahlii to other materials, including glass, graphite and glassy carbon, the latter two representing conductive materials often used as electrodes in bio-electrochemical systems, was tested. These materials were placed vertically in a tube to simulate the orientation of the electrode in most bio-electrochemical setups. Also on these materials with a vertical orientation, C. ljungdahlii was able to form viable biofilms with the addition of NaCl to the medium (Fig 4B). The vertical biofilms grown on the different materials slightly differed in surface coverage, but there was no significant difference in their maximum biofilm thickness (Table 2). Remarkably, even vertically, the salt-induced biofilms were rather thick.
Composition of the C. ljungdahlii biofilm matrix
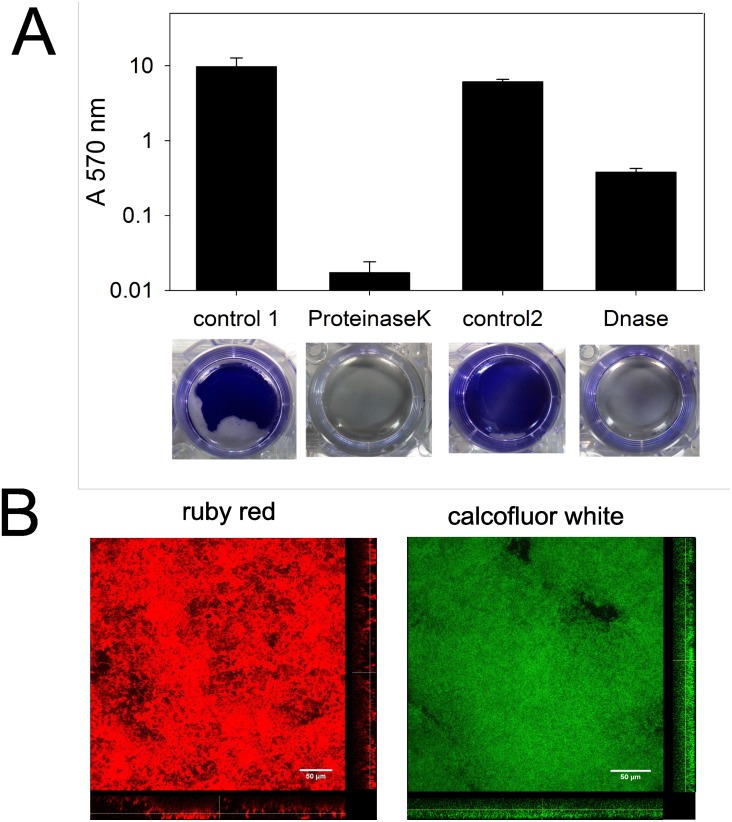
The composition of the C. ljungdahlii biofilm matrix was examined similarly as described before [23, 27, 28]. Enzyme treatment using proteinaseK or DNaseI was used to investigate the involvement of respectively extracellular proteins and DNA. Hereto, biofilms were grown in well plates and after the required incubation time, the biofilms were treated with enzyme and the remaining biofilm was quantified using the crystal violet assay. Both enzymes caused the complete degradation of the biofilms, as the attachment was one to three orders of magnitude lower than in the controls without enzyme treatment (Fig 5A). In addition, specific dyes were used to investigate the role of extracellular proteins and polysaccharides in C. ljungdahlii biofilms. Hereto, biofilms were grown in chamber slides and were stained after the required incubation time with SYPRO ruby red biofilm matrix stain or calcofluor white. Ruby red labels most classes of proteins, including glycoproteins, phosphoproteins, lipoproteins, calcium binding proteins and fibrillar proteins and clearly stained the biofilm matrix of C. ljungdahlii (Fig 5B, left). In addition, calcofluor white, which specifically stains the beta-1,3 and beta-1.4 bindings of polysaccharides [22], was found to stain the biofilm (Fig 5B, right). These results show that extracellular proteins, polysaccharides and DNA are all involved in the biofilm matrix of C. ljungdahlii.
Fig 5. Investigation of the biofilm composition.
A) Effect of enzyme treatment. C. ljungdahlii biofilms were grown in well plates by adding 200 mM NaCl to the medium. After 2 days of incubation, the supernatant of all wells was removed and the biofilms were washed twice with PBS buffer. PBS buffer without (control 1) or with 0.2 mg·mL-1 proteinaseK or reaction buffer (10 mM Tris HCl pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2) without (control 2) or with 2 U·mL-1 DNaseI was added to the wells (n = 3). After one hour of enzyme treatment at 37°C, the crystal violet assay (described in text) was performed. B) Confocal laser scanning microscopy images of component-specific stained C. ljungdahlii biofilms. C. ljungdahlii was grown in chamber slides with the addition of 200 mM NaCl to the medium and, after 2 days of incubation, the biofilms were stained as described in the text with SYPRO ruby red biofilm matrix stain (left) or calcofluor white (right). The scale bars are 50 μm long.
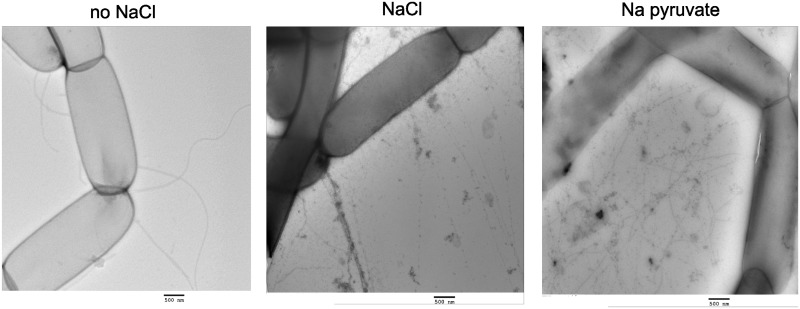
To further examine the type of extracellular proteins involved in the biofilm formation, TEM was used to visualize the cell appendages of C. ljungdahlii. Without the addition of salt to the medium, long and curvy flagella were observed, but no pilus-like structures, often involved in biofilm formation, could be seen (Fig 6, left). In contrast, with the addition of 200 mM sodium chloride to the medium, less flagella were present, but networks of pilus-like fibers could be observed between some closely located cells (Fig 6, middle). Also with sodium pyruvate as substrate instead of fructose, these fibers were present and even much more pronounced, as the observed networks of pilus-like appendages were very dense and seen between almost all cells (Fig 6, right). The observed fibers likely play a role in binding cells together, rather than in binding cells to a surface, as the addition of sodium pyruvate led to the formation of large aggregates, but not to attachment (Fig 1). The observed pilus-like fibers were mostly seen covered by apparent debris and were often more than 10 μm long with a diameter of 6 to 10 nm and were therefore clearly thinner than flagella (20–25 nm diameter).
Fig 6. TEM images of C. ljungdahlii appendages in different growth conditions.
C. ljungdahlii was grown in tubes without (left) or with (middle) the addition of 200 mM NaCl to the medium. In addition, C. ljungdahlii was grown with 200 mM sodium pyruvate, while fructose was omitted from the medium (right). After 2 days of incubation, cells were harvested and grids were prepared as described in the text. The scale bars are 500 nm.
