Abstract
Background
Clinical relevance of nontuberculous mycobacteria (NTM) is increasing worldwide including in Saudi Arabia. A high species diversity of NTM’s has been noticed in a recent study. However, the identification in diagnostic laboratories is mostly limited to common species. The impact of NTM species diversity on clinical outcome is so far neglected in most of the clinical settings.
Methodology/Principal Findings
During April 2014 to September 2015, a nationwide collection of suspected NTM clinical isolates with clinical and demographical data were carried out. Primary identification was performed by commercial line probe assays. Isolates identified up to Mycobacterium species level by line probe assays only were included and subjected to sequencing of 16S rRNA, rpoB, hsp65 and 16S-23S ITS region genes. The sequence data were subjected to BLAST analysis in GenBank and Ez-Taxon databases. Male Saudi nationals were dominated in the study population and falling majorly into the 46–59 years age group. Pulmonary cases were 59.3% with a surprising clinical relevance of 75% based on American Thoracic Society guidelines. Among the 40.7% extra-pulmonary cases, 50% of them were skin infections. The identification revealed 16 species and all of them are reporting for the first time in Saudi Arabia. The major species obtained were Mycobacterium monascence (18.5%), M. cosmeticum (11.1%), M. kubicae (11.1%), M. duvalli (7.4%), M.terrae (7.4%) and M. triplex (7.4%). This is the first report on clinical relevance of M. kubicae, M. tusciae, M.yongonense, M. arupense and M.iranicum causing pulmonary disease and M. monascence, M. duvalli, M. perigrinum, M. insubricum, M. holsaticum and M. kyorinense causing various extra-pulmonary diseases in Saudi Arabia. Ascites caused by M. monascence and cecum infection by M. holsaticum were the rarest incidents.
Conclusions/Significance
To the first time in the country, clinical significance of various rare NTM’s are well explored and the finding warrants a new threat to the Saudi Arabian clinical settings.
Author Summary
Nontuberculous mycobacteria (NTM) are ubiquitous in nature and they are opportunistic pathogens. In the last decade, infections caused by NTM’s increased—around the world in immune-suppressed and immune-competent individuals and Saudi Arabia is not an exception. Developments in diagnostic technologies increased the identification of several new or rare species of NTM’s. Indeed, the species diversity of NTM has a direct impact on clinical outcome and therapies. Saudi Arabian clinics so far report only the common species of NTM’s and rare species are mostly neglected due to the lack of proper infrastructure or ignorance. To the first time in the country, an exploration on the existence of clinically relevant rare NTM species was conducted on a nationwide level. The findings showed a huge diversity of rare NTM species causing both pulmonary and extra-pulmonary diseases. Clinical relevance of pulmonary infection based on American Thoracic Society guidelines was confirmed as an aggressive 75%, which is really alarming. Interestingly, 16 species of NTM’s were isolated in the study, and all of them are reporting for the first time in country. Overall finding shows Saudi Arabia faces serious threat from rare NTM species with high clinical significance and warrants the immediate need for more advanced infrastructure.
Introduction
In the last decade, the prevalence of pulmonary and extra-pulmonary diseases caused by nontuberculous mycobacteria (NTM) has been increased [1–7]. This elevation in case rates, whether it is a real emergence or due to the development of advanced diagnostic tools is still unclear. On the other hand, the elevation in immunosuppressive conditions including infectious or non-infectious diseases and therapies contribute considerably in this phenomenon. To date, more than 140 species of NTM’s have been described from different sources with varying pathogenicity and almost 50 species were identified in the last 8 years alone [8]. However, in literature only a small number of reports are available about the new species as their role in clinical microbiology is largely undetermined. Mostly, the species defined as “rare” will remain unrecognized or misidentified due to the lack of proper resources, lack of knowledge or ignorance [8]. The clinical characteristics of diseases caused by the rare or new NTM’s are still not fully established. The advancement in technologies such as genome sequencing, line probe assays, high performance liquid chromatography (HPLC) and matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) to identify the NTM species increased the detection of rare and new NTM species. However, accessibility to such tools in resource poor settings is a major concern for timely identification. Thus, the species level identification is mostly neglected regardless of its importance in clinical outcome.
