Abstract
In the RV144 gp120 HIV vaccine trial, decreased transmission risk was correlated with antibodies that reacted with a linear epitope at a lysine at position 169 (K169) in the HIV-1 envelope (Env) second variable (V2) loop. The K169 V2 response was restricted to antibodies bearing Vλ rearrangements that expressed aspartic acid/glutamic acid (ED motif) in CDR L2. The AE.A244 gp120 in AIDSVAX B/E also bound to the unmutated ancestor of a V2-glycan broadly neutralizing antibody (bnAb), but this antibody type was not induced in the RV144 trial. Here we sought to determine if immunodominance of the V2 linear epitope could be overcome in the absence of human Vλ rearrangements. We immunized IGH and IGκ-humanized mice with the AE.A244 gp120 Env. In these mice the V2 antibody response was focused on a linear epitope that did not include K169. V2 antibodies were isolated that utilized the same human VH gene segment as an RV144 V2 antibody, but paired with a mouse lambda light chain. Structural characterization of one of these V2 antibodies revealed how the linear V2 epitope could be engaged despite the lack of an ED motif encoded in the mouse repertoire. Thus, in spite of the absence of the human Vλ locus in these humanized mice, the dominance of Vλ pairing with human VH for HIV-1 Env V2 recognition resulted in human VH pairing with mouse lambda light chains instead of allowing otherwise subdominant V2-glycan bnAbs to develop.
INTRODUCTION
The RV144 ALVAC/AIDSVAX B/E gp120 vaccine trial demonstrated an estimated 31% efficacy (1) and epidemiologic data indicated that efficacy was highest when the envelope (Env) of infecting virus matched the vaccine Env at lysine residue at position 169 (K169) in the second variable region (V2) of gp120 (2). Additionally Env V2-reactive antibodies were shown to be a correlate of reduced transmission risk in RV144 vaccinees (3). The importance of antibodies that recognize the V2 epitope at K169 was further underscored when four antibodies isolated from RV144 subjects that recognized the K169 V2 determinant were shown to mediate killing of CD4+ T cells infected by primary isolate HIV-1 strains by ADCC (4). Despite use of two different Vλ gene segments (Vλ3-10 and Vλ6-57), all four RV144-derived antibodies contained a germline glutamic acid-aspartic acid (ED) motif in their respective light chain second complementarity determining regions (CDR L2). The crystal structure of two of these antibodies, CH58 and CH59, in complex with V2 peptides revealed that the ED motif formed stabilizing salt bridges with two lysine residues in the V2 loop, including with K169 (4).
Recognition at K169 by antibodies with the CDR L2 ED motif was also a hallmark of the HIV-1 Env V2 response elicited by three independent rhesus macaque HIV-1 immunization regimens including two regimens that utilized RV144 immunogens (5). Rhesus macaque antibodies targeted to the V2 K169 determinant predominately utilized (66%) light chains containing the macaque Vλ gene segment orthologous to human Vλ3-10; this ortholog is the only VL gene in rhesus that contains an CDR L2 ED motif (5). We concluded that the phylogenetic conservation of V gene segments that contain the Vλ3-10-like CDR L2 ED motif implies a fitness advantage in pathogen recognition by the primate adaptive immune system (5). That the CDRL2 ED motif was independently used by V2 K169 antibodies in multiple subjects, different species, and following distinct immunization protocols strongly implies that V2 K169 recognition is limited by a restricted set of paratopic structural solutions (4, 5).
Another group of antibodies that bind to V2 at K169 are the V2-glycan broadly neutralizing antibodies (bnAbs) (6, 7); these bnAbs bind an epitope on the Env that includes both glycans at the N156 and N160 positions as well as the V2 polypeptide chain (7, 8). V2-glycan antibodies arise during infection but, to date, have not been induced by vaccination (9, 10). Induction of a V2-glycan bnAb is a preferred vaccine response because bnAbs have been shown to be protective in non-human primate infection models (11). A component in the RV144 vaccine, AE.A244 gp120, also expressed an epitope bound by mature V2-glycan bnAbs and the V2-glycan bnAb CH01 germline unmutated ancestor (UA) (4). Thus, in the RV144 vaccine trial, in spite of the vaccine immunogen expressing two different types of V2 epitopes that involve K169, one for a linear ADCC epitope and one for a bnAb V2-glycan epitope, only the linear V2 peptide antibody response was dominant.
