Abstract
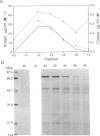
Copper-resistant strains of Pseudomonas syringae pathovar tomato accumulate copper and develop blue colonies on copper-containing media. Three of the protein products of the copper-resistance operon (cop) were characterized to provide an understanding of the copper-resistance mechanism and its relationship to copper accumulation. The Cop proteins, CopA (72 kDa), CopB (39 kDa), and CopC (12 kDa), were produced only under copper induction. CopA and CopC were periplasmic proteins and CopB was an outer membrane protein. Leader peptide sequences of CopA, CopB, and CopC were confirmed by amino-terminal peptide sequencing. CopA, CopB, and CopC were purified from strain PT23.2, and their copper contents were determined. One molecule of CopA bound 10.9 +/- 1.2 atoms of copper and one molecule of CopC bound 0.6 +/- 0.1 atom of copper. The Cop proteins apparently mediate sequestration of copper outside of the cytoplasm as a copper-resistance mechanism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Cooksey D. A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987 Feb;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Characterization of a Copper Resistance Plasmid Conserved in Copper-Resistant Strains of Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1987 Feb;53(2):454–456. doi: 10.1128/aem.53.2.454-456.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erardi F. X., Failla M. L., Falkinham J. O., 3rd Plasmid-encoded copper resistance and precipitation by Mycobacterium scrofulaceum. Appl Environ Microbiol. 1987 Aug;53(8):1951–1954. doi: 10.1128/aem.53.8.1951-1954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber N., Nachshon I. Localization of cholinesterase in Pseudomonas aeruginosa strain K. J Gen Microbiol. 1980 Mar;117(1):279–283. doi: 10.1099/00221287-117-1-279. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978 Oct;136(1):381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988 Sep;170(9):4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt A., Rossi A., Ladenstein R., Huber R., Bolognesi M., Gatti G., Marchesini A., Petruzzelli R., Finazzi-Agró A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini. Analysis of the polypeptide fold and a model of the copper sites and ligands. J Mol Biol. 1989 Apr 5;206(3):513–529. doi: 10.1016/0022-2836(89)90498-1. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins. FEBS Lett. 1991 Feb 11;279(1):73–78. doi: 10.1016/0014-5793(91)80254-z. [DOI] [PubMed] [Google Scholar]
- Rio D. C., Laski F. A., Rubin G. M. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986 Jan 17;44(1):21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- Rouch D., Camakaris J., Lee B. T., Luke R. K. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985 Apr;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Effect of ethylenediaminetetraacetic acid, Triton X-100, and lysozyme on the morphology and chemical composition of isolate cell walls of Escherichia coli. J Bacteriol. 1971 Oct;108(1):553–563. doi: 10.1128/jb.108.1.553-563.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Tetaz T. J., Luke R. K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J. T. Copper resistance in bacteria. Microbiol Sci. 1987 Jan;4(1):29–31. [PubMed] [Google Scholar]
- Wood P. M. Periplasmic location of the terminal reductase in nitrite respiration. FEBS Lett. 1978 Aug 15;92(2):214–218. doi: 10.1016/0014-5793(78)80757-1. [DOI] [PubMed] [Google Scholar]