RNA sequencing results
The gene expression of C. ljungdahlii with and without salt addition to the medium was analyzed to obtain mechanistic insight into its response to salt stress and biofilm formation. Hereto, cells were grown in well plates with and without the addition of 200 mM NaCl to the medium (n = 6). Wells without NaCl addition were harvested after 1 day of incubation, while wells with NaCl addition were harvested after 2 days, to ensure that cells in both conditions would be in their exponential growth phase (Fig 2B). Three replicate wells were harvested for RNA extraction, while the other three replicates were used to assess biofilm formation using the crystal violet assay. At the time of harvesting, the attachment with NaCl addition was two orders of magnitude higher and the optical density six times lower than without NaCl addition (Fig A in S2 File), confirming that with NaCl addition the cells formed a biofilm, while they were planktonic without NaCl addition.
General gene expression
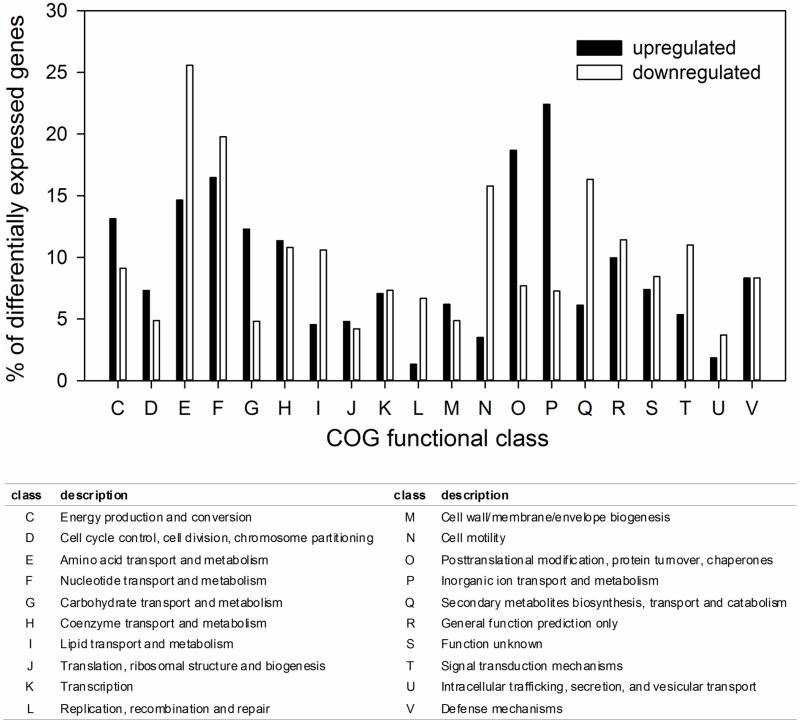
The reference genes gyrA, fotl and rho [29] were not differentially expressed between the planktonic and the biofilm condition (results not shown), illustrating that the normalization between the different treatments was performed well. A total of 403 genes (9.4% of the genes of the C. ljungdahlii genome) were significantly upregulated in the biofilm versus the planktonic condition (log2 FC >1 and q-value < 0.001) and 103 genes (2.4%) were even more than fivefold upregulated (log2 FC > 2.32) (S1 File). In addition, a total of 417 genes (9.7%) were significantly downregulated with the addition of NaCl compared to without NaCl addition (log2 FC <-1 and q-value < 0.001) and 102 genes (2.4%) were even more than five fold downregulated (log2 FC < -2.32). A first overview of the function of the differentially expressed genes was obtained using the cluster of orthologous groups (COG) classification (Fig 7). This analysis showed that the genes involved in amino acid transport and metabolism (class E) and nucleotide transport and metabolism (class F) were strongly differentially expressed. In addition, there was a strong upregulation of the genes involved in inorganic ion transport and metabolism (class P), carbohydrate transport and metabolism (class G) and posttranslational modification, protein turnover and chaperones (class O). Strong downregulation was found for the genes involved in cell motility (class N) and secondary metabolites biosynthesis, transport and catabolism (class Q).
Fig 7. Distribution of up- and downregulated genes in the biofilm versus the planktonic condition over the different COG functional classes.
Regulators
Several transcriptional regulators have been reported to affect biofilm formation or attachment by C. difficile and perfringens [23, 24, 30–35]. For this reason, the expression of these regulators in C. ljungdahlii was compared between the biofilm and planktonic condition (Table 3). The only significantly differentially expressed regulator was lexA, which was downregulated with a log2 FC of -1.83. In addition, the spo0A gene showed an upregulated trend and the putative ccpA gene (CLJU_c01900) a downregulated trend. Other regulators known to affect biofilm formation in other Clostridium species, including luxS, ctrAB, codY, agrB and abrB, were not differentially expressed. In addition to the regulators known to be involved in C. difficile or perfringens biofilm formation, also many other transcriptional regulators were significantly up- or downregulated (Table A in S2 File).
Table 3. Overview of the expression of transcriptional regulators known to affect biofilm in other Clostridium species.
a Expression 1 reflects the gene expression of the planktonic cells (no NaCl addition). b Expression 2 reflects the gene expression of the biofilm cells (NaCl addition). c log2 FC expresses the gene expression of the biofilm versus the planktonic cells. d A star marks a q-value (false discovery rate) lower than 0.001. Genes in bold are significantly differentially expressed (q-value < 0.001 and log2 FC <-1 or >1).
| Name | Locus tag | Annotation | Expression 1a | Expression 2b | log2 FCc | qd | reference |
|---|---|---|---|---|---|---|---|
| ccpA | CLJU_c01900 | LacI family transcriptional regulator | 934 | 469 | -0.99 | * | [32] |
| ctrAB | CLJU_c06150/60 | transcriptional regulator | 169 | 114 | -0.57 | [33] | |
| spo0A | CLJU_c11220 | Spo0A-like protein | 181 | 319 | 0.82 | * | [23, 24, 33] |
| codY | CLJU_c13010 | transcriptional repressor CodY | 580 | 488 | -0.25 | [35] | |
| lexA | CLJU_c21030 | LexA repressor | 358 | 101 | -1.83 | * | [31] |
| luxS | CLJU_c23480 | S-ribosylhomocysteinase | 1023 | 918 | -0.16 | [23, 34] | |
| - | CLJU_c27530 | AgrB-like protein | 0 | 2 | 0.00 | [34] | |
| - | CLJU_c28480 | AgrB-like protein | 14 | 9 | -0.64 | [34] | |
| abrB | CLJU_c41700 | AbrB family transcriptional regulator | 4 | 5 | 0.00 | [33] |
Motility, cell appendages and sporulation
Several genes involved in the biosynthesis of flagella, i.e. mainly those encoding for flagellin proteins, the hook-filament junction, the filament cap and the flagellar motor proteins (KEGG), were significantly downregulated in the biofilm versus the planktonic condition (Table B in S2 File). This is in agreement with our TEM observations of less flagella for the biofilm cells compared to the planktonic cells (Fig 6) and is a common finding for biofilms [36]. In addition, several genes involved in chemotaxis were significantly downregulated in the biofilm cells (Table C in S2 File). Two genes clusters of putative type IV pili biosynthesis genes were identified using PilFind [37], including a pil operon (CLJU_c10900 to 10940) and a tad operon (CLJU_c28790 to 28890). These genes were not expressed in the planktonic nor in the biofilm condition (Table D in S2 File), even though pilus-like appendages were observed for cells grown with NaCl addition (Fig 6). Only pilT, the gene encoding for twitching motility and involved in the retraction of type IV pili, was expressed in both conditions and was significantly downregulated in the biofilm versus the planktonic cells. In several gram-negative bacterial species, pilT gene deletions resulted in hyperpiliation and improved biofilm formation [38, 39]. In contrast, C. perfringens is the only species for which the pilT gene was found to be essential for biofilm formation [22]. Most sporulation genes were not expressed in the biofilm nor in the planktonic condition (results not shown). This is consistent with the lack of spores observed in the studied conditions, as well as the rare sporulation reported for C. ljungdahlii in general [1].