In this study we investigated whether the immunodominance of the non-neutralizing linear V2 epitope would diminish in the absence of Vλ gene segments carrying the CDR L2 ED motif. Is the HIV-1 Env V2 K169 determinant intrinsically immunodominant or is the high frequency of antibody responses to this determinant controlled by the antibody repertoire? To address this question, we immunized humanized VelocImmune® mice with the RV144 immunogens (1). VelocImmune® mice carry the complete IGH and IGK variable loci in place of the endogenous Igh and Igk variable loci, respectively (12, 13). These humanized animals retain the endogenous Igλ loci, but in contrast to humans and macaques, none of the mouse Vλ gene segments have a CDR L2 with the ED motif (14). Consequently, even though these mice primarily (≥90%) utilize human Igκ VJ rearrangements to produce B-cell antigen receptors (BCRs) and antibody responses, they cannot produce the germline ED motif BCRs that dominate human and macaque antibody responses to the V2 K169 epitope (4, 5). Remarkably, we found that even though VelocImmune® mice lacked Vλ gene segments with the CDR L2 ED motif, they mounted robust humoral responses against the HIV-1 Env V2 that were dominated by chimeric antibodies composed of human VDJ heavy chain rearrangements paired with mouse λ light chain rearrangements. However, K169 was not included in the epitope of these V2 antibodies which was consistent with our previous findings that the ED motif is necessary for precise recognition of K169. We conclude that the immunodominance of linear V2 epitopes is largely due to intrinsic factors and that the promotion of rare antibody responses to the V2-glycan bnAb epitope cannot be achieved simply by modulation of the available Ig light chain repertoire.
MATERIALS AND METHODS
Immunization of humanized mice
Three Regeneron VelocImmune® mice were immunized by intraperitoneal injection with 25 µg of AE.A244 gp120Δ11(15) and 80 µl of the adjuvant STR8S-C 9 times at weeks 0, 2, 5, 7, 9, 11, 14, 16, and 18. Mouse RE503 was then fusion boosted at week 30, mouse RE504 was fusion boosted at week 40, and mouse RE505 was immunized a tenth time at week 40 then fusion boosted at week 57. The STR8S-C adjuvant was formulated as previously described (16) with concentrations of TLR agonists modified for mouse (0.5 mg/mL of R848, 0.2 mg/mL of CpG oligodeoxynucleotides).
Mice were housed in the Duke University vivarium in a pathogen-free environment with 12-h light/dark cycles at 20–25°C. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use committee at Duke University Medical Center under the National Institutes of Health/PHS Policy on Humane Care and Use of Laboratory Animals.
Antibody isolation
Mouse serum was screened for binding to AE.A244 gp120Δ11(15) and A244 V1V2 tags (4). B cell hybridomas were then generated according to methods described previously (17). Two mice, RE504 and RE505 yielded successful hybridoma fusions whereas a third mouse RE503 did not, due to technical issues. Hybridoma supernatants were then assayed for AE.A244 gp120Δ11, AE.A244 V1V2 tags binding and neutralization to virus isolates AE.92TH023 or AE.CM244. A total number of 8.57 × 10^7 splenocytes were harvested from mouse RE504. 16 96 well plates (with 80 wells containing cells) with ~66,000 splenocytes per well were then screened. 132 wells were positive for A244 gp120Δ11 and/or AE.A244 V1V2 tags binding and/or neutralization. After secondary screening to confirm reactivity, 57 wells repeated. A representative set of wells were then selected based on highest reactivity to put into limiting dilutions to confirm activity from a single clone. The resulting five clones were then purified according to methods described previously (17) and yielded antibodies RE504-46, RE504-60, RE504-97, RE504-117, and RE504-125. A total number of 6.27 × 10^7 splenocytes were harvested from mouse RE505. 11 96 well plates (with 80 wells containing cells) with ~71,000 splenocytes per well were then screened. 75 wells were positive for A244 gp120Δ11 and/or AE.A244 V1V2 tags binding and/or neutralization. A representative set of wells were then selected based on reactivity and secondary screening for binding was performed on this selected set to confirm reactivity. Those clones that repeated were put into limiting dilutions. The resulting seven clones were then purified and yielded antibodies RE505-11, RE505-22, RE505-58, RE505-70, RE505-23, RE505-27, and RE505-33.
ELISA binding and epitope mapping
ELISA binding assays were performed as previously described (18). V2 antibody epitopes were mapped by alanine scanning mutagenesis of the A244 V2 peptide (LRDKKQKVHALFYKLDIVPIED). Amino acid positions were included in the epitope if the log of the area under the binding curve (LogAUC) of the alanine mutated V2 peptide was reduced below 25% of the LogAUC value of the binding to wildtype V2 peptide.
Bio-Layer Interferometry
BLI measurements were made using the ForteBio OctetRed 96 instrument and streptavidin sensors. Data analysis was performed using the ForteBio evaluation software. The peptide sensors were prepared by incubating streptavidin sensor tips in biotinylated V2 peptide, AE.A244 171 (LRDKKQKVHALFYKLDIVPIED), at 5µg/mL for 5 minutes. The sensors tips were then washed in PBS for 60s prior to obtaining a baseline. Affinity measurements were made by dipping the peptide sensors in RE505Fab and CH59Fab ranging in concentration from 0.5ug/mL to 10ug/mL for 600s with a subsequent dissociation length of 600s. A non-HIV Env specific Fab 7968 was used as a negative control to subtract out non-specific binding to peptide sensors. Using the Fortebio evaluation software, subtracted binding curves for each Fab were fit using a 1:1 Langmuir model to obtain the association and dissocation rates constants, ka and kd, and the dissociation constant, KD.
