Abstract
Bacterial-host attachment by means of bacterial adhesins is a key step in host colonization. Phase variation (reversible on-off switching) of the type 1 fimbrial adhesin of Escherichia coli involves a DNA inversion catalyzed by FimB (switching in either direction) or FimE (mainly on-to-off switching). fimB is separated from the divergent yjhATS operon by a large (1.4 kbp) intergenic region. Short (≈28 bp) cis-active elements (regions 1 and 2) close to yjhA stimulate fimB expression and are required for sialic acid (Neu5Ac) sensitivity of its expression [El-Labany, S., Sohanpal, B. K., Lahooti, M., Akerman, R. & Blomfield, I. C. (2003) Mol. Microbiol. 49, 1109-1118]. Here, we show that whereas NanR, a sialic acid-response regulator, binds to region 1, NagC, a GlcNAc-6P-responsive protein, binds to region 2 instead. The NanR- and NagC-binding sites lie adjacent to deoxyadenosine methylase (Dam) methylation sites (5′-GATC) that are protected from modification, and the two regulators are shown to be required for methylation protection at regions 1 and 2, respectively. Mutations in nanR and nagC diminish fimB expression, and both fimB expression and FimB recombination are inhibited by GlcNAc (3- and >35-fold, respectively). Sialic acid catabolism generates GlcNAc-6-P, and whereas GlcNAc disrupts methylation protection by NagC alone, Neu5Ac inhibits the protection mediated by both NanR and NagC as expected. Type 1 fimbriae are proinflammatory, and host defenses enhance the release of both Neu5Ac and GlcNAc by a variety of mechanisms. Inhibition of type 1 fimbriation by these amino sugars may thus help balance the interaction between E. coli and its hosts.
Keywords: type 1 fimbriae
Bacterial-host attachment plays a central role in colonization and is often crucial in pathogenesis. Escherichia coli produce a variety of fimbrial adhesins that allow attachment to specific host receptors. However, whereas many adhesins are more restricted in their distribution, type 1 fimbriae (fim) are produced by most pathogenic and commensal strains alike (1, 2).
Although the role of type 1 fimbriate E. coli in the intestinal tract is poorly defined, the adhesin is a virulence factor in urinary tract infections (2-7). Type 1 fimbriae are proinflammatory, stimulating release of IL-6, IL-8, and tumor necrosis factor α, and they act synergistically with other bacterial products such as lipopolysaccharide (8-12). The adhesin is sufficient for invasion of uroepithelial cells and may contribute to the etiology of the chronic inflammatory diseases, Crohn's disease, and interstitial cystitis (13-16).
As with the expression of many adhesins, fim is controlled by phase variation that results in a mixture of expressing (fimbriate) and nonexpressing (afimbriate) bacteria. Phase variation in bacteria is determined by various mechanisms including insertion and deletion of short sequence elements by mismatch repair, deoxyadenosine methylase (Dam) methylation-dependent alternative nucleoprotein complexes, and DNA rearrangements (17). Phase variation of fim is associated with inversion of a short (≈300 bp) DNA element that contains a promoter for the structural genes (18, 19). Inversion of the fim element involves two tyrosine family recombinase proteins, FimB and FimE, and is subject to elaborate control producing both (i) a relatively low frequency of switching from the afimbriate (off) to fimbriate (on) phase (between 10-4 and 10-3 per cell per generation), and (ii) control by environmental signals, including temperature, the branched-chain amino acids (particularly leucine) and alanine, and Neu5Ac (20-22). Whereas the amino acids stimulate fim phase variation in both directions, Neu5Ac inhibits switching from the off to on phase specifically (21, 22). Neu5Ac inhibits fimB expression, and thus exerts a selective effect on the inversion by inhibiting FimB, but not FimE, recombination (21).
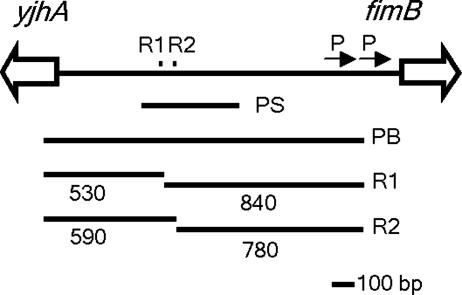
fimB is separated from the divergently transcribed yjhATS operon by one of the largest (1.4 kbp) intergenic regions in E. coli. Recently it was shown that two sequence elements, termed regions 1 and 2, situated >500 bp upstream of the fimB promoters in the fimB-yjhATS intergenic region, stimulate the recombinase genes expression in cis (ref. 21 and Fig. 1). When fimB is moved to an ectopic location, deletion of regions 1 and 2 only affects FimB recombination if sequences proximal to yjhA are included in the construct. Thus, regions 1 and 2 may control fimB expression by antagonizing a cis-active regulatory element or elements near yjhA. Region 1 includes, and region 2 lies adjacent to, a Dam methylation site that is unmethylated in a significant proportion of cells, indicating the presence of stable nucleoprotein structures (21, 25). Neu5Ac inhibits methylation protection at both 5′-GATC sites, suggesting that factor binding to regions 1 and 2 is suppressed by the amino sugar (21).
Fig. 1.