Salt stress
The highest upregulated gene encodes for a Na+/H+ antiporter (CLJU_c40350, log2 FC of 6.63) (Table E in S2 File). This demonstrates that C. ljungdahlii coped with the NaCl stress by pumping Na+ out of the cell. Also several other Na+ and other cation antiporters and symporters were significantly up- or downregulated (Table E in S2 File).
Stress response in general is known to imply the upregulation of genes encoding for stress proteins and chaperones, which are involved in the refolding, export or degradation of improperly folded proteins [40]. As expected, several of those genes known for C. ljunghdahlii were significantly upregulated (Table F in S2 File). The highest upregulation was recorded for the gene encoding for the carbon starvation protein A (CLJU_c37340, log2 FC 4.2), while also several heat shock proteins and chaperones were significantly upregulated. In addition, rubrerythrin, a non-haem Fe protein reported to be involved in the response of C. ljungdahlii to oxygen stress [41], was upregulated.
A common bacterial strategy to overcome osmotic stress is the accumulation of compatible solutes or osmoprotectants in the cytoplasm to increase the intracellular osmolarity [40]. Betaine is the preferred osmoprotectant for many bacteria and the whole gene cluster encoding for the degradation of betaine to acetate [10] was significantly downregulated in the biofilm versus the planktonic cells (Table G in S2 File). Betaine cannot be synthesized by most microorganisms and is usually transported into the cell [40]. Yeast extract, present is the TYF medium used in this study, is a known source of betaine [42]. The downregulation of the degradation of betaine, therefore, suggests that C. ljungdahlii imported betaine and accumulated it in its cytoplasm as an osmoprotectant.
In addition, a gene cluster encoding for peptide ABC transporters (CLJU_c22540-22670), which was significantly upregulated in the biofilm versus the planktonic condition (Table H in S2 File), is potentially involved in the osmotic stress response. The import of peptides and their subsequently cleavage into amino acids is a strategy used by several bacteria to overcome osmotic stress [43, 44], since mainly the amino acid proline, but also glutamate and glutamine, can act as osmoprotectants in addition to betaine [40]. Both tryptone and yeast extract, present in the used TYF medium, are sources of peptides. The uptake of peptides for osmoregulation by C. ljungdahlii can also explain the strong upregulation of a peptidase (pepT2, CLJU_c19310, log2 FC of 6.0), as well the significant upregulation of a Xaa-Pro dipeptidase (pepQ2, CLJU_c29640) (Table H in S2 File), potentially cleaving the imported peptides. Alternatively, the upregulated peptide ABC transporters (Table H in S2 File) could play a role in the secretion of signaling peptides, which are also of importance for stress response and biofilm formation by gram-positive bacteria [45]. Interestingly, a cluster of homologous genes was also upregulated in biofilm cells of C. acetobutylicum [46]. So, the involvement of the peptide ABC transporters in biofilm formation is another possibility.
Biofilm formation
Another strongly upregulated gene encodes for an alanine racemase (alr3, CLJU_c40390, log2 FC 5.27) (Table I in S2 File). This enzyme is part of the pathway converting L-alanine over D-alanine into D-alanyl-d-alanine and also the genes encoding the other enzymes for this pathway were strongly upregulated (Table I in S2 File). In addition, the biosynthesis of UDP-N-acetylglucosamine was significantly upregulated (Table I in S2 File). Both D-alanyl-d-alanine and UDP-N-acetylglucosamine are precursor for peptidoglycan synthesis. These findings might suggest that C. ljungdahlii strengthened its cell wall as another strategy to cope with the osmotic stress resulting from the NaCl addition. Some of the peptidoglycan biosynthesis genes showed an upregulated trend, however, most of those genes were not differentially expressed (Table J in S2 File).
Alternatively, the upregulation of D-alanyl-d-alanine and UDP-N-acetylglucosamine biosynthesis can be related to biofilm formation. UDP-N-acetylglucosamine is also the precursor for poly-N-acetylglucosamine, which is an extracellular polysaccharide essential for biofilm formation of many bacteria, including Staphylococcus and Bacillus species [47–49]. The biosynthesis of this polysaccharide in Staphylococcus is encoded by the ica operon, which contains a glycosyltransferase, a deacetylase and an acetyl transferase [50]. Homologous genes were not found in the genome of C. ljungdahlii, but several genes encoding enzymes with similar functions were upregulated in the biofilm versus the planktonic cells (Table I in S2 File), indicating that poly-N-acetylglucosamine can also play a role in the biofilm formation by C. ljungdahlii.
In addition, D-alanine is potentially involved in biofilm formation. A common modification of cell wall teichoic acids is their esterification with D-alanine [51]. This modification gives a positive charge to the outer cell surface and is essential for the biofilm formation of Staphylococcus species [48]. The upregulation of the D-alanine metabolism, therefore, suggests an involvement in biofilm formation, although no homologues of the genes responsible for teichoic acid esterification with D-alanine (dlt operon in Staphylococcus) could be found in the C. ljungdahlii genome.
Nucleotide metabolism
Many genes of the purine metabolism were strongly upregulated (Table K in S2 File). These include mostly the genes involved in the pathway from ribose till inosine monophosphate (IMP), while the genes encoding the further conversion of IMP were not differentially expressed. In addition, the genes encoding an ABC transporter for the uptake of ribose, the precursor of the purine metabolism, were strongly upregulated (Table K in S2 File). In contrast, many genes of the pyrimidine metabolism were significantly downregulated (Table K in S2 File). These genes were mainly involved in the biosynthesis of uridine monophosphate (UMP) from glutamine, while its further conversion was not differentially expressed. Interestingly, similar results were previously reported for C. acetobutylicum exposed to metabolite stress from acetate, butyrate or butanol [52, 53]. Alsaker et al. [52] explained the upregulation of the purine metabolism by the fact that one of its intermediates (aminoimidazole ribotide, AIR) is needed for the biosynthesis of thiamine (vitamin B1), for which some genes were upregulated in their study. In this study, however, the biosynthesis of thiamine was not differentially expressed (results not shown). Moreover, in both studies the pathway past AIR till IMP was also upregulated. For this reason, the role of the differential expression of the nucleotide metabolism in stress response remains unknown.