Neutralization
Neutralization activity of antibodies was measured in the TZM-bl cell-based neutralization assay as described previously (19).
Peptide Microarray
Peptide microarray analysis was performed with modifications from a previously-reported protocol using a Tecan HS4000 Hybridization Workstation (20, 21). Peptide libraries consisting of 15-mers overlapped by 12 amino acids were printed onto glass slides, covering the full length of consensus gp160 Env from clades A, B, C, D, CRF01_AE, and CRF02_AG. Sequences for all peptides in the library were previously published (21, 22). Monoclonal antibodies were tested at 40 µg/ml with the exception of antibodies 505-11 and 505-22 which were tested at 60 µg/ml. Signal intensity of each spot was defined as the median 645 nm foreground measurement after background subtraction; defined as the belt located 3× the diameter around each spot. Positivity criteria for epitope binding are: more than 2 peptides in the epitope bind >99th percentile binding intensity of all 2058 peptides in the array library. mAbs with only peptide regions known to have higher non-specific binding in the top 99th percentile were defined as negative for linear epitope binding.
Protein production
RE505-22 Fab was recombinantly produced as previously described for other anti-HIV antibodies (4, 5,23, 24). Briefly, chains were generated by PCR using light and heavy chain genes as templates with appropriate primer pairs, and cloned into pcDNA3.1/hygro (25). Recombinant Fabs were produced in 293T cells by transient co-transfection with heavy chain and light chain gene plasmids. Recombinantly expressed Fab was captured with CaptureSelect Fab lambda affinity matrix (BAC) using 10 mM sodium phosphate pH 7.2, 150 mM NaCl, 0.05% azide as the loading buffer and 100 mM glycine pH 2.4, 150 mM NaCl, 0.05% azide as the eluting buffer. RE505-22 Fab was further purified via size exclusion chromatography on a Superdex 200pg 26/60 column (GE Life Sciences) in a buffer of 10 mM HEPES pH 7.2, 50 mM NaCl, 0.02% azide. Peak Fab-containing fractions were pooled, concentrated, and buffer exchanged as necessary.
Crystallography
Purified RE505-22 Fab was brought to a concentration of 12.9 mg/ml in ddH2O. A gp120164–182 peptide bearing an acetylated N-terminus and amidated C-terminus was dissolved in ddH2O to 100 mg/ml. Fab and peptide were mixed in a 1:3 respective molar ratio and diluted to a total protein concentration of 14.2 mg/ml. Crystals were observed over a reservoir of 0.2 M NaF, 20% PEG 3350 at a temperature of 20 °C. Crystals were cryoprotected by soaking briefly in mother liquor supplemented with 10% ethylene glycol then cryocooled in liquid nitrogen. Diffraction data were collected at SER-CAT with an incident beam of 1 Å in wavelength. The data were reduced in HKL-2000 (26). Matthews analysis suggested two Fab molecules in the asymmetric unit (27). The structure was phased by molecular replacement in PHENIX (28) using source models chosen by high sequence homology: the heavy chain of anti-HIV antibody CH59 (PDB:4HPY) (4) and the light chain of an antibody against parathyroid hormone-related protein (PDB:3FFD) (29). Rebuilding and real-space refinements were done in Coot (30) with reciprocal space refinements in PHENIX (31) and validations in MolProbity (32). During the rebuilding and refinement process, significant positive peaks were observed in the difference electron density maps in the vicinity of the paratope thus were interpreted to represent bound peptide; however, neither a single conformation nor even a single tracing of the peptide backbone could be confidently established in the structural model. Neither unaveraged omit maps nor phase improvement using non-crystallographic symmetry averaging improved the density of the peptides. Additional datasets on isomorphous crystals also failed to resolve any bound peptides. Difference fo-fc maps calculated at lower resolution (3.0 Å) still showed contiguous tracings with strong positive peaks, ruling out the possibility that the features could be explained as a network of water molecules or other ions present in the crystallization experiment. Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5KG9. Structural image rendering and RMSD calculations were performed using PyMOL (Version 1.8, Schrödinger).