The fimB-yjhA intergenic region. Region (R)1 and R2 (small squares) lie close to 5′-GATC sequences protected from Dam methylation (21). The arrows and P mark the two known fimB promoters (23, 24). PS represents the size and position of the PCR product (Fim1-Fim2) used for EMSA and DNase I footprinting, and PB represents the PCR product used as a probe in Southern hybridization analysis. The length of DNA fragments (bp) generated after digestion with a combination of HpyCH4 IV and MboI are also shown.
Sialic acids play a pivotal role in molecular recognition, and sialylation of cell surfaces controls both constitutive and humoral defenses (26-29). These amino sugars are restricted to higher animals and their pathogens, and Neu5Ac levels rise during inflammation (27, 30, 31). Thus, particularly in sialidase-negative bacteria like E. coli, Neu5Ac could be a key signal within the host milieu (21, 31). A mutant containing the invertible element locked in the on orientation is more pathogenic in a mouse model for cystitis, and thus phase variation of fim can affect the host-parasite relationship (32). Here, we investigate the regulatory factors and signals required for the inhibition of FimB recombination in response to sialic acid. It is shown that regions 1 and 2 interact with different regulatory proteins to provide alternative Neu5Ac-responsive pathways controlling fimB, and it is proposed that such regulation helps balance the interaction between E. coli and its hosts.
Materials and Methods
Bacterial Strains, Plasmids, Media, Growth, and Assay Conditions. Bacterial strains were all derivatives of E. coli K-12 MG1655 (33). Allelic exchange of WT sequences was carried out as reported (34, 35). Intermediate strains containing deletions of the yjhA-fimB intergenic region, nanR or pdhR, replaced by a sacB-kanr cassette, were transformed with derivatives of the temperature-sensitive plasmid, pMAK705 (35). Mutations in region 1 (Rm1; 5′-CTTTATACCTGTTA in the WT altered to 5′-GGATCCTGGACAAT at the positions underlined) and in region 2 (Rm5; 5′-TTGCAATTCGTGTC altered to 5′-GGATCCAAGCACAG) were described (21). P1 transduction, using P1vir, was performed as reported (36). Media included LB broth and LB and sucrose agar (LB agar containing 6% sucrose but lacking sodium chloride) (35, 36). Rich-defined medium (RD) is minimal 3-(N-morpholino)propanesulfonic acid media (37) supplemented with 10 mM thiamine, 0.4% glycerol, bases, vitamin B supplement, amino acids, and Neu5Ac or GlcNAc as indicated. Reagents were obtained from Sigma unless indicated otherwise. Lactose MacConkey agar (Difco) was used to distinguish Lac+ from Lac- bacteria. Liquid cultures were grown aerobically at 37°C, and culture densities were monitored spectrophotometrically at 420 or 600 nm. For β-galactosidase assays, cells were grown in RD media at 37°C to an OD600 of 0.2, and assays were conducted as described (38). FimB recombination was measured as reported by using the fimA-lacZ reporter strains indicated in the text (22). At least five duplicate cultures were examined after growth for ≈22 generations at 37°C with rapid aeration.
DNA Manipulations. Plasmid DNA was isolated by using a kit (Qiagen, Valencia, CA), and chromosomal DNA was prepared as described (39). Restriction enzymes (Promega or New England Biolabs) and thermostable DNA polymerases (Boehringer Mannheim) used in PCR were utilized according to the manufacturers' instructions. Deletion mutations, used to replace WT sequences, were constructed by using standard PCR techniques (39) and cloned into pMAK705 (34). DNA sequencing was performed by the Advanced Biotechnology Centre, Imperial College, London. For in vitro DNA-binding analysis, a 438-bp PCR product (Fim1-Fim2), corresponding to positions 4537413-4537850 on the MG1655 genome, was synthesized by using PCR with either oligo labeled with [γ-32P] ATP and polynucleotide kinase. Southern hybridization was performed as described (21). A 1.3-kb PCR product (Fig. 1) was labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham Pharmacia Biosciences) by using Ready to go labeling beads (Amersham Pharmacia Biosciences). Hybridization was performed at 65°C for 16-18 h, and the blot was washed to high stringency with 0.1× SSC and 0.1% SDS before exposure to Hyperfilm ECL (Amersham Pharmacia Biosciences).
Electrophoretic Mobility-Shift Assay (EMSA) and DNase I Footprinting.Labeled DNA (1-3 nM) was incubated with various concentrations of NanR or NagC at room temperature for 10 min in a buffer containing 50 mM Hepes, 100 mM K glutamate (pH 8.0), and 0.5 mg/ml BSA. For EMSA, the samples (8 μl) were fractionated by electrophoresis through 5% (wt/vol) native acrylamide gels before drying and autoradiography. For DNase I footprinting, complexes (40 μl) were treated with DNase I (4 μl, 0.1 μg/ml) for 1 min at 37°C, and the reaction was stopped by addition of 100 μl of phenol, 200 μl of 0.4 M sodium acetate (pH 5.0), 2.5 mM EDTA, and 20 μg/μl sonicated DNA. Samples were phenol extracted, ethanol-precipitated, and analyzed on 6% (wt/vol) denaturing acrylamide gels (40).