Amino acid metabolism
Many genes involved in the biosynthesis or conversion of amino acids were differentially expressed. Three very strongly upregulated genes encode for a threonine synthase (CLJU_c40360, log2 FC 6.55), a 2-iminobutanoate/2-iminopropanoate deaminase (CLJU_c40400, log2 FC 6.61) and another enamine/imine deaminase (CLJU_c40370, log2 FC 5.95) (Table L in S2 File). These genes are likely involved in the conversion of homoserine over threonine to oxobutyrate. Oxobutyrate is a precursor for the biosynthesis of isoleucine, but the biosynthesis of this compound, as well as of the other branched amino acids valine and leucine, was downregulated in the biofilm versus planktonic cells (Table L in S2 File). Alternatively, oxobutyrate can be converted to propanoyl-CoA or propionate and can be an end product of C. ljungdahlii [5]. In addition, the enamine/imine deaminases could play a role in the degradation of toxic cell metabolites [54]. So far, it remains unclear how these genes are involved in the biofilm formation or response to salt stress by C. ljungdahlii.
In addition, the genes encoding the biosynthesis of histidine were all significantly upregulated in the biofilm versus the planktonic cells (Table L in S2 File). Also the regulator of the histidine metabolism (hisZ) was significantly upregulated (Table A in S2 File). In contrast, the conversion of histidine to glutamate was strongly downregulated (Table L in S2 File). Previously, the upregulation of histidine synthesis was already shown after an alkaline shock in C. difficile [55] and metabolite stress in C. acetobutylicum [52]. The histidine metabolism forms an intermediate for the purine biosynthesis, which was also upregulated as described above (Table K in S2 File).
Furthermore, the genes involved in the biosynthesis of cysteine from serine were significantly upregulated (Table L in S2 File). Similar findings were reported for biofilm cells and after metabolite stress in C. acetobutylicum [46, 52].
In contrast, the genes involved in the synthesis of tryptophan were strongly downregulated, while the formation of chorismate, tyrosine and phenylalanine were not differentially expressed (Table L in S2 File). Similar findings were reported for C. acetobutylicum exposed to metabolite stress [52, 53] and a recent study demonstrated that tryptophan acts as an incompatible solute inhibiting growth of several bacterial species during osmotic stress [56].
In addition, the genes responsible for the biosynthesis of arginine were strongly downregulated (Table L in S2 File). Arginine is a precursor for the pyrimidine metabolism, which was also downregulated as described above (Table K in S2 File). Differential expression of arginine biosynthesis genes was also found with metabolite stress in C. acetobutylicum [53], while their upregulation was shown for C. acetobutylcum biofilm cells [46].
Finally, several amino acid transporters were differentially expressed (Table M in S2 File). There was a high upregulation for two amino acid permeases (CLJU_c19320, log2 FC of 5.85, and CLJU_c24250, log2 FC of 5.13), but it is unknown which amino acid they transport.
Substrate utilization
The putative fructose uptake genes [10] were not differentially expressed, but almost all genes encoding for the glycolysis from fructose to pyruvate and its further conversion to acetate were significantly upregulated (Table N in S2 File). In contrast, the gene adhE1 (CLJU_c16510), mainly responsible for the production of ethanol by C. ljungdahlii [13], was strongly downregulated. Also a gene encoding a putative aldehyde oxidoreductase (CLJU_c24130), which was involved in the increased ethanol production after oxygen stress [41], was significantly downregulated (Table N in S2 File). These results are in contrast to what was expected, since stress causes a slower metabolism, while ethanol production by C. ljungdahlii is often induced by stress from, for instance, a low pH or oxygen exposure [41, 57]. Our findings, therefore, might reflect different growth characteristics of the planktonic and the biofilm cells. An upregulation of the glycolysis pathway in comparison to planktonic cells was already reported for biofilm cells of C. acetobutylicum [46], but in the latter study biofilm formation was not induced by stress. Alternatively, our results can be explained by the different time points the cells were harvested (after 1 day of incubation for the planktonic cells and 2 days for the biofilm). Harvesting the cells at the same time point, however, would have made the comparison of the gene expression between the two conditions impossible, since no biofilm growth was observed after 1 day of incubation (results not shown), while at day 2 the planktonic cells would have reached their stationary phase (Fig 2B).
Iron and molybdate uptake
Many genes encoding iron and iron complex transporters were significantly upregulated in the biofilm versus the planktonic cells (Table O in S2 File). In addition, an iron uptake regulation protein (fur3, CLJU_c16280) was upregulated, while another one (fur5, CLJU_c37710) was significantly downregulated (Table A in S2 File). Also several Fe containing proteins were upregulated (Table O in S2 File). Furthermore, the genes encoding molybdate ABC transporters were significantly upregulated, while also a molybdenum cofactor biosynthesis family protein was strongly upregulated (CLJU_c17110, log2 FC of 4.0) (Table O in S2 File). The upregulation of genes encoding for iron uptake and iron containing proteins was previously already reported for C. acetobutylicum exposed to butyrate stress [52]. Similar genes were also upregulated in biofilm cells of C. acetobutylicum [46]. Iron and molybdenum containing enzymes play an important role in the metabolism of C. ljunghdahlii [10] and the upregulation of the glycolysis (described above, Table N in S2 File) could also explain the upregulation of Fe and molybdate containing proteins and transporters.
Translation
Many tRNAs and several of their synthetases were significantly upregulated in the biofilm versus the planktonic cells (Table P in S2 File). This demonstrates that translation in the biofilm cells was enhanced, which is likely also related to the upregulation of the glycolysis (described above, Table N in S2 File). In addition, several genes involved in the biosynthesis of putrescine were upregulated, while genes encoding a putrescine/spermidine ABC transporter were downregulated (Table Q in S2 File). Polyamines, such as putrescine, are known to be important for translation and protein synthesis and their positive effect on translation by Escherichia coli was even larger under heat stress [58].
Others
Interestingly, several of the highest upregulated genes are located in few gene clusters, in which also some yet unidentified genes were predicted by the transcriptome analysis (Table R in S2 File). The highest upregulated cluster (CLJU_c40350-40400) contains several genes which were discussed above and are likely involved in various processes such as Na+ export, biofilm formation and amino acid metabolism. However, the potential role in salt stress response or biofilm formation of some other highly upregulated genes, such as a gene encoding for nucleoside-5'-diphosphate-sugar dehydratase (CLJU_c37840) and a dihydrodipicolinate synthase (dapA1, CLJU_c04300) (Table R in S2 File), remains unknown. Moreover, it should be noted that 86 of the upregulated genes and 107 of the downregulated genes are annotated as hypothetical proteins of which the function is still completely unknown.