Sequence analysis
A multi-step approach was used to search in mammalian species for candidate orthologs of VL3-10 that included the CDR L2 ED motif. First we used BLAST (33)] with the rhesus IGLV3-10 ortholog, IGLV3-17 (5), as the query against the RefSeq genomic database (34)]. The top 5000 hits from the BLAST search were retained and the highest similarity to IGLV3-10 for each species was included in the dataset of orthologs. To account for potential allelic differences at the ED positions, if a suboptimal BLAST hit within a species included the ED motif, that hit was retained in the dataset of candidate orthologs for that species. If no ED motif was found within VL3-10 candidate orthologs, we applied a second step utilizing the BLAT web server (35, 36) to find additional potentially orthologous alleles within fully sequenced mammalian genomes. Two BLAT queries were constructed, one using as the query sequence the non-ED containing hit from the BLAST run described above, and one using as the query sequence the rhesus IGLV3-17 sequence. If any hits from the BLAT runs resulted in sequences that included the CDR L2 ED motif, they were included in the dataset. If no hits for a species included the CDR L2 ED motif, the hit with the highest similarity to VL3-17 was included for comparison. A multiple sequence alignment was created from the dataset using the MAFFT program (37).
RESULTS
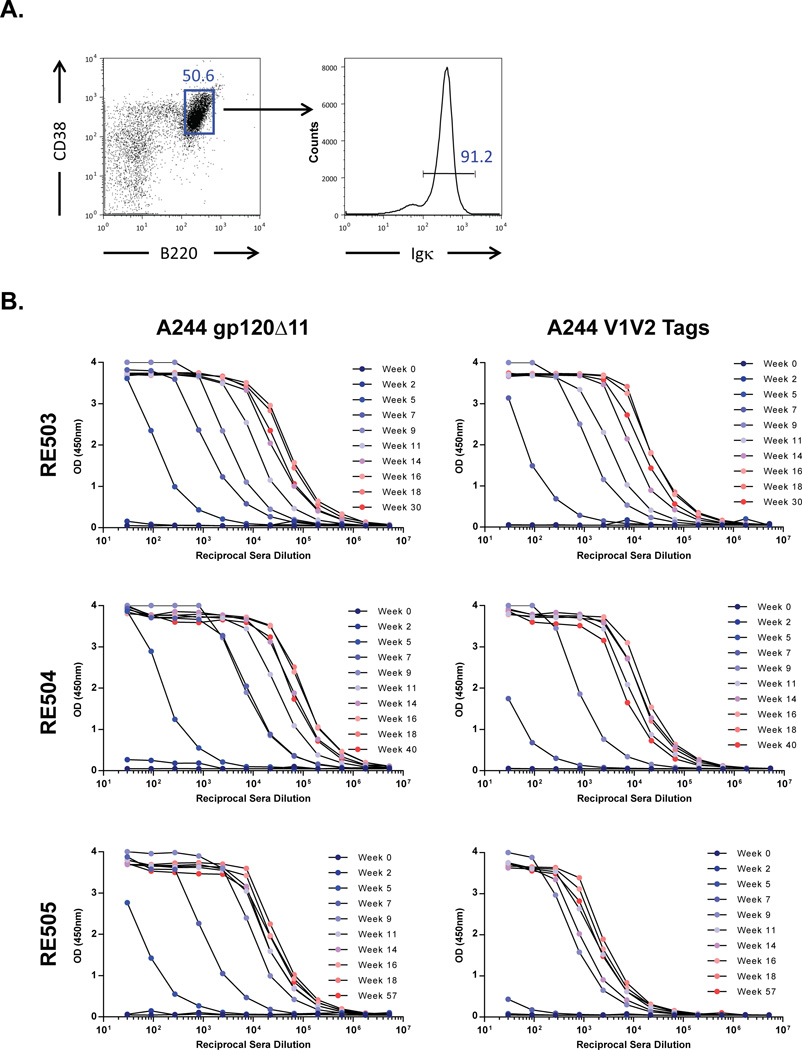
To characterize the light chain kappa and lambda usage in the naïve B cell repertoire in VelocImmune® mice, we isolated CD38+B220+ naïve, splenic B cells and determined that 91.2% of this population bore kappa light chains (Fig. 1A). This observation is virtually identical to the ≈95% expected kappa usage for laboratory mice (38). We subsequently immunized three VelocImmune® mice by intraperitoneal injection 9 times over 18 weeks with an HIV-1 Env gp120 identical to AE.A244 gp120, a gp120 component of the RV144 HIV vaccine (15). Mice were rested for 30 weeks and given one or two boost immunizations by intraperitoneal injection. 17 days after boosting, we quantified serum IgG that bound the A244 gp120 immunogen and a recombinant Env fragment that contained the V1 and V2 loops, termed A244 V1V2 tags (4). We observed serum IgG binding to both A244 gp120 and A244 V1V2 tags in all three mice (Fig. 1B). Next, in two mice (RE504 and RE505) we generated B cell hybridomas from splenocytes to identify and profile monoclonal antibodies (mAbs) representative of serum IgG activity. Hybridoma supernatants were assayed for binding to A244 gp120 and A244 V1V2 tags and HIV-1 neutralization. We selected mAbs that bound A244 gp120, A244 V1V2 tags and/or neutralized HIV-1 AE.92TH023.
Fig. 1. Characterization of light chain usage in naïve mice and sera response in immunized mice.