Following the global trend of NTM prevalence, Saudi Arabia also reports with an increasing numbers of NTM diseases [7, 9]. In 2013, Varghese et al. reported in the first national level study a highly diverse population of clinically relevant NTM’s with the potential of causing pulmonary and extra-pulmonary diseases [9]. Interestingly, a new species of pathogenic mycobacteria also has been identified from the country named M. riyadhense [10]. However, the diagnosis of NTM’s is mostly limited to the common species only in majority of the laboratories, because of the lack of infrastructure. Therefore, there is no data available on the existence of rare NTM species in the country so far.
To explore the diversity of rare NTM species with clinical relevance in the Saudi Arabian clinical setting, a prospective analysis on a nationwide isolate collection has been designed. Sequencing of 16S rRNA, rpoB, hsp65 and 16S-23S ITS region genes were carried out to identify the species. Clinical significance of pulmonary isolates in the study was determined by applying the criteria based on American Thoracic Society (ATS) guidelines for NTM pulmonary diseases [11]. The species diversity and clinical significance of each isolates have been evaluated.
Materials and Methods
Study population
This study has been conducted as part of the first national NTM surveillance survey of Saudi Arabia. The duration of collection was 18 months, from April 2014 to September 2015. All the suspected NTM isolates from different mycobacterial diagnostic laboratories were collected and transferred to the Mycobacteriology Research Section of King Faisal Specialist Hospital and Research Centre, Riyadh. The demographical and clinical data were collected by using standard data collection forms without keeping any patients identifiers. Pulmonary cases were defined as clinically relevant based on ATS guidelines [11]. Briefly, a minimum of two positive cultures from separate sputum samples or at least one positive culture from bronchial wash, lavage or one positive culture from trans-bronchial or other lung biopsy showing mycobacterial- histopathological features were considered as clinically relevant to define NTM pulmonary disease.
Primary identification and enrollment
Isolates were maintained on Lowenstein Jensen slants and modified Middle Brook 7H9 medium (Becton Dickinson, USA). The genomic DNA was extracted by using the QIAamp DNA Mini kit (Qiagen, Germany). The primary screening to identify the NTM’s was carried out by commercially available line probe assay kit- Genotype MTBC (Hain Life science, Germany). The non-MTBC isolates were initially identified with Genotype Mycobacterium CM kit (Hain Life science, Nehren, Germany) and unidentified isolates were further tested with Genotype Mycobacterium AS kit (Hain Lifescience, Nehren Germany).
Isolates which were detected by the Genotype Mycobacterium AS assay up to genus level (Mycobacterium species) only were included in the study as “unidentified” species. This study has been reviewed and approved by the Office of Research Affairs in King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia.
Sequencing analysis
Sequencing assay was carried out by using the BigDye Terminator cycle sequencing chemistry kits (Applied Biosystems, CA, USA) in DNA analyzer 3730 (Applied Biosystems, CA, USA). The first attempt of identification was based on a 645-655bp hyper variable region of the 16S rRNA and a 342bp region of rpoB genes based on previously standardized protocol [12, 13]. Isolates which could not be identified by 16S rRNA and rpoB gene sequencing were subjected to further sequencing of highly conservative regions of two more genes hsp65 (439bp) and 16S-23S ITS region (480bp) according to previously validated primers [14–16].
Data analysis
The line probe assay test strips were scanned with Genoscan (Hain Lifescience, Germany) and the results were interpreted with the Blotrix software (Hain Lifescience, Germany). The sequence base calling and assembly were carried out by using Sequence Analysis software v5.3.1 (Applied Biosystems, USA) and Lasergene core suite 12 (DNA STAR, WI, USA) respectively. Assembled sequences were subjected to BLAST analysis in NCBI GenBank and EzTaxon (http://www.ezbiocloud.net/identify; 16S rRNA Based Database) online data bases [17]. A stringent similarity index of ≥99–100% was kept with the type strain in GenBank and EzTaxon. Statistical data analysis was carried out by using SPSS V20.0 software package (IBM, USA).