Results
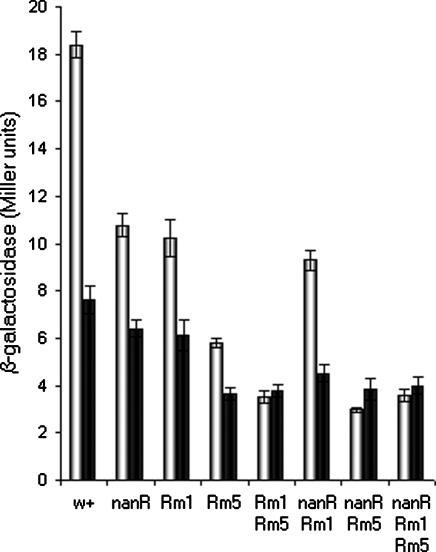
NanR Is a Positive Regulator of fimB Expression. NanR is a repressor of the nan operon, and growth on Neu5Ac prevents repression (40). To determine whether NanR controls fimB, a ΔnanR mutation of strain BGEC905 (MG1655 ΔlacZYA FimB-LacZ) was first constructed by allelic exchange (strain KCEC357). The mutation contains a deletion of the entire nanR ORF (from 3371332 to 3372124). As expected, the level of β-galactosidase produced by the mutant was diminished relative to the WT (Fig. 2). However, the effect observed was modest (≈1.7-fold), and the mutant remained partially sensitive to Neu5Ac (Fig. 2). Therefore, NanR cannot account fully for the effects of Neu5Ac on fimB expression.
Fig. 2.
The effect of ΔnanR, region 1 (Rm1) and region 2 (Rm5) mutations on the β-galactosidase produced by FimB-LacZ fusion in the absence (white bars) and presence (black bars) of sialic acid. The WT and mutant strains indicated were grown in RD glycerol medium to an OD600 of ≈0.2 at 37°C with rapid aeration before sampling, and β-galactosidase activity was measured as described (38).
Region 1 contains a conserved 27-bp element also found at the nan promoter (21). To determine whether the effect of NanR on fimB expression depends on region 1 alone, double mutants containing mutations in both nanR and region 1 (ΔnanR Rm1) and nanR and region 2 (ΔnanR Rm5) were constructed and characterized (Fig. 2). The two Rm mutations contain substitutions of 13 and 14 bp, respectively (ref. 21; see also Materials and Methods). Whereas the region 1 mutant had little additional effect on fimB expression in the ΔnanR background, the region 2 mutant does. Moreover, whereas any combination of mutations in region 1 and nanR remain sensitive to Neu5Ac, those expected to affect both regions 1 and 2 are insensitive to the amino sugar. Thus, loss of interaction with region 1, but not with region 2, accounts for the effect of NanR on fimB expression. Furthermore, the residual effects of Neu5Ac on fimB expression in the ΔnanR background are likely to be mediated by changes in another regulator that interacts with region 2. In addition, loss of region 1 activity, either by means of mutation of the DNA sequence per se, or by loss of NanR, produced a compound effect when combined with the mutation in region 2. Thus, regions 1 and 2 apparently function separately to stimulate fimB expression.
The Effects of pdhR and nagC on fimB Expression. Catabolism of Neu5Ac generates pyruvate and GlcNAc-6-P (41). The nucleotide sequence of region 2 contains an element (5′-AATTcGTNNNACaAAaT) that shows partial dyad symmetry homologous to (5′-AATTGGTNNNACCAATT) thought to be important for DNA binding of the pyruvate-responsive regulator, PdhR (42). Moreover, region 2 also contains a sequence that shows homology (5′-tgcAaTT(N9)AAATAtG) to a binding consensus for the GlcNAc-6-P-responsive protein, NagC (5′-STTATTT(N9)AAATAAS) (43). To determine whether these regulators control fimB expression, the effect of mutations in the corresponding genes was tested on the FimB-LacZ fusion. Although the pdhR mutant had little effect (20% decrease; data not shown), fimB expression decreased >4-fold in the nagC mutant (20.8 ± 0.7 to 4.6 ± 0.4 Miller unit in the WT and mutant, respectively). Thus, NagC is a positive regulator of fimB expression and could participate in the control of the recombinase genes expression in response to sialic acid.
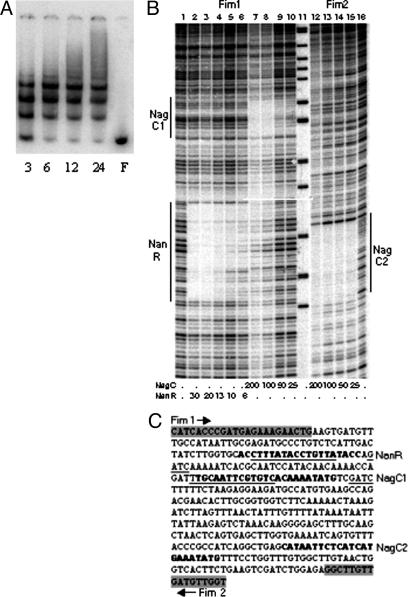
The Interaction of NanR and NagC with Regions 1 and 2 in Vitro. To determine whether NanR and NagC bind to regions 1 and 2, the interaction of the proteins with a 438-bp PCR product encompassing both elements (Fig. 1) was investigated by EMSA and by DNase I footprinting (Fig. 3).