A) Naïve B cells (CD38+B220+) derived from the spleen of a VelocImmune® mouse (left panel) expressed 91.2% kappa light chains (right panel). A representative plot of two independent experiments is shown. B) Sera in all three immunized mice (RE503 to RE505) were observed to be reactive to A244 gp120 (left column) and A244 V1V2 tags (right column), an Env fragment construct that included the V2 epitope. Sera reactivity increased over time in each mouse for both antigens. Line marker colors correspond to immunization time points from blue (early) to red (late).
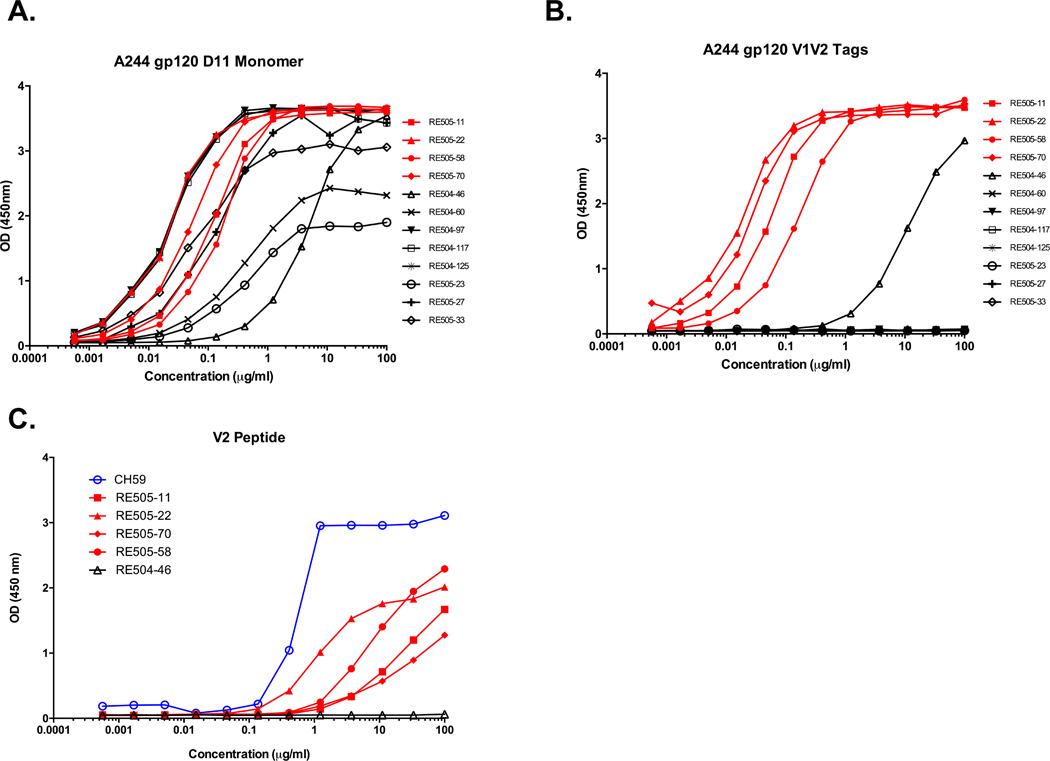
Twelve antibodies from this set were successfully cloned and purified for further characterization. Five of the twelve gp120-reactive mAbs bound A244 V1V2 tags (Fig. 2, A and B) and binding was not dependent on the presence of V2 glycans (Supplemental Fig. S1A). Of these, four bound V2 peptides (Fig. 2C, and Supplemental Table I). These V2-binding antibodies were isolated from mouse RE505 and shared the same VH gene segment (VH3-9) used by the human mAbs CH59 and HG107, recovered from RV-144 vaccinees that recognize the K169 V2 epitope. Remarkably, the four Velocimmune® V2 antibodies did not bear human kappa light chains but rather mouse λ light chains encoded by rearrangements of the mouse Vλ3-1 gene segment (Table I).
Fig. 2. Chimeric humanized mouse antibodies bind a gp120 V2 linear epitope.
Binding of the isolated antibodies to A) A244 gp120 and B) an A244 V1V2 construct. C) Only the four chimeric humanized mouse antibodies (red curves) bound V2 peptide. Binding was measured by ELISA and values reported as optical density (OD) at 450nm or the natural log of the area under the binding curve (LogAUC).
Table 1.