Results
Demographical and clinical summary
During the study period, 510 suspected NTM clinical isolates were collected and subjected to line probe assay identification. Of the total, 27 isolates met the inclusion criteria and enrolled for further analysis. Demographical summary of the study subjects showed 22 (81.5%) of the enrolled cases were Saudi nationals with a male (77.8%) gender domination. Interestingly, the age group of the study subjects showed a predominance of 46–59 years (48.2%). Seven cases out of 27 had a previous history of tuberculosis and 3 were reactive to HIV antigens. The major comorbidities noticed among the study subjects were rheumatoid arthritis (18.5%), malignancies (18.5%) and diabetes (14.8%). Considerable percentage of Chronic Obstructive Pulmonary Disease (COPD), asthma and bronchiectasis also were observed (Table 1).
Table 1. Study summary of 27 rare NTM isolates from Saudi Arabia.
| Isolate | Species | 1Type Strain/ 16S rRNA Similarity % | 2Type Strain/ rpoB Similarity % | Age/Gender | Nationality | Specimen type3 | Clinical relevance4 | Treatment5 | Underlying Conditions6 |
|---|---|---|---|---|---|---|---|---|---|
| 1. | M.arupense | NR_043588/99.6 | JN571215/99.0 | 30/F | Saudi | BAL2 | Yes | RIF, EMB, CLR | HL,COPD,HIV |
| 2. | M.cosmeticum | NR_025787/ 99.8 | AY262742/ 96.8 | 39/M | Saudi | sputum1 | No | - | AS,PTB,HIV |
| 3. | M.cosmeticum | NR_025787/ 99.0 | AY262742/ 96.0 | 48/M | Saudi | sputum7 | Yes | - | Lung Transplant |
| 4. | M.cosmeticum | NR_025787/ 99.2 | AY262742/ 96.6 | 59/M | Yemen | skin3 | - | DM | |
| 5. | M.duvalii | NR_026073/99.1 | AY544907/99.1 | 56/F | Saudi | sputum1 | No | - | NHL,RA |
| 6. | M.duvalii | NR_026073/99.7 | AY544907/99.3 | 63/M | Saudi | peritoneum1 | - | CF,AK | HL,CAPD |
| 7. | M.holsaticum | NR_028945/99.4 | AY859705/98.6 | 47/M | Saudi | cecum2 | - | EMB,OF | DM,UC,RA |
| 8. | M.insubricum | NR_125525/99.3 | EU605693/90.0 | 52/M | Eritrea | skin1 | - | - | PTB |
| 9. | M.iranicum | KU861842/99.6 | JQ90669809/98.9 | 51/M | Saudi | BAL2 | Yes | - | PTB |
| 10. | M.kubicae | HM022200/99.3 | 69/F | Saudi | BAL1 | Yes | RA | ||
| 11. | M.kubicae | HM022200/99.7 | 49/M | Saudi | sputum1 | No | - | PTB | |
| 12. | M.kubicae | NR_025000/99.1 | 51/F | Saudi | sputum3 | Yes | - | DM,CVD,RA | |
| 13. | M.kyorinense | NR_041663/98.7 | JQ717032/99.2 | 14/M | Saudi | lymphnode1 | EMB, CLR | HL | |
| 14. | M.monascence | NR_041723/99.5 | HM229793/99. | 68/M | Saudi | sputum6 | Yes | - | PTB,AS |