Fig. 3.
In vitro binding of NanR and NagC to regions 1 and 2. (A) EMSA with NanR was carried out with the protein concentrations indicated (3-24 nM). NanR was absent in F. (B) DNase I footprinting with NanR and NagC. DNA was labeled at Fim1 (lanes 1-10) or Fim2 (lanes 12-16) and incubated with the nanomolar concentrations of proteins indicated. Region 1 (NanR), region 2 (NagC1), and NagC2 are indicated. (C) Sequence of the DNA used. The position and orientation of primers Fim1 and Fim2 is highlighted and marked by arrows. The sequences in bold are the consensus sequence matches for NanR and NagC contained within the regions protected from DNase I digestion are shown. The position of the replacement mutations Rm1 (region 1 and NanR-binding site) and Rm5 (region 2 and NagC1-binding site), as well as the corresponding 5′-GATC sequences that are protected from Dam methylation, are underlined.
Region 1 contains, with two mismatches, three direct repeats of the hexanucleotide sequence (5′-GGTATA) separated by 2-3 bp that characterize the binding site for NanR at nan (40). NanR binds to the fimB-yjhA region by EMSA to produce the same ladder pattern of three complexes reported for nan (Fig. 3A). Furthermore, NanR protects, from digestion with DNase I, a region of 30 bp that coincides with the hexanucleotide repeats (Fig. 3B, lanes 2-6). As expected, mutation Rm1 within region 1, which disrupts the NanR-binding consensus considerably, inhibited NanR binding to region 1 (data not shown) (21, 40). Thus, NanR binds with high affinity to region 1 in a way analogous to its binding at nan (40).
DNase I footprinting demonstrated that NagC binds to two sites, one site corresponds to region 2 as described above (Fig. 3B, lanes 7-10, NagC1) and the other to an additional site centered 212 bp downstream (Fig. 3B, lanes 12-15, NagC2). In a control experiment, mutation Rm5 within region 2 eliminated NagC binding to NagC1 as anticipated (data not shown). NagC2 includes the sequence 5′-CATA AT TCTCATCATGAAATATG, which matches well the consensus for the NagC-binding site (43). In EMSA, NagC produced a low-mobility complex that did not enter a 5% acrylamide gel (data not shown), indicating that it may form a large looped DNA-protein complex with the region (44). NagC sites occur in pairs normally, and cooperative binding between sites is necessary for regulation (43). However, because NagC2 is occupied at a concentration at which NagC1 remains unbound in vitro (Fig. 3B), cooperativity between the two elements is not apparent. Moreover, a deletion that includes NagC2 has little effect on fimB expression (21). Thus, although NagC bound to NagC1 at region 2 is likely to control fimB expression, the function of NagC2 is unclear.
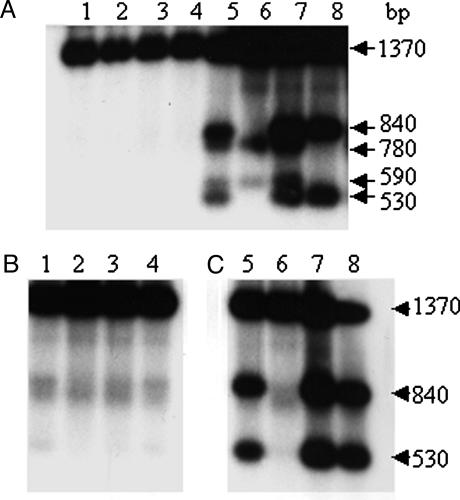
Protection from Dam Methylation at Regions 1 and 2 in Vivo. The 5′-GATC (Dam) sites at regions 1 and 2 become more fully methylated in the presence of Neu5Ac (21). To determine whether NanR and NagC are methylation-blocking factors, DNA isolated from cells grown to exponential phase in RD medium was digested with a combination of MboI (which only cuts unmethylated 5′-GATC sequences) and HypCH4IV and examined by Southern hybridization (Figs. 1 and 4). The HypCH4IV fragment examined contains six 5′-GATC sites, but only digestion at the third (region 1) and fourth (region 2) sites is detected in the WT, as shown by the appearance of bands of ≈530 bp plus 840 bp and 590 bp plus 780 bp, respectively (Fig. 4A, lane 5). As predicted, nanR (lane 6) and nagC (lane 8) mutations lead to loss of protection at regions 1 and 2, respectively, whereas the pdhR mutation (lane 7) had no effect.
Fig. 4.
Methylation protection of regions 1 and 2 by Southern blot hybridization analysis. The analysis included WT (BGEC905, lanes 1 and 5), ΔnanR (KCEC357, lanes 2 and 6), ΔpdhR (KCEC231, lanes 3 and 7), and ΔnagC (KCEC505, lanes 4 and 8) strains grown at 37°C in RD glycerol medium (A) and RD glycerol containing 3 mM Neu5Ac (B, lanes 1-4), or 3 mM GlcNAc (C, lanes 5-8). Chromosomal DNA was digested with HpyCH4 IV (A, lanes 1-4) or a combination of HpyCH4 IV and MboI (A, lanes 5-8 and B, lanes 1-8), and hybridized with a 32P-labeled PCR product (Fig. 1; PB) as described (21).