Discussion
Biofilm formation is part of the salt stress response of C. ljungdahlii
The results presented in this study show that biofilm formation by C. ljungdahlii is induced by the addition of NaCl to the medium (Figs 1, 2A and 4A). Higher ionic strengths are known to reduce the repulsion between a bacterial cell and a material surface, which are both typically negatively charged [59]. The induction of C. ljungdahlii biofilm formation by NaCl, however, was not just a physical effect, but rather a biological response to the experienced stress. This was clear from the NaCl concentrations that triggered biofilm formation, as they adversely impacted the growth rate (Fig 2B). In addition, several other stress factors induced phenotypes showing attachment or aggregation (Table 1). Moreover, the gene expression analysis clearly showed that the general stress response system was triggered in the biofilm condition (many stress proteins and chaperones were upregulated, Table F in S2 File), while also several specific mechanisms to overcome the osmotic stress from the NaCl addition were activated. These include the Na+ transport out of the cell (Table E in S2 File), the potential strengthening of the cell wall (Table J in S2 File), as well as the accumulation of osmoprotectants in the cytoplasma. The latter was clear from the downregulation of the betaine degradation (Table G in S2 File) and also the upregulation of peptide transporters and peptidases (Table H in S2 File). In addition, the differential expression of the amino acid and nucleotide metabolism (Tables K and L in S2 File) seems to be related to stress response, since similar gene expression patterns were found for C. acetobutylicum exposed to metabolite stress [52, 53], even though their exact role in the stress response remains largely unknown. Finally, also biofilm formation was likely a mechanism to protect the cells from the high salt environment.
As far as we know, our study is the first to show that salt stress induces biofilm formation in a Clostridium species. For C. difficile, the effect of 300 mM NaCl was already tested, but this addition resulted in a decreased biofilm formation [23]. For both C. difficile and C. perfringens, however, it has been shown that biofilms offer protection against an adverse environment, such as the exposure to antibiotics and oxygen [23, 24, 28]. In addition, for other gram-positives such as Staphylococcus species and Listeria monocytogenes, it is well known that salt stress induces biofilm formation [60–63] and, for S. aureus and S. epidermidis, insights into the regulatory mechanism triggered by NaCl are available [64, 65].
Regulation of the stress response and biofilm formation by C. ljungdahlii
Several transcriptional regulators have been reported to affect biofilm formation or attachment by C. difficile and perfringens, including LexA, Spo0A, CcpA, LuxS, CtrAB, CodY, AgrB and AbrB [23, 24, 30–35]. Our study found that of these regulators, only LexA was significantly differentially expressed and downregulated in the biofilm versus the planktonic condition (Table 3). LexA is the global transcriptional repressor of the SOS network responding to DNA damage [40]. The putative DNA repair genes in C. ljungdahlii were not differentially expressed (results not shown), but besides the core SOS genes, LexA also affects the expression of genes encoding for various other functions. For instance, a mutation in the lexA gene led to reduced flagellar motility and increased biofilm formation in Pseudomonas aeruginosa [66], as well as in C. difficile [31]. For this reason, the lexA gene seems an interesting target to further study the regulation of biofilm formation in C. ljungdahlii, as well as to engineer a C. ljungdahlii strain with an improved attachment.
In addition, the spo0A gene showed an upregulated trend in the biofilm condition, even though its expression was not significantly different (Table 3). Spo0A is the master regulator for sporulation, but it also directly or indirectly affects the transcription of many other genes [67]. In Bacillus and Clostridium species, Spo0A is involved in stress response and biofilm formation. Metabolite stress in C. acetobutylicum, for instance, caused the upregulation of spo0A, while similarly as in our study, sporulation genes were not differentially expressed [52]. In addition, spo0A deletion mutants of Bacillus subtilis and C. difficile are deficient in biofilm formation [23, 24, 68]. Therefore, the potential role of spo0A in the C. ljungdahlii biofilm formation warrants further investigation.
Furthermore, our gene expression analysis found a downregulated trend (not significantly different) for the putative catabolite control protein CcpA (Table 3). CcpA regulates the response to carbohydrate availability and is required for biofilm formation of B. subtilis, S. aureus and C. perfringens [22, 69, 70].
Finally, many other transcriptional regulators, which are not known to be involved in stress response or biofilm formation, were differentially expressed (Table A in S2 File), while the C. ljungdahlii regulators triggered by NaCl remain unknown. Therefore, in-depth future investigations will be required to unravel the regulatory mechanism behind C. ljungdahlii biofilm formation triggered by salt stress.
Composition of the C. ljungdahlii biofilm matrix
Our experimental analysis showed that the C. ljungdahlii biofilms were composed of extracellular polysaccharides, proteins, as well as DNA (Fig 5), similarly as was previously reported for C. difficile and C. perfringens biofilms [23, 27, 28]. Our gene expression analysis showed that the involved extracellular polysaccharides could be poly-N-acetylglucosamines, as the biosynthesis of their precursor, UDP-N-acetylglucosamine, was upregulated, as well as some potentially involved polysaccharide biosynthesis genes (Table I in S2 File). Poly-N-acetylglucosamines are known to play a major role in the biofilm formation of various species, including the gram-positives Staphylococcus and Bacillus [47–49]. In addition, the gene expression analysis showed the strong upregulation of the D-alanine metabolism (Table I in S2 File). D-alanine is used for esterification of cell wall teichoic acids, giving them a positive charge and improving attachment to the negatively charged surface of most materials [51]. This modification is important for biofilm formation by several species, including Staphylococcus [48, 51], and could also play a role in the C. ljungdahlii biofilm formation.
The extracellular DNA present in the C. ljungdahlii biofilm matrix was most likely the result of the lysis of some of the cells [71]. In Staphylococcus biofilms, this cell lysis is enhanced by autolysin [48, 72], but the autolysin gene of C. ljungdahlii (CLJU_c04300) was not differentially expressed in the biofilm versus the planktonic condition (results not shown).
The extracellular proteins involved in the C. ljungdahlii biofilm matrix could be the observed pilus-like fibers (Fig 6). Network of these fibers were observed between some of the cells with, but not without, the addition of NaCl to the medium, suggesting an inducible role in the C. ljungdahlii biofilm formation. Type IV pili were already attributed a role in the aggregation of C. difficile [73, 74] and the biofilm formation of C. perfringens [22, 75]. For wild type C. perfringens and C. difficile, pili have only been shown for cells grown on agar plates [73, 75, 76], suggesting that also for these species the expression of pili depends on the growth conditions. Besides type IV pili, also other types of pili have been found for gram-positive bacteria, including amyloid fibers for Bacillus species [77, 78] and covalently bound pilins for Streptococcus species [79]. Two gene clusters of putative type IV pili biosynthesis genes were identified in the genome of C. ljungdahlii, but none of those genes were expressed in the planktonic nor in the biofilm condition (Table D in S2 File). Nevertheless, pilT, the gene involved in the retraction of type IV pili, was expressed in both conditions and was significantly downregulated in the biofilm versus the planktonic cells (Table D in S2 File). This indicates that other, still unannotated, genes could encode for the type IV pili biosynthesis or, alternatively, that the observed fibers are another type of pili. For this reason, the identification of the observed fibers will need a closer examination. Hereto, sodium pyruvate could be used as substrate instead of fructose, since this led to a more pronounced production of the pilus-like structures (Fig 6).
In general, more research will be required to further unravel the molecular composition of the C. ljungdahlii biofilm matrix and the genetic encoding of its biosynthesis.