| 15. | M.monascence | NR_041723/99.3 | KU361327/99.0 | 52/M | Saudi | sputum2 | Yes | - | COPD, BE |
| 16. | M.monascence | NR_041723/99.0 | HM229793/99.0 | 2/F | Saudi | BAL1 | Yes | - | COPD |
| 17. | M.monascence | NR_041723/99.6 | KU361327/99.6 | 52/M | Saudi | Ascitic fluid | - | COPD, IBD | |
| 18. | M.monascence | NR_041723/99.4 | HM229793/98.4 | 63/M | Saudi | Lymphnode | - | BE | |
| 19. | M.novocastrense | NR_029208/99.2 | AY859704/98.9 | 74/M | Saudi | sputum2 | Yes | - | BE,RA |
| 20. | M.perigrinum | NR_114447/99.1 | AY147166/98.7 | 48/M | Sudan | skin1 | - | - | PTB |
| 21. | M.marinum | NR_113366/ 99.2 | CP000854/ 98.2 | 33/M | Sudan | skin1 | RIF,CLR | - | |
| 22. | M.terrae | NR_117886/99.0 | JN571264/98.9 | 68/M | Saudi | sputum1 | No | AK,CF | COPD, DM |
| 23. | M.terrae | NR_115677/99.1 | AY544967/99.0 | 49/M | Nigeria | skin | - | PTB,AS | |
| 24. | M.triplex | NR_117226/99.1 | AY544970/99.0 | 69/M | Saudi | BAL | Yes | - | CVD,AS |
| 25. | M.triplex | NR_117226/99.5 | KF856622/98.7 | 48/F | Saudi | lymphnode | AK, EMB,CLR | HIV | |
| 26. | M.tusciae | NR_117885/99.3 | 64/M | Saudi | sputum3 | Yes | INH, EMB,RIF | BE, HCA | |
| 27. | M.yongonense | KF_224994/99.8 | JF271829/99.1 | 16/M | Saudi | BAL2 | Yes | - | COPD,NHL |
1 16S rRNA gene closest match- Gene bank accession number.
2 rpoB gene closest match- Gene bank accession number.
3 Number shows how many times the species was isolated from the patient.
4 ATS and IDSA NTM pulmonary disease criteria met.
5 Drugs (on use while collecting isolates)- EMB- Ethambutol; CLR- Clarithromycin; AK-Amikacin; RIF-Rifampicin; CF-Ciprofloxacin, OF-Ofloxacin.
6 HL-Hodgkin Lymphoma; DM- Diabetes Mellitus, NHL- Non-Hodgkins Lymphoma; UC-Ulcerative Colitis; RA- Rheumatoid Arthritis; PTB- Previous Tuberculosis; BE- Bronchiectasis; COPD- Chronic Obstructive Pulmonary Disease; IBD- Inflammatory Bowel Disease; HCA- Hepatocellular Carcinoma; AS- Asthma; CAPD- Continuous Ambulatory Peritoneal Dialysis, CVD- Cardio Vascular Diseases, INH-Isoniazid.
Diversity of NTM species
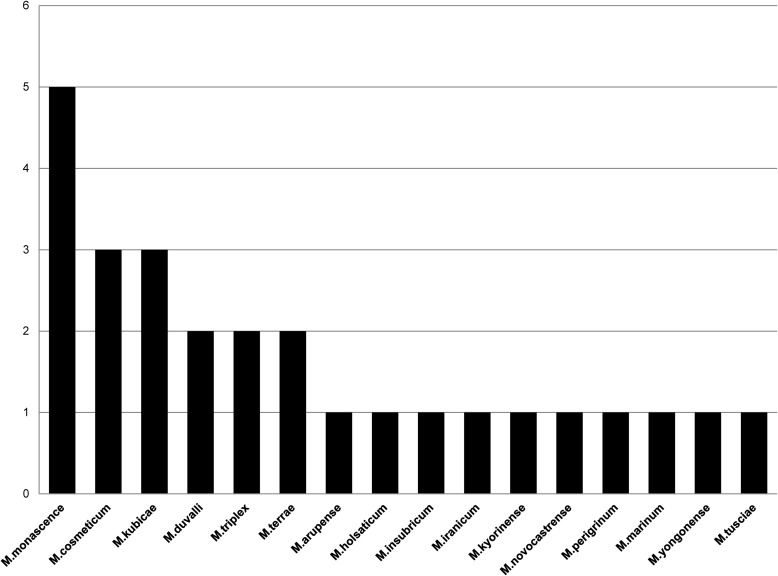
Analysis of 16S rRNA, rpoB, hsp65 and 16-23S ITS region genes, showed an extreme diversity of NTM species distributed to 16 species. The majorly detected species were M. monascence (18.5%), M. cosmeticum (11.1%), M.kubicae (11.1%), M. duvalii (7.4%), M. triplex (7.4%) and M. terrae (7.4%). Rest of the 10 species were reported with one case each (3.7%) (Fig 1).