Neu5Ac leads to loss of protection at both regions 1 and 2 in the WT, as reported (21), as well as in the mutants (Fig. 4B). The observation that NagC is required for methylation protection at region 2 indicated that GlcNAc would stimulate methylation of this site specifically, and this was found to be the case (Fig. 4C, lanes 5-7). Thus, the loss of methylation protection at both regions 1 and 2 in the presence of Neu5Ac is explained by the fact that the Neu5Ac generates the inducing signal for NanR, and its metabolism produces GlcNAc-6-P, the inducing signal for NagC (41).
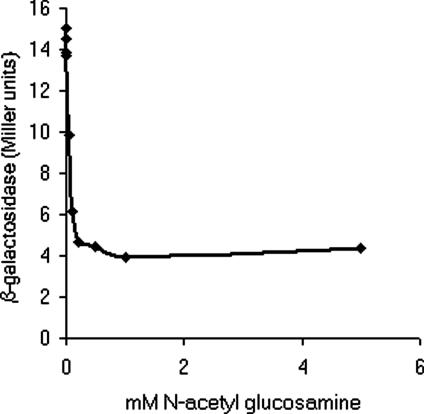
The Effect of GlcNAc on fimB Expression and FimB Recombination. The identification of NagC as an activator of fimB expression indicated that the expression of the recombinase would be inhibited by GlcNAc. As expected, expression of the FimB-LacZ fusion was depressed almost 3-fold in the presence of saturating amounts (≈0.3 mM) of GlcNAc (Fig. 5). Likewise, the rate of FimB catalyzed off-to-on inversion was reduced >35-fold in its presence [40.8 × 10-4 per cell per generation (n = 8; range of 160-3.4 × 10-4) to 1.1 × 10-4 per cell per generation (n = 8; range 2.1-0.4 × 10-4)]. Because only FimB catalyses off-to-on recombination at a detectable rate (19), GlcNAc inhibits the phase variation of type 1 fimbriation from the afimbriate-to-fimbriate phase.
Fig. 5.
The effect of GlcNAc on fimB expression. The β-galactosidase produced by strain BGEC905 (FimB-LacZ) in the presence of various concentrations of GlcNAc was measured as described (38). The bacteria were grown in RD glycerol medium to an OD600 of ≈0.2 at 37°C with rapid aeration before sampling.
Discussion
The phase variation of type 1 fimbriation in E. coli (fim) is controlled by an intricate regulatory network (reviewed in ref. 20). The off-to-on phase variation of fim is suppressed by Neu5Ac, and it was proposed that this is a response to the activation of host defenses (21). Here trans-active factors required for the regulation of fimB by Neu5Ac are identified, and the off-to-on phase variation of fim is also shown to be suppressed by GlcNAc. Type 1 fimbriae are proinflammatory and are a known (urinary tract infections) or suspected (Crohn's disease) virulence factor, yet they are also produced by many nonpathogenic strains of E. coli. N-acetyl-β-glucosaminidase is a lysosomal enzyme, and high levels of this enzyme characterize upper urinary tract infections (45, 46). Furthermore, levels of GlcNAc rise during inflammation (47). Thus, the results presented here contribute to our understanding of the signals and factors likely to affect the relationship between E. coli and its hosts in commensal and pathogenic interactions alike.
According to our current model, fimB transcription is repressed by a distant cis-active silencer situated proximal to yjhA, and “antirepressor” factors, binding to regions 1 and 2 antagonize, this effect (21). In addition, it was also proposed that Neu5Ac inhibits fimB expression by diminishing antirepressor binding to regions 1 and 2 (21). The demonstration here that NanR and NagC are the factors that interact with regions 1 and 2, respectively, supports and extends this model. Mutations in region 1 have less effect on fimB expression then do those in region 5 (21), and, as expected, mutations in nanR have less effect than those in nagC. Metabolism of Neu5Ac generates GlcNAc-6-P (41), the inducing signal for NagC, and it would therefore be expected to prevent suppression from both regions 1 and 2. Surprisingly, however, GlcNAc has a stronger effect on both fimB expression and FimB recombination than does Neu5Ac (ref. 21 and this work). This raises the possibility that NagC bound to region 2 is only partially inactivated by sialic acid, or that the regulator plays a more complex role in the control of fimB.
NagC operators characteristically occur in pairs, so that cooperative binding to two sites through DNA looping is necessary for regulation (43). Thus, NagC could form a more extensive looped DNA structure than that formed by NanR binding to three adjacent sites, and this might at least in part, contribute to NagC being more effective as an antirepressor than NanR for fimB expression. Although the nature of the inhibitory effect exerted by the yjhA proximal sequences is not yet understood, we hypothesize that NagC and NanR somehow interrupt long-range cis-acting repression by forming alternative incompatible nucleoprotein structures.
yjhA encodes an outer membrane channel that can facilitate Neu5Ac uptake (G. Condemine, personal communication), and this gene is also repressed strongly by NanR, an effect which is modulated by NagC binding to the NagC1 site. Region 1, shown here to be a NanR-binding site, in fact overlaps the yjhA promoter (ref. 31 and G. Condemine, personal communication). Thus the same factors, NanR and NagC, control both fimB and yjhA expression. However, whereas NagC exerts a stronger regulatory effect on fimB than NanR, the converse is true for yjhA. Although it is unclear to us why it should be, it is apparent that the expression of yjhA and fimB are coordinated.