Implications for C. ljungdahlii applications
A good understanding of biofilm formation by C. ljungdahlii is important for applications using this bacterium. Most syngas fermentation setups are based on planktonic growth and the formation of a biofilm would adversely impact the operation of such systems, although some reactor designs depend on immobilized bacteria [14]. In addition, microbial electrosynthesis will benefit from the formation of a biofilm on the electrode [15, 17]. Many microbial electrosynthesis studies have tried to stimulate attachment to the electrode by the addition of extra substrate before poising of the electrode or during the initial stages of the reactor operation [5, 7, 80, 81]. The results presented in this study suggest that the addition of NaCl to the medium could be a more effective strategy to obtain attachment to the electrode. This study showed that NaCl not only induced biofilm formation on glass and plastic, but also on conductive materials, such as graphite and glassy carbon, even when placed with a vertical orientation, as often used in bio-electrochemical setups (Fig 4B). Besides the induction of biofilm formation, the addition of salt also offers the advantage of reducing the resistivity and therefore enhancing the energetic efficiency of an electrochemical cell [82, 83]. Furthermore, this study showed that an established biofilm can be removed by lowering the salt concentration in the medium (Fig 3), offering possibilities for the cleaning of microbial electrosynthesis reactors as well as syngas fermentors.
It should be noted that all experiments in this study used a rich medium to grow biofilms, which is not typically used for gas fermentations and microbial electrosynthesis. However, the addition of NaCl to a defined medium (DSMZ 879, with fructose as substrate) also triggers biofilm formation by C. ljungdahlii (results not shown). In addition, it has to be emphasized that the induction of C. ljungdahlii biofilms requires the addition of precise amounts of NaCl, as NaCl concentrations below the optimal lead to planktonic growth, while concentrations higher than the optimal cause severe stress (Fig 2) and will possibly strongly decrease the production.
The feasibility of inducing C. ljungdahlii biofilms by NaCl for practical applications remains to be assessed, but also other strategies for an improved attachment could be developed. The further investigation of the C. ljungdahlii biofilm forming mechanisms and their regulation, for instance, could lead to an engineered strain with an improved attachment. In addition, insights into the molecular mechanisms of attachment will help the design of new (electrode) materials facilitating better adhesion to their surface [84, 85]. All these strategies are expected to improve biofilm formation, which will offer possibilities to immobilize biomass for syngas fermentations, as well as enhance the rate and efficiency of microbial electrosynthesis.
Supporting Information
(XLSX)
(DOC)
Acknowledgments
We want to thank prof. Robert Bellin (College of the Holy Cross) and Geert Meesen (UGent) for their help with confocal microscopy and Dale Callaham and Louis Raboin (UMass) for their help with electron microscopy. We thank dr. Jessica Smith (UMass) for her advice on performing RNA sequencing and dr. Hugo Roume, dr. Alberto Scoma and dr. Amanda Luther (UGent) for the advice on the transcriptomics data analysis. We thank Muktak Aklujkar (Townson University) for his corrections of the C. ljungdahlii genome annotations.
Data Availability
The obtained sequences are deposited at the NCBI SRA under accession number SRP094966.
Funding Statement
JP was funded by a postdoctoral grant of the Belgian American Educational Foundation (BAEF) and by the Special Research Fund (BOF) of the Ghent University. DRL was supported by an Office of Naval Research Grant (N000141310549). KR is supported by the European Research Council, starter grant ELECTROTALK. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.Tanner RS, Miller LM, Yang D. Clostridium ljungdahlii sp. nov., an acetogenic species in Clostridial rRNA homology group I. International Journal of Systematic Bacteriology. 1993;43(2):232–6. 10.1099/00207713-43-2-232 [DOI] [PubMed] [Google Scholar]
- 2.Henstra AM, Sipma J, Rinzema A, Stams AJM. Microbiology of synthesis gas fermentation for biofuel production. Current Opinion in Biotechnology. 2007;18(3):200–6. 10.1016/j.copbio.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 3.Mohammadi M, Najafpour GD, Younesi H, Lahijani P, Uzir MH, Mohamed AR. Bioconversion of synthesis gas to second generation biofuels: A review. Renewable & Sustainable Energy Reviews. 2011;15(9):4255–73. [Google Scholar]
- 4.Bengelsdorf FR, Straub M, Duerre P. Bacterial synthesis gas (syngas) fermentation. Environmental Technology. 2013;34(13–14):1639–51. [DOI] [PubMed] [Google Scholar]
- 5.Nevin KP, Hensley SA, Franks AE, Summers ZM, Ou J, Woodard TL, et al. Electrosynthesis of organic compounds from carbon dioxide Is catalyzed by a diversity of acetogenic microorganisms. Applied and Environmental Microbiology. 2011;77(9):2882–6. 10.1128/AEM.02642-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philips J, Verbeeck K, Rabaey K, Arends JBA. Electron transfer mechanisms in biofilms In: Scott K, Yu E, editors. Microbial electrochemical and fuel cells: Fundamentals and applications: Elsevier; 2016. [Google Scholar]
- 7.Nevin KP, Woodard TL, Franks AE, Summers ZM, Lovley DR. Microbial electrosynthesis: Feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. Mbio. 2010;1(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabaey K, Rozendal RA. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nature Reviews Microbiology. 2010;8(10):706–16. 10.1038/nrmicro2422 [DOI] [PubMed] [Google Scholar]
- 9.Lovley DR, Nevin KP. Electrobiocommodities: powering microbial production of fuels and commodity chemicals from carbon dioxide with electricity. Current Opinion in Biotechnology. 2013;24(3):385–90. 10.1016/j.copbio.2013.02.012 [DOI] [PubMed] [Google Scholar]
- 10.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(29):13087–92. 10.1073/pnas.1004716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leang C, Ueki T, Nevin KP, Lovley DR. A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Applied and Environmental Microbiology. 2013;79(4):1102–9. 10.1128/AEM.02891-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueki T, Nevin KP, Woodard TL, Lovley DR. Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii. Mbio. 2014;5(5):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee A, Leang C, Ueki T, Nevin KP, Lovley DR. Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii. Applied and Environmental Microbiology. 2014;80(8):2410–6. 10.1128/AEM.03666-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munasinghe PC, Khanal SK. Biomass-derived syngas fermentation into biofuels: Opportunities and challenges. Bioresource Technology. 2010;101(13):5013–22. 10.1016/j.biortech.2009.12.098 [DOI] [PubMed] [Google Scholar]
- 15.Pons L, Delia ML, Bergel A. Effect of surface roughness, biofilm coverage and biofilm structure on the electrochemical efficiency of microbial cathodes. Bioresource Technology. 2011;102(3):2678–83. 10.1016/j.biortech.2010.10.138 [DOI] [PubMed] [Google Scholar]
- 16.Patil SA, Arends JBA, Vanwonterghem I, van Meerbergen J, Guo K, Tyson GW, et al. Selective enrichment establishes a stable performing community for microbial electrosynthesis of acetate from CO2. Environ Sci Technol. 2015;49(14):8833–43. 10.1021/es506149d [DOI] [PubMed] [Google Scholar]