Fig 1. Species diversity of rare NTM’s isolated in Saudi Arabia.
The figure shows the overall diversity of 27 rare NTM isolates from pulmonary and extra-pulmonary samples.
Site of infection and clinical relevance
The major site of infection observed in the study was pulmonary (59.3%). Clinical relevance of pulmonary isolates based on the ATS guidelines was dominating (75%). Of the 16 pulmonary cases, four cases did not qualify the clinical relevance guidelines and thus considered as colonization. Clinically relevant diseases were caused by M. arupense, M. cosmeticum, M. iranicum, M. kubicae, M. monascence, M. novocatrense, M. tusciae and M. yongonense (Table 1).
On the other hand, extra-pulmonary involvement was found with 11 (40.7%) cases. Among extra-pulmonary cases, skin (45.5%) was the most affected site of infection followed by lymphnode (27.3%). M. insubricum, M. perigrinum and M. marinum were found exclusively causing granuloma or sepsis. Interestingly, all these five skin infections were observed among non-Saudi patients. Cecum infection by M. holsaticum and ascites caused by M. monascence were the rarest incidents in this study (Table 1).
Discussion
For the first time in Saudi Arabia, the existence of rare NTM species has been explored on a nationwide collection of clinical isolates. The findings showed a strong presence of clinically relevant NTM rare species in the Saudi Arabian clinical settings. The species diversity of rare NTM’s causing both pulmonary and extra-pulmonary diseases was huge (16 species). To date, all of these 16 species with clinical relevance are reporting for the first time in the country.
Demographical analysis showed predominance of male Saudi nationals and mainly the age group 46–59 years and a similar domination had been noticed in a recent nationwide study of NTM prevalence [9]. Clinical data showed various comorbidities existed among the study group. Rheumatoid arthritis and various malignancies were the major problems followed by diabetes. There were no studies so far analyzed the reasons behind the predominance of male Saudi nationals towards the NTM disease. Perhaps, there were some speculations like higher rate of consanguinity existed in the community which leads to several genetic susceptibility diseases, increasing rate of immunosuppressive therapies and various malignancies in the geographical region [18–20]. Indeed, the confounding factors for this predominance need further detailed scientific exploration.
In this study, pulmonary diseases caused by rare NTM species were predominant with higher clinical relevance (75%). Of the 16 identified species, 11 species except M. insubricum, M. kyorinense, M. holsaticum, M. perigrinum and M. marinum were isolated from respiratory samples. Of the 11 species, except M. duvalii and M. terrae all the others reported with clinically relevant diseases. Moreover, most of these species are isolated very rarely from clinical specimens with relevance so far around the world [21]. Interestingly, to date, M. tusciae, M. yongonense, M. novocastrens and M. monascence pulmonary diseases were reported in hardly 2–3 publications. Therefore, these findings are important as it shows and confirms the growing problems with rare NTM species in Saudi Arabia as well for the first time in the Gulf Cooperation Council (GCC) states also [22] [21, 23, 24]. The higher clinical relevance established after consulting the ATS guidelines shows the increasing potential of NTM respiratory diseases in the country. Supportively, NTM respiratory diseases caused by various species have been observed in a recent nationwide study in the country [9]. In the current study majority of the isolation was from sputum samples, except six bronchial washes. The frequency of isolation from sputum was peaked up to 7 occasions for a case of M. cosmeticum. The higher frequency shows the increasing potential of NTM’s as en establishing pathogen in the clinical settings. Most of the NTM species isolated in the current study causing pulmonary diseases are not only rare in Saudi Arabia but also around the globe. In concordance to the current findings, previous global studies showed an increasing prevalence of NTM’s and particularly the domination of pulmonary isolates. The existence of numerous rare or new species was observed in most of the large level studies [5–7, 21]. The increased clinical relevance is a warranting key to be vigilant on the pathogenicity and potential of NTM’s to cause confirmed diseases rather than colonization.