The Rm1 mutation studied here should disrupt transcription initiation of yjhA, as well as diminish NanR binding (21, 40). However, a mutation of yjhA does not affect fimB expression in the absence (21) or presence (data not shown) of sialic acid. Region 1, nanR and region 1-nanR double mutants have similar effects on fimB, and thus neither YjhA, nor transcription directed toward yjhA, apparently affects the recombinase genes expression under the conditions studied here.
Dam methylation protection at regions 1 and 2 is apparently mutually exclusive, suggesting that NanR and NagC each form alternative, stable nucleoprotein complexes with their cognate-binding sites (21, 25). If NanR is less effective at enhancing fimB expression than NagC, as it seems to be, then the recombinase genes expression must therefore be controlled by phase variation. In support of this idea, we note that the frequency of FimB recombination varies between replicates considerably in the absence of the amino sugars (ref. 21 and this work). However, variable FimB-LacZ-expressing colonies are not seen on indicator media (data not shown), and methylation protection is detected at both sites in DNA isolated from single clones. Accordingly, if phase variation does occur, switching must happen at high frequency (>10-2 per cell per generation; ref. 22). fimE is regulated by the fim invertible element, and hence, like the Pap adhesin regulators PapB and PapI, both of the fim regulatory proteins are probably controlled by phase variation as well (48, 49).
The phase variation of many adhesins in E. coli and Salmonella is controlled by Dam methylation (reviewed in ref. 49), and fimB expression is altered in a dam mutant (50). Thus, Dam could affect fimB expression by inhibiting NanR and NagC binding to their cognate sites as it can do for Lrp and OxyR (49, 51). However, mutation of the 5′-GATC site adjacent to region 2 does not affect fimB expression (21), and under the conditions studied here, the recombinase genes expression is actually decreased in a dam mutant (data not shown). Therefore even if Dam methylation of regions 1 and/or 2 does affect fimB expression, the effects of methylation are unlikely to be accounted for by this alone.
Most strains of E. coli do not synthesize Neu5Ac, and nor is the amino sugar required for viability (31). In contrast, GlcNAc is essential, and the bacterium uses GlcNAc obtained from the environment or produces the phosphorylated derivative by de nova synthesis or from compounds like sialic acid. E. coli recycles cell wall material, including GlcNAc (52, 53). Thus levels of GlcNAc-6-P might change not only during growth on Neu5Ac or GlcNAc, but also when the balance of cell wall synthesis to recycling is altered. The dual control of fimB by Neu5Ac and GlcNAc should integrate signals from the environment with those originating within the cell. Peptidoglycan recycling may provide a means of monitoring the condition of the cell envelope (54), and fimB and GlcNAc-6-P could be part of such a regulatory circuit.
A commensal strain containing the fim invertible element locked in the orientation colonized the mouse large intestine poorly (55), and in the same model, GlcNAc and Neu5Ac enhance the early stages of E. coli colonization (56). On the other hand, constitutive expression of type 1 fimbriae in uropathogens can increase virulence (32). Temperatures >37°C, low levels of leucine, high osmolarity, Neu5Ac, and GlcNAc all inhibit FimB-catalyzed off-to-on phase variation (21, 22, 57). The biosynthesis of the branched-chain amino acids is restricted by oxidative stress, and OmpR protects E. coli from host defence peptides (58, 59). Furthermore, both Neu5Ac and GlcNAc levels increase during inflammation. Thus, the factors that regulate the off-to-on phase variation of fim could each signal activation of host defenses to the bacterium, to suppress expression of the adhesin, and, hence, help limit inflammation. GlcNAc is an antiinflammatory, mediating effects on both nitric oxide and IL-6 production, and shows promise in the treatment of Crohn's disease (60, 61). Inhibition of the expression of bacterial factors such as type 1 fimbriation could contribute to the efficacy of this and other amino sugars as antiinflammatory agents.
Acknowledgments
We thank Guy Condemine, Eric Vimr, Marjan van der Woude, Shadi Shafei, and Gary Robinson for discussions; Kathryn Kalivoda and Eric Vimr (University of Illinois at Urbana-Champaign, Urbana), for purified NanR and Charles Bell and Mitchell Lewis, (University of Pennsylvania School of Medicine, Philadelphia) for purified NagC. This work was supported by Wellcome Trust Grant 066447/Z/01/Z, (to I.C.B. and B.K.S.), Biotechnology and Biological Sciences Research Council Grant 96/P12206 (to I.C.B. and M.L.) and by a Biotechnology and Biological Sciences Research Council studentship to the University of Kent (to S.E.).
Author contributions: B.K.S., S.E.-L., J.A.P., and I.C.B. designed research; B.K.S., S.E.-L., and J.A.P. performed research; B.K.S., S.E.-L., M.H., and J.A.P. contributed new reagents/analytic tools; B.K.S., S.E.-L., J.A.P., and I.C.B. analyzed data; and B.K.S., J.A.P., and I.C.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Rm1 or Rm5, region 1 or 5 mutation; EMSA, electrophoretic mobility-shift assay; RD, rich-defined medium; Dam, deoxyadenosine methylase.