Immunogenetics of VelocImmune Mouse Antibodies
| Name | VH | JH | CDR H3 Lengtha |
VH Mut Freqb |
Isotype | VL | JL | CDR L3 Lengtha |
VL Mut Freqb |
|---|---|---|---|---|---|---|---|---|---|
| Antibodies isolated from mouse RE504 | |||||||||
| RE504-46 | 3-30 | 4 | 15 | 4.6% | IgG1 | VK1-9 | 4 | 9 | 3.1% |
| RE504-60 | 3-23 | 2 | 18 | 4.2% | IgG2 | VK3-20 | 5 | 8 | 1.5% |
| RE504-97 | 4-59 | 4 | 14 | 6.3% | IgG2 | VK4-1 | 4 | 9 | 2.2% |
| RE504-117 | 3-33 | 4 | 8 | 3.3% | IgG2 | VK2-24 | 4 | 9 | 0.0% |
| RE504-125 | 4-39 | 6 | 11 | 7.6% | IgG1 | VK1-5 | 5 | 8 | 2.7% |
| Antibodies isolated from mouse RE505 | |||||||||
| RE505-11 | 3-9 | 4 | 9 | 7.3% | IgG1 | Vλ3-01* | 2 | 13 | 4.7% |
| RE505-22 | 3-9 | 4 | 9 | 4.9% | IgG1 | Vλ3-01* | 2 | 13 | 3.1% |
| RE505-58 | 3-9 | 4 | 9 | 5.2% | IgG1 | Vλ3-01* | 2 | 13 | 3.1% |
| RE505-70 | 3-9 | 4 | 9 | 7.3% | IgG1 | Vλ3-01* | 2 | 13 | 3.9% |
| RE505-23 | 3-48 | 4 | 14 | 5.2% | IgG1 | VK3-11 | 2 | 10 | 1.5% |
| RE505-27 | 3-21 | 6 | 17 | 9.0% | IgG1 | VK1-6 | 2 | 9 | 4.2% |
| RE505-33 | 3-23 | 4 | 18 | 5.2% | IgG2 | VK3-20 | 4 | 8 | 1.9% |
CDR3 length calculated using IMGT definition of CDR3 after invariant cysteine and before invariant tryptophan
Mutation frequency calculated as percentage of nucleotide mutations in V gene segment ending at second invariant cysteine
Underlined names denote antibodies have been inferred to be clonally related
The usage of endogenous mouse lambda light chains was unexpected given the low frequency of lambda light chain usage in the VelocImmune® mouse (Fig. 1A). Mouse Vλ3-1 does not have a CDR L2 ED motif but rather has a Lys-Asp pair (KD) in the CDR L2, and is the mouse Vλ gene with the highest sequence identity to human Vλ3-10 (Supplemental Table II). In addition, the CDR L2 in the murine Vλ3-1 gene segment was 4 residues longer than in human Vλ3-10. The four V2 antibodies induced in the VelocImmune® mouse were clonally related (Table I). Thus, this clonal lineage represents expansion of an improbable human heavy, mouse lambda chimeric antibody response that shared similarities with the human RV144 vaccine-induced V2 mAb CH59 (4) despite the VelocImmune® mouse lacking a light chain repertoire with a germline-encoded ED motif. Given these antibodies lacked an ED motif, we next asked how the V2 linear epitope could be engaged without it. To address this question, we turned next to characterizing the V2 epitopes of these antibodies as well as structural analysis.
V2 recognition by ED motif-containing antibodies is mediated by electrostatic interaction via salt bridge formation between the ED motif in the CDR L2 and two positively charged lysines, including K169, in the V2 loop (4, 5). The substitution of a positively charged lysine for a negatively charged glutamate in the composition of the CDR L2 of the chimeric V2 antibodies described in this study has the potential to alter the electrostatic interaction component of V2 recognition. To characterize changes in how the four chimeric antibodies may recognize the V2 loop with a CDR L2 that lacks the ED motif, we next performed epitope mapping by ELISA with a set of alanine-scanning mutants of an AE.A244 V2 peptide (LRDKKQKVHALFYKLDIVPIED). Compared to the human ED-motif containing V2 antibodies induced in the RV144 trial, the chimeric mice V2 antibodies elicited in VelocImmune® mice were markedly less sensitive to K169A mutation, and recognized an epitope shifted away from K169 towards the C-terminus of the V2 loop (Table II, Supplemental Fig. 1B–E). The structure of CH59 in complex with a V2 peptide showed that the C-terminal half of the V2 was primarily contacted by the heavy chain whereas the N-terminal half, marked by multiple positively-charged lysines, was contacted by the light chain, specifically by salt-bridges formed with the CDRL2 ED-motif (4). The epitope mapping of RE505-22 is consistent with a similar mode of recognition as CH59, whereby usage of a VH3-9 rearranged heavy chain mediates interaction with the C-terminal half of the V2. However, because of a lack of an ED-motif in the RE505-22 light chain, N-terminal V2 recognition is altered and K169 is not fully engaged by complementary electrostatic interactions in RE505-22. Such an alteration of the V2 recognition could reduce overall binding of RE505-22 and indeed we observed that RE505-22 binds with nearly an order of magnitude less affinity to the V2 peptide than CH59 (Supplemental Fig. 1F–H).
Table II.