In the current study, 11 cases of extra-pulmonary diseases were observed with a predominance of skin infections (50%) caused by M. cosmeticum, M. insubricum, M. perigrinum, M. marinum and M. terrae. The M. perigrinum and M. marinum were isolated from two immigrant patients from the Western coast of the country, where the major fishing harbors located with considerable number of foreign fishermen. The skin infection caused by M. marinum and M. perigrinum among people who work in fisheries or swimming pool maintenance is generally reported elsewhere [25, 26]. The rarest cases in the study were the ascites caused by M. monascence and cecum infection caused by M. holsaticum. To our knowledge, this might be the first cases of the same reporting globally. On the other hand, lymphadenitis caused by M. kyorinense and M. triplex are rare manifestations which have been reported in only two or three cases so far globally and the current study also found a case of each [27, 28]. The infections caused by M. duvalii and M. monascence in patients undergoing peritoneal dialysis are reported for the first time in Saudi Arabia and in GCC nations. Such cases are rarely reported around the world [8].
Conclusion
As the first report on the existence of rare NTM species in Saudi Arabia, the findings showed an alarming diversity of clinically relevant NTM’s causing both pulmonary and extra-pulmonary diseases. The diagnostic facilities in the country requires more advanced infrastructure to identify the rare species on time, as it influence the final clinical outcome and treatment adversely.
Sequence deposit accession numbers
The following 16S rRNA gene sequences have been deposited in GenBank/DDBJ/EMBL data bases. M. monacense (KY287007), M. iranicum (KY287008), M. kubicae (KY287009), M. cosmeticum (KY287010), M. duvalii (KY287011), M. terrae (KY287012), M. arupense (KY287013) and M. novocastrense (KY287014).
Acknowledgments
We are thankful to Ms. Mary Grace Fernandez and Ms. Fatima Alsafar for their technical assistance.
Data Availability
The following 16S rRNA gene sequences have been deposited in GenBank/DDBJ/EMBL data bases: M.monacense (KY287007), M.iranicum (KY287008), M.kubicae (KY287009), M.cosmeticum (KY287010), M.duvalii (KY287011), M.terrae (KY287012), M.arupense (KY287013) and M.novocastrense (KY287014).
Funding Statement
This study has been funded by King Abdulaziz City for Science and Technology (NSTIP), Riyadh, Saudi Arabia under grant code 12-MED-3172-20. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kendall BA, Winthrop KL. Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Seminars Resp Crit care Med. 2013;34(1):87–94. [DOI] [PubMed] [Google Scholar]
- 2.Thomson RM, on behalf of the NTMwgatQTBCC, Queensland Mycobacterial Reference L. Changing Epidemiology of Pulmonary Nontuberculous Mycobacteria Infections. Emerg Infect Dis. 2010;16(10):1576–83. 10.3201/eid1610.091201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morimoto K, Iwai K, Uchimura K, Okumura M, Yoshiyama T, Yoshimori K, et al. A steady increase in nontuberculous mycobacteriosis mortality and estimated prevalence in Japan. Annals Am Thoracic Soc. 2014;11(1):1–8. [DOI] [PubMed] [Google Scholar]
- 4.Falkinham JO 3rd. Epidemiology of infection by nontuberculous mycobacteria. Clin Mcrobiol Rev. 1996;9(2):177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Werf MJ, Ködmön C, Katalinić-Janković V, Kummik T, Soini H, Richter E, et al. Inventory study of non-tuberculous mycobacteria in the European Union. BMC Infect Dis. 2014;14(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Casabona N, Bahrmand AR, Bennedsen J, Thomsen VO, Curcio M, Fauville-Dufaux M, et al. Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int J Tuberc Lung Dis. 2004;8. [PubMed] [Google Scholar]
- 7.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Euro Resp J. 2013;42(6):1604–13. [DOI] [PubMed] [Google Scholar]