References
- 1.Johnson, J. R., Moseley, S. L., Roberts, P. L. & Stamm, W. E. (1988) Infect. Immun. 56, 405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloch, C. A., Stocker, B. A. & Orndorff, P. E. (1992) Mol. Microbiol. 6, 697-701. [DOI] [PubMed] [Google Scholar]
- 3.McCormick, B. A., Franklin, D. P., Laux, D. C. & Cohen, P. S. (1989) Infect. Immun. 57, 3022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connell, H., Agace, W., Klemm, P., Schembri, M., Mårild, S. & Svanborg, C. (1996) Proc. Natl. Acad. Sci. USA 93, 9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langermann, S., Palaszynski, S., Barnhart, M., Auguste, G., Pinkner, J. S., Burlein, J., Barren, P., Koenig, S., Leath, S., Jones, C. H., et al. (1997) Science 276, 607-611. [DOI] [PubMed] [Google Scholar]
- 6.Martindale, J., Stroud, D., Moxon, E. R. & Tang, C. M. (2000) Mol. Microbiol. 37, 1293-1305. [DOI] [PubMed] [Google Scholar]
- 7.Bahrani-Mougeot, F. K., Buckles, E. L., Lockatell, C. V., Hebel, J. R., Johnson, D. E., Tang, C. M. & Donnenberg, M. S. (2002) Mol. Microbiol. 45, 1079-1093. [DOI] [PubMed] [Google Scholar]
- 8.Malaviya, R., Ikeda, T., Ross, E. & Abraham, S. N. (1996) Nature 381, 77-80. [DOI] [PubMed] [Google Scholar]
- 9.Schilling, J. D., Martin, S. M., Hunstad, D. A., Patel, K. P., Mulvey, M. A., Justice, S. S., Lorenz, R. G. & Hultgren S. J. (2003) Infect. Immun. 71, 1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godaly, G., Frendeus, B., Proudfoot, A., Svensson, M., Klemm, P. & Svanborg, C. (1998) Mol. Microbiol. 30, 725-735. [DOI] [PubMed] [Google Scholar]
- 11.Samuelsson, P., Hang, L., Wullt, B., Irjala, H. & Svanborg, C. (2004) Infect. Immun. 72, 3179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goetz, M. B. (1989) J. Infect. Dis. 159, 533-542. [DOI] [PubMed] [Google Scholar]
- 13.Martinez, J. J., Mulvey, M. A., Schilling, J. D., Pinkner, J. S. & Hultgren, S. J. (2000) EMBO J. 19, 2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malaviya, R., Ikeda, T., Ross, E. A., Jakschik, B. A. & Abraham, S. N. (1995) Am. J. Ther. 2, 787-792. [DOI] [PubMed] [Google Scholar]
- 15.Boudeau, J., Barnich, N. & Darfeuille-Michaud, A. (2001) Mol. Microbiol. 39, 1272-1284. [DOI] [PubMed] [Google Scholar]
- 16.Schilling, J. D., Mulvey, M. A. & Hultgren, S. J. (2001) Urology 57, 56-61. [DOI] [PubMed] [Google Scholar]
- 17.Hallet, B. (2001) Curr. Opin. Microbiol. 4, 570-581. [DOI] [PubMed] [Google Scholar]
- 18.Abraham, J. M., Freitag, C. S., Clements, J. R. & Eisenstein, B. I. (1985) Proc. Natl. Acad. Sci. USA 82, 5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClain, M. S., Blomfield, I. C., Eberhardt, K. J. & Eisenstein, B. I. (1993) J. Bacteriol. 175, 4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomfield, I. C. (2001) Adv. Microb. Physiol. 45, 1-49. [DOI] [PubMed] [Google Scholar]
- 21.El-Labany, S., Sohanpal, B. K., Lahooti, M., Akerman, R. & Blomfield, I. C. (2003) Mol. Microbiol. 49, 1109-1118. [DOI] [PubMed] [Google Scholar]
- 22.Gally, D. L., Bogan, J. A., Eisenstein, B. I. & Blomfield, I. C. (1993) J. Bacteriol. 175, 6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donato, G. M., Lelivelt, M. J. & Kawula, T. H. (1997) J. Bacteriol. 179, 6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwan, W. R., Seifert, H. S. & Duncan, J. L. (1994) Mol. Gen. Genet. 242, 623-630. [DOI] [PubMed] [Google Scholar]
- 25.Braaten, B. A., Blyn, L. B., Skinner, B. S. & Low, D. A. (1991) J. Bacteriol. 173, 1789-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vimr, E. & Lichtensteiger, C. (2002) Trends Microbiol. 10, 254-257. [DOI] [PubMed] [Google Scholar]
- 27.Sillanaukee, P., Ponnio, M. & Jaaskelainen, I. P. (1999) Eur. J. Clin. Invest. 29, 413-425. [DOI] [PubMed] [Google Scholar]
- 28.Crocker, P. R. & Varki, A. (2001) Trends Immunol. 22, 337-342. [DOI] [PubMed] [Google Scholar]
- 29.Sakarya, S., Rifat, S., Zhou, J., Bannerman, D. D., Stamatos, N. M., Cross, A. S. & Goldblum, S. E. (2004) Glycobiology 14, 481-494. [DOI] [PubMed] [Google Scholar]
- 30.Nakano, K., Kuboniwa, M., Nakagawa, I., Yamamura, T., Nomura, R., Okahashi, N., Oshima, T. & Amano, A. (2004) Oral Microbiol. Immunol. 19, 205-209. [DOI] [PubMed] [Google Scholar]
- 31.Vimr, E. R. Kalivoda, K. A., Deszo, E. L. & Steenbergen, S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunther, N. W., IV, Snyder, J. A., Lockatell, V., Blomfield I., Johnson, D. E. & Mobley, H. L. (2002) Infect. Immun. 70, 3344-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guyer, M. S., Reed, R. R., Steitz, J. A. & Low, K. B. (1981) Cold Spring Harbor Symp. Quant. Biol. 45, 135-140. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P. & Kushner, S. R. (1989) J. Bacteriol. 171, 4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomfield, I. C., Vaughn, V., Rest, R. F. & Eisenstein, B. I. (1991) Mol. Microbiol. 5, 1447-1457. [DOI] [PubMed] [Google Scholar]
- 36.Silhavy, T. J., Berman, M. L. & Enquist, L. W. (1984) Experiments with Gene Fusions (Cold Spring Harbor Lab. Press, Plainview, NY).