- 17.Jourdin L, Yang L, Flexer V, Keller J, Freguia S. Biologically-induced hydrogen production drives high rate / high efficiency microbial electrosynthesis of acetate from carbon dioxide. Chemelectrochem. 2016. [Google Scholar]
- 18.Zhang F, Ding J, Shen N, Zhang Y, Ding Z, Dai K, et al. In situ hydrogen utilization for high fraction acetate production in mixed culture hollow-fiber membrane biofilm reactor. Appl Microbiol Biotechnol. 2013;97(23):10233–40. 10.1007/s00253-013-5281-3 [DOI] [PubMed] [Google Scholar]
- 19.Pantaleon V, Bouttier S, Soavelomandroso AP, Janoir C, Candela T. Biofilms of Clostridium species. Anaerobe. 2014;30:193–8. 10.1016/j.anaerobe.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 20.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environmental Microbiology. 2013;15(10):2631–41. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Research. 2014;42(D1):D581–D91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga JJ, Therit B, Melville SB. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infection and Immunity. 2008;76(11):4944–51. 10.1128/IAI.00692-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. Journal of Bacteriology. 2013;195(3):545–55. 10.1128/JB.01980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. Plos One. 2012;7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology. 2004;2(2):95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 26.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Research. 2013;41(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenyuk EG, Laning ML, Foley J, Johnston PF, Knight KL, Gerding DN, et al. Spore formation and toxin production in Clostridium difficile biofilms. Plos One. 2014;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlebois A, Jacques M, Archambault M. Biofilm formation of Clostridium perfringens and its exposure to low-dose antimicrobials. Front Microbiol. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Tan Y, Yang X, Chen X, Li F. Evaluation of Clostridium ljungdahlii DSM 13528 reference genes in gene expression studies by qRT-PCR. J Biosci Bioeng. 2013;116(4):460–4. 10.1016/j.jbiosc.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 30.Li JH, Freedman JC, McClane BA. NanI sialidase, CcpA, and CodY work together to regulate epsilon toxin production by Clostridium perfringens type D strain CN3718. Journal of Bacteriology. 2015;197(20):3339–53. 10.1128/JB.00349-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter BM, Cartman ST, Minton NP, Butala M, Rupnik M. The SOS response master regulator LexA is associated with sporulation, motility and biofilm formation in Clostridium difficile. Plos One. 2015;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez M, Huang IH, Ohtani K, Grau R, Shimizu T, Sarker MR. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. Journal of Bacteriology. 2008;190(1):48–60. 10.1128/JB.01407-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obana N, Nakamura K, Nomura N. A sporulation factor is involved in the morphological change of Clostridium perfringens biofilms in response to temperature. Journal of Bacteriology. 2014;196(8):1540–50. 10.1128/JB.01444-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal JE, Shak JR, Canizalez-Roman A. The CpAL quorum sensing system regulates production of hemolysins CPA and PFO to build Clostridium perfringens biofilms. Infection and Immunity. 2015;83(6):2430–42. 10.1128/IAI.00240-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JH, Ma ML, Sarker MR, McClane BA. CodY Is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. Mbio. 2013;4(5):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazazzera BA. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Current Opinion in Microbiology. 2005;8(2):222–7. 10.1016/j.mib.2005.02.015 [DOI] [PubMed] [Google Scholar]
- 37.Imam S, Chen Z, Roos DS, Pohlschroeder M. Identification of surprisingly diverse Type IV Pili, across a broad range of gram-positive bacteria. Plos One. 2011;6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rollefson JB, Levar CE, Bond DR. Identification of genes involved in biofilm formation and respiration via mini-himar transposon mutagenesis of Geobacter sulfurreducens. Journal of Bacteriology. 2009;191(13):4207–17. 10.1128/JB.00057-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang P, Burrows LL. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. Journal of Bacteriology. 2003;185(7):2374–8. 10.1128/JB.185.7.2374-2378.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White D, Drummond J, Fuqua C. The physiology and biochemistry of prokaryotes . fourth edition ed. New York: Oxford University Press; 2012. [Google Scholar]
- 41.Whitham JM, Tirado-Acevedo O, Chinn MS, Pawlak JJ, Grunden AM. Metabolic response of Clostridium ljungdahlii to oxygen exposure. Applied and Environmental Microbiology. 2015;81(24):8379–91. 10.1128/AEM.02491-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson DE, Noll D, Roberts MF, Menaia J, Boone DR. Detection of the osmoregulator betaine in methanogens Applied and Environmental Microbiology. 1990;56(2):563–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaprasis A, Brill J, Thuering M, Wuensche G, Heun M, Barzantny H, et al. Osmoprotection of Bacillus subtilis through import and proteolysis of proline-containing peptides. Applied and Environmental Microbiology. 2013;79(2):576–87. 10.1128/AEM.01934-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amezaga MR, Davidson I, McLaggan D, Verheul A, Abee T, Booth IR. The role of peptide metabolism in the growth of Listeria Monocytogenes ATCC-23074 at high osmolarity Microbiology-Uk. 1995;141:41–9. [DOI] [PubMed] [Google Scholar]
- 45.Li Y-H, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors. 2012;12(3):2519–38. 10.3390/s120302519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D, Xu J, Wang Y, Chen Y, Shen X, Niu H, et al. Comparative transcriptomic analysis of Clostridium acetobutylicum biofilm and planktonic cells. Journal of Biotechnology. 2016;218:1–12. 10.1016/j.jbiotec.2015.11.017 [DOI] [PubMed] [Google Scholar]
- 47.Cerca N, Jefferson KK. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. Fems Microbiology Letters. 2008;283(1):36–41. 10.1111/j.1574-6968.2008.01142.x [DOI] [PubMed] [Google Scholar]
- 48.Gotz F. Staphylococcus and biofilms. Molecular Microbiology. 2002;43(6):1367–78. [DOI] [PubMed] [Google Scholar]
- 49.Roux D, Cywes-Bentley C, Zhang YF, Pons S, Konkol M, Kearns DB, et al. Identification of poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. Journal of Biological Chemistry. 2015;290(31):19261–72. 10.1074/jbc.M115.648709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Frontiers in Cellular and Infection Microbiology. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown S, Maria JPS Jr., Walker S. Wall teichoic acids of gram-positive bacteria. In: Gottesman S, editor. Annual Review of Microbiology, Vol 67 Annual Review of Microbiology. 672013. p. 313–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alsaker KV, Paredes C, Papoutsakis ET. Metabolite stress and tolerance in the production of biofuels and chemicals: Gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng. 2010;105(6):1131–47. 10.1002/bit.22628 [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Venkataramanan KP, Huang H, Papoutsakis ET, Wu CH. Transcription factors and genetic circuits orchestrating the complex, multilayered response of Clostridium acetobutylicum to butanol and butyrate stress. Bmc Systems Biology. 2013;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Downs DM, Ernst DC. From microbiology to cancer biology: the Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Molecular Microbiology. 2015;96(2):211–9. 10.1111/mmi.12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emerson JE, Stabler RA, Wren BW, Fairweather NF. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. Journal of Medical Microbiology. 2008;57(6):757–64. [DOI] [PubMed] [Google Scholar]