- 8.Tortoli E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin Microbiol Rev 2014;27(4):727–52. 10.1128/CMR.00035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varghese B, Memish Z, Abuljadayel N, Al-Hakeem R, Alrabiah F, Al-Hajoj SA. Emergence of clinically relevant Non-Tuberculous Mycobacterial infections in Saudi Arabia. PLoS Neglect Trop Dis. 2013;7(5):e2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ingen J, Al-Hajoj SA, Boeree M, Al-Rabiah F, Enaimi M, de Zwaan R, et al. Mycobacterium riyadhense sp. nov., a non-tuberculous species identified as Mycobacterium tuberculosis complex by a commercial line-probe assay. Int J Syst Evolut Microbiol. 2009;59(Pt 5):1049–53. [DOI] [PubMed] [Google Scholar]
- 11.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 12.Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol. 2002;118(5):796–801. 10.1309/HN44-XQYM-JMAQ-2EDL [DOI] [PubMed] [Google Scholar]
- 13.Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Chae GT, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol. 1999;37(6):1714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNabb A, Eisler D, Adie K, Amos M, Rodrigues M, Stephens G, et al. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J Clin Microbiol. 2004;42(7):3000–11. 10.1128/JCM.42.7.3000-3011.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31(2):175–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frothingham R, Wilson KH. Sequence-based differentiation of strains in the Mycobacterium avium complex. J Bacteriol. 1993;175(10):2818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Eolut Microbiol. 2007;57(Pt 10):2259–61. [DOI] [PubMed] [Google Scholar]
- 18.Global Burden of Disease Cancer C. THe global burden of cancer 2013. JAMA Oncology. 2015;1(4):505–27. 10.1001/jamaoncol.2015.0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warsy AS, Al-Jaser MH, Albdass A, Al-Daihan S, Alanazi M. Is consanguinity prevalence decreasing in Saudis?: a study in two generations. African Health Sciences. 2014;14(2):314–21. 10.4314/ahs.v14i2.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almoallim HM, Alharbi LA. Rheumatoid arthritis in Saudi Arabia. Saudi Med J. 2014;35(12):1442–54. [PMC free article] [PubMed] [Google Scholar]
- 21.Lima CA, Gomes HM, Oelemann MA, Ramos JP, Caldas PC, Campos CE, et al. Nontuberculous mycobacteria in respiratory samples from patients with pulmonary tuberculosis in the state of Rondonia, Brazil. Memorias do Instituto Oswaldo Cruz. 2013;108(4):457–62. 10.1590/S0074-0276108042013010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shojaei H, Hashemi A, Heidarieh P, Hosseini N, Daei Naser A. Chronic pulmonary disease due to Mycobacterium monacense infection: the first case from Iran. Annals Lab Med. 2012;32(1):87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tortoli E, Mariottini A, Pierotti P, Simonetti TM, Rossolini GM. Mycobacterium yongonense in pulmonary disease, Italy. Emerg Infect Dis. 2013;19(11):1902–4. 10.3201/eid1911.130911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shojaei H, Hashemi A, Heidarieh P, Naser AD. Mycobacterium novocastrense-associated pulmonary and wound infections. Emerg Infect Dis. 2011;17(3):550–1. 10.3201/eid1703.101400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamijo F, Uhara H, Kubo H, Nakanaga K, Hoshino Y, Ishii N, et al. A Case of Mycobacterial Skin Disease Caused by Mycobacterium peregrinum, and a Review of Cutaneous Infection. Case reports Dermatol. 2012;4(1):76–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolly HW Jr, Seabury JH . INfections with mycobacterium marinum. Arch Dermatol. 1972;106(1):32–6. [PubMed] [Google Scholar]
- 27.Ohnishi H, Yonetani S, Matsushima S, Wada H, Takeshita K, Kuramochi D, et al. Mycobacterium kyorinense infection. Emerg Infect Dis. 2013;19(3):508–10. 10.3201/eid1903.12-0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruso G, Angotti R, Molinaro F, Benicchi E, Cerchia E, Messina M. Cervical Lymphadenitis by Mycobacterium triplex in an Immunocompetent Child: Case Report and Review. Indian J Microbiol. 2013;53(2):241–4. 10.1007/s12088-013-0367-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following 16S rRNA gene sequences have been deposited in GenBank/DDBJ/EMBL data bases: M.monacense (KY287007), M.iranicum (KY287008), M.kubicae (KY287009), M.cosmeticum (KY287010), M.duvalii (KY287011), M.terrae (KY287012), M.arupense (KY287013) and M.novocastrense (KY287014).