- 37.Neidhardt, F. C., Bloch, P. L. & Smith, D. F. (1974) J. Bacteriol. 119, 736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 39.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G. & Struhl, K. (1987) Current Protocols in Molecular Biology (Wiley, New York).
- 40.Kalivoda, K. A., Steenbergen, S. M., Vimr, E. R. & Plumbridge, J. (2003) J. Bacteriol. 185, 4806-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plumbridge, J. & Vimr, E. R. (1999) J. Bacteriol. 181, 47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quail, M. A. & Guest, J. R. (1995) Mol. Microbiol. 15, 519-529. [DOI] [PubMed] [Google Scholar]
- 43.Plumbridge, J. (2001) Nucleic Acids Res. 29, 506-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plumbridge, J. & Kolb, A. (1991) J. Mol. Biol. 217, 661-679. [DOI] [PubMed] [Google Scholar]
- 45.Aronson, N. N., Jr., & Kuranda, M. J. (1989) FASEB J. 3, 2615-2622. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Cuartero, A., Lopez-Fernandez, A. & Perez-Blanco, F. (1998) Eur. Urol. 33, 348-350. [DOI] [PubMed] [Google Scholar]
- 47.Kriat, M., Vion-Dury, J., Fayre, R., Maraninchi, D., Harle, J. R., Confort-Gouny, S., Sciaky, M., Fontanarava, E., Viout, P. & Cozzone, P. J. (1991) Biochimie 73, 99-104. [DOI] [PubMed] [Google Scholar]
- 48.Sohanpal, B. K., Kulasekara, H. D., Bonnen, A. & Blomfield, I. C. (2001) Mol. Microbiol. 42, 483-494. [DOI] [PubMed] [Google Scholar]
- 49.Hernday, A., Braaten, B. & Low, D. (2004) Adv. Exp. Med. Biol. 547, 83-89. [DOI] [PubMed] [Google Scholar]
- 50.Oshima, T., Wada, C., Kawagoe, Y., Ara, T, Maeda, M., Masuda, Y., Hiraga, S. & Mori, H. (2002) Mol. Microbiol. 45, 673-695. [DOI] [PubMed] [Google Scholar]
- 51.Bolker, M. & Kahmann, R. (1989) EMBO J. 8, 2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodell, E. W. (1985) J. Bacteriol. 163, 305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park, J. T. (2001) J. Bacteriol. 183, 3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobs, C., Huang, L.-J., Bartowsky, E., Normark, S. & Park, J. T. (1994) EMBO J. 13, 4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCormick, B. A., Klemm, P., Krogfelt, K. A., Burghoff, R. L., Pallesen, L., Laux, D. C. & Cohen P. S. (1993) Microb. Pathog. 14, 33-43. [DOI] [PubMed] [Google Scholar]
- 56.Chang, D. E., Smalley, D. J., Tucker, D. L., Leatham, M. P., Norris, W. E., Stevenson, S. J., Anderson, A. B., Grissom, J. E., Laux, D. C., Cohen, P. S., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwan, W. R., Lee, J. L., Lenard, F. A., Matthews, B. T. & Beck, M. T. (2002) Infect. Immun. 70, 1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imlay, J. A. (2003) Annu. Rev. Microbiol. 57, 395-418. [DOI] [PubMed] [Google Scholar]
- 59.Prohinar, P., Forst, S. A., Reed, D., Mandic-Mulec, I. & Weiss, J. (2002) Mol. Microbiol. 43, 1493-1504. [DOI] [PubMed] [Google Scholar]
- 60.Shikhman, A. R., Kuhn, K., Alaaeddine, N. & Lotz, M. (2001) J. Immunol. 166, 5155-5160. [DOI] [PubMed] [Google Scholar]
- 61.Salvatore, S., Heuschkel, R., Tomlin, S., Davies, S. E., Edwards, S., Walker-Smith, J. A., French, I. & Murch, S. H. (2000) Aliment Pharmacol. Ther. 14, 1567-1579. [DOI] [PubMed] [Google Scholar]