- 56.Koseki S, Nakamura N, Shiina T. Growth inhibition of Listeria monocytogenes, Salmonella enterica, and Escherichia coli O157:H7 by D-tryptophan as an incompatible solute. Journal of Food Protection. 2015;78(4):819–24. 10.4315/0362-028X.JFP-14-374 [DOI] [PubMed] [Google Scholar]
- 57.Martin ME, Richter H, Saha S, Angenent LT. Traits of selected Clostridium strains for syngas fermentation to ethanol. Biotechnol Bioeng. 2016;113(3):531–9. 10.1002/bit.25827 [DOI] [PubMed] [Google Scholar]
- 58.Terui Y, Higashi K, Tabei Y, Tomitori H, Yamamoto K, Ishihama A, et al. Enhancement of the synthesis of RpoE and StpA by polyamines at the level of translation in Escherichia coli under heat shock conditions. Journal of Bacteriology. 2009;191(17):5348–57. 10.1128/JB.00387-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geesey GG, Wigglesworth-Cooksey B, Cooksey KE. Influence of calcium and other cations on surface adhesion of bacteria and diatoms: a review. Biofouling. 2000;15(1–3):195–205. 10.1080/08927010009386310 [DOI] [PubMed] [Google Scholar]
- 60.Rachid S, Cho S, Ohlsen K, Hacker J, Ziebuhr W. Induction of Staphylococcus epidermidis biofilm formation by environmental factors: The possible involvement of the alternative transcription factor sigB In: Emody L, Pal T, Hacker J, BlumOehler G, editors. Genes and Proteins Underlying Microbial Urinary Tract Virulence: Basic Aspects and Applications. Advances in Experimental Medicine and Biology. 485 2000. p. 159–66. [DOI] [PubMed] [Google Scholar]
- 61.Moretro T, Hermansen L, Holck AL, Sidhu MS, Rudi K, Langsrud S. Biofilm formation and the presence of the intercellular adhesion locus ica, among staphylococci from food and food processing environments. Applied and Environmental Microbiology. 2003;69(9):5648–55. 10.1128/AEM.69.9.5648-5655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen A, Larsen MH, Ingmer H, Vogel BF, Gram L. Sodium chloride enhances adherence and aggregation and strain variation influences invasiveness of Listeria monocytogenes strains. Journal of Food Protection. 2007;70(3):592–9. [DOI] [PubMed] [Google Scholar]
- 63.Pan Y, Breidt F Jr., Gorski L. Synergistic effects of sodium chloride, glucose, and temperature on biofilm formation by Listeria monocytogenes serotype 1/2a and 4b strains. Applied and Environmental Microbiology. 2010;76(5):1433–41. 10.1128/AEM.02185-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim Y, Jana M, Luong TT, Lee CY. Control of glucose- and NaCI-induced biofilm formation by rbf in Staphylococcus aureus. Journal of Bacteriology. 2004;186(3):722–9. 10.1128/JB.186.3.722-729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knobloch JKM, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. Journal of Bacteriology. 2001;183(8):2624–33. 10.1128/JB.183.8.2624-2633.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chellappa ST, Maredia R, Phipps K, Haskins WE, Tao W. Motility of Pseudomonas aeruginosa contributes to SOS-inducible biofilm formation. Research in Microbiology. 2013;164(10):1019–27. 10.1016/j.resmic.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 67.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, et al. The Spo0A regulon of Bacillus subtilis. Molecular Microbiology. 2003;50(5):1683–701. [DOI] [PubMed] [Google Scholar]
- 68.Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Molecular Microbiology. 2001;42(5):1199–209. [DOI] [PubMed] [Google Scholar]
- 69.Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. Journal of Bacteriology. 2003;185(6):1951–7. 10.1128/JB.185.6.1951-1957.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seidl K, Goerke C, Woiz C, Mack D, Berger-Baechi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infection and Immunity. 2008;76(5):2044–50. 10.1128/IAI.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flemming HC, Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8(9):623–33. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 72.Qin ZQ, Ou YZ, Yang LA, Zhu YL, Tolker-Nielsen T, Molin S, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology-Sgm. 2007;153:2083–92. [DOI] [PubMed] [Google Scholar]
- 73.Piepenbrink KH, Maldarelli GA, de la Pena CFM, Mulvey GL, Snyder GA, De Masi L, et al. Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins. Journal of Biological Chemistry. 2014;289(7):4334–45. 10.1074/jbc.M113.534404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bordeleau E, Purcell EB, Lafontaine DA, Fortier L-C, Tamayo R, Burrus V. Cyclic Di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. Journal of Bacteriology. 2015;197(5):819–32. 10.1128/JB.02340-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. Type IV pili-dependent gliding motility in the gram-positive pathogen Clostridium perfringens and other Clostridia. Molecular Microbiology. 2006;62(3):680–94. 10.1111/j.1365-2958.2006.05414.x [DOI] [PubMed] [Google Scholar]
- 76.Borriello SP, Davies HA, Barclay FE. Detection of fimbriae amongst strains of Clostridium difficile. Fems Microbiology Letters. 1988;49(1):65–7. [Google Scholar]
- 77.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(5):2230–4. 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caro-Astorga J, Perez-Garcia A, de Vicente A, Romero D. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front Microbiol. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nature Reviews Microbiology. 2006;4(7):509–19. 10.1038/nrmicro1443 [DOI] [PubMed] [Google Scholar]
- 80.Kuroda M, Watanabe T. CO2 reduction to methane and acetate using a bio-electro reactor with immobilized methanogens and homoacetogens on electrodes Energy Conversion and Management. 1995;36(6–9):787–90. [Google Scholar]
- 81.Su M, Jiang Y, Li D. Production of acetate from carbon dioxide in bioelectrochemical systems based on autotrophic mixed culture. J Microbiol Biotechnol. 2013;23(8):1140–6. [DOI] [PubMed] [Google Scholar]
- 82.Liu H, Cheng SA, Logan BE. Power generation in fed-batch microbial fuel cells as a function of ionic strength, temperature, and reactor configuration. Environ Sci Technol. 2005;39(14):5488–93. [DOI] [PubMed] [Google Scholar]
- 83.Lefebvre O, Tan Z, Kharkwal S, Ng HY. Effect of increasing anodic NaCl concentration on microbial fuel cell performance. Bioresource Technology. 2012;112:336–40. 10.1016/j.biortech.2012.02.048 [DOI] [PubMed] [Google Scholar]
- 84.Zhang T, Nie H, Bain TS, Lu H, Cui M, Snoeyenbos-West OL, et al. Improved cathode materials for microbial electrosynthesis. Energy & Environmental Science. 2013;6(1):217–24. [Google Scholar]
- 85.Guo K, Prevoteau A, Patil SA, Rabaey K. Engineering electrodes for microbial electrocatalysis. Current Opinion in Biotechnology. 2015;33:149–56. 10.1016/j.copbio.2015.02.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOC)
Data Availability Statement
The obtained sequences are deposited at the NCBI SRA under accession number SRP094966.