V2 Epitopes of Selected Antibodies and RV144 VH3-9 Antibodies
| Antibody | Species | VH | VL | V2 Epitope |
|---|---|---|---|---|
| RE505-11 | VelocImmune Mouse |
3-9 | λ3-1* | Q170, K171, H173, L175, F176, Y177, K178 |
| RE505-22 | VelocImmune Mouse |
3-9 | λ3-1* | Q170, K171, H173, L175, F176, Y177, K178 |
| RE505-58 | VelocImmune Mouse |
3-9 | λ3-1* | Q170, K171, H173, L175, F176, Y177, K178 |
| RE505-70 | VelocImmune Mouse |
3-9 | λ3-1* | Q170, K171, H173, L175, F176, Y177, K178 |
| CH59 | Human | 3-9 | λ3-10 | K169, H173, F176, Y177 |
| HG107 | Human | 3-9 | λ3-10 | K169, H173, F176 |
V2 epitope residues were defined as residues where LogAUC was reduced to <25% of wild type peptide binding in ELISA assay when mutated to alanine.
VH and VL gene segments are from human repertoire unless otherwise noted.
Antibody utilized endogenous mouse lambda V gene segments
RE505-22 weakly neutralized the easy-to-neutralize (tier 1) Clade AE virus 92TH023.6 in the TZM-bl neutralization assay but failed to neutralize a difficult-to-neutralize tier 2 Clade AE virus CM244 or other Clade C viruses (Supplemental Table III). This pattern of neutralization breadth for RE505-22 was similar to CH59 (4), however RE505-22 was observed to be an order of magnitude less potent neutralizer of 92TH023.6.
Structural comparison between human/mouse V2 and human V2 antibodies
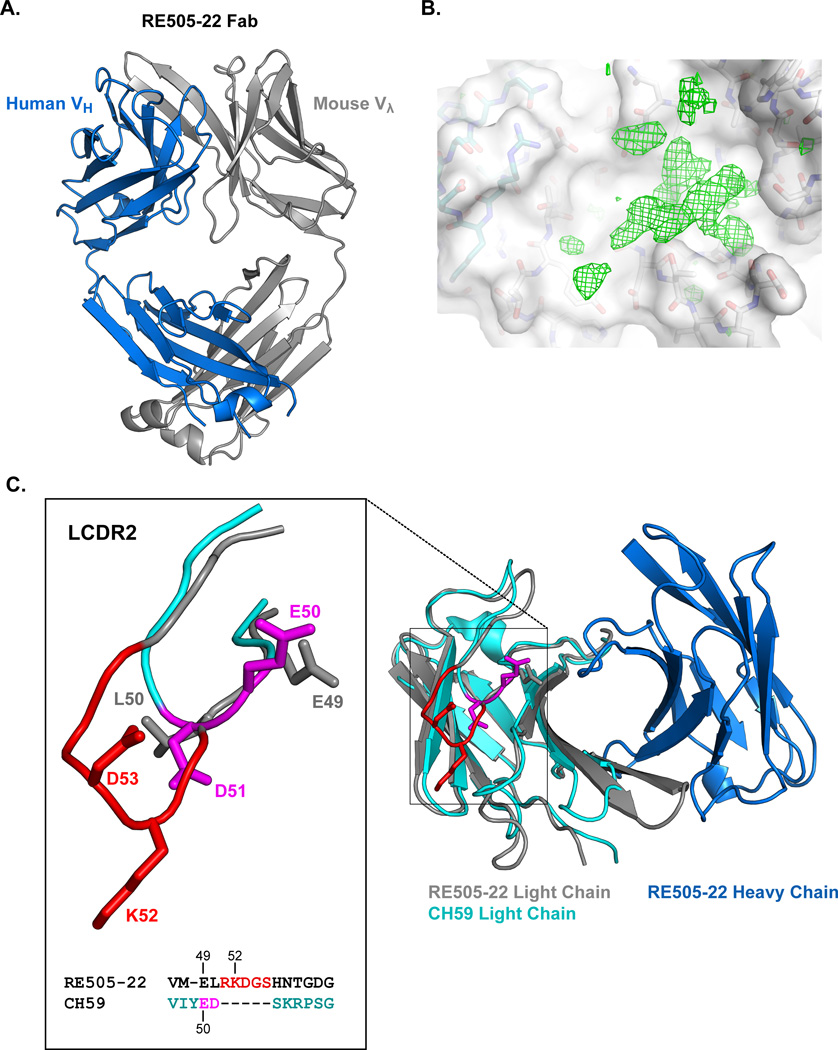
To define the structural differences in the CDR L2 orientation of the human Ig knock-in mouse V2 antibodies, we determined the structure of Fab RE505-22. We co-crystallized RE505-22 with V2 peptide (ELRDKKQKVHALFYKLDIV) and the antibody structure was solved to 2.3 Å resolution (Fig. 3A and Table III). While the presence of the V2 peptide in the asymmetric unit was confirmed in the difference electron density map (Fig. 3B), the peptide was disordered and its structural coordinates could not be resolved. The RE505-22 antibody Fab structure therefore represents the bound antibody conformation. The conformation of RE505-22 used for recognition of V2 could be compared to the bound conformation of the highly similar human V2 antibody CH59 (Fig. 3C). Because of their high sequence similarity due to shared VH3-9 usage, the structure of the RE505-22 H-chain was nearly identical to that of the CH59 heavy chain (0.32Å Cα RMSD). Pairing with different light chains, however, resulted in key structural differences between RE505-22 and CH59, with the largest differences observed in the light chain CDRs (Fig. 3C). Of particular interest to V2 recognition is the CDR L2 which in CH59 contains the ED motif that forms salt bridges with lysines K169 and K171 in the N-terminal half of the V2 peptide. In RE505-22, the CDR L2 is 4 residues longer than the CDR L2 of CH59 due to the usage of mouse Vλ3-1 which encodes for a CDR L2 with a length of 7 amino acids. Interestingly, Vλ3-1 is the only gene segment in the mouse light chain repertoire of this length. All other CDR L2s in the mouse light chain repertoire are of 3 amino acids in length. The pairing of the only VL gene segment with a long CDR L2 could be the result of selection such that the lysine 52 (K52) of the KD pair in the RE505-22 CDR L2 is oriented away from the critical like-charged lysines in the V2 epitope. Superposition of the RE505-22 and CH59 light chains demonstrated that the K52 and D53 (KD) do not share the structural positions of the ED motif of CH59 (Fig. 3C inset). Therefore, if the antibodies recognize V2 in the same orientation, K52 may be positioned far enough away from K169 and K171 that repulsive forces would be limited. This model of RE505-22 interaction is consistent with the evidence that the presence of K52 does not completely abrogate V2 binding. In addition, it is consistent with data that a K52E mutation introduced in RE505-22 to substitute in the ED motif did not improve binding or neutralization and did not alter the epitope from wildtype RE505-22 (Supplemental Fig. 1I, Supplemental Table III).
Fig. 3. Structure of the bound conformation of RE505-22.
A) Structure of the bound conformation of the RE505-22 Fab. Heavy chain colored blue and light chain colored gray. B) The difference electron density map in the paratope of the RE505-22 Fab shows strong peaks and contiguous features; nonetheless, bound peptide could not be built into the structure model with confidence. Light and heavy chain carbons are colored as in (A) behind a semitransparent surface (white). The electron density map (green mesh) is contoured at 3sigma level. C) Superposition of the CH59 light chain (cyan) onto the RE505-22 Fv domain light chain (gray) showed structural differences in the CDRs. A four residue insertion alters the conformation of the RE505-22 CDR L2 (inset) such that the positively charged K52 is oriented away from key positively charged V2 lysines (K169 and K171). E49 in RE505-22 adopts a similar position and orientation as E50 in CH59 which interacts with K171 in the V2 in the CH59-V2 complex. A sequence alignment of the CDR L2 region in RE505-22 and CH59 reflective of this structural alignment is shown at bottom of inset.
Table III.
Structural data collection and model refinement statistics.
| Data collection1 | |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 72.18, 76.75, 184.29 |
| α, β, γ (°) | 90, 90, 90 |
| No. of total reflections | 261869 |
| No. of unique reflections | 46019 |
| Resolution (Å)2 | 50-2.30 (2.34-2.30) |
| Rmerge (%) | 11.3 (57.3) |
| <I/σI> | 17.9 (2.3) |
| Completeness (%) | 99.7 (98.8) |
| Redundancy | 5.7 (4.9) |
| Model refinement | |
| No. reflections | 44809 |
| Rwork/Rfree(%) | 24.0 / 28.1 (32.2 / 35.7) |
| No. atoms / Average B values | |
| Protein | 6434 / 57.5 |
| Water | 37 / 39.6 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.753 |
| Ramachandran favored (%) | 95.9 |
| Ramachandran allowed (%) | 4.1 |
| Ramachandran outliers (%) | 0 |
Data were collected from a single crystal.
Highest resolution shell is shown in parentheses.
The superposition of the CDR L2s of CH59 and RE505-22 also revealed that E50 of the ED motif in CH59 is close in position to E49 of RE505-22 which was not apparent from our sequence alignments first performed in the absence of the RE505-22 structural information. E50 of CH59 forms a salt bridge with K171 of V2, thus similar positioning of E49 would suggest an explanation for the inclusion of K171 in the RE505-22 epitope (Table II). It is notable that Vλ3-1 is also the only mouse Vλ gene segment with a negatively charged residue encoded at position 49. The CDR L2 of RE505-22 lacks a negatively charged residue in the corresponding position to D51 of CH59, the residue that forms a salt bridge with K169, thus providing a rationale for why K169 is not included in the RE505-22 epitope (Table II).
Presence of the CDR L2 ED motif in mammals other than the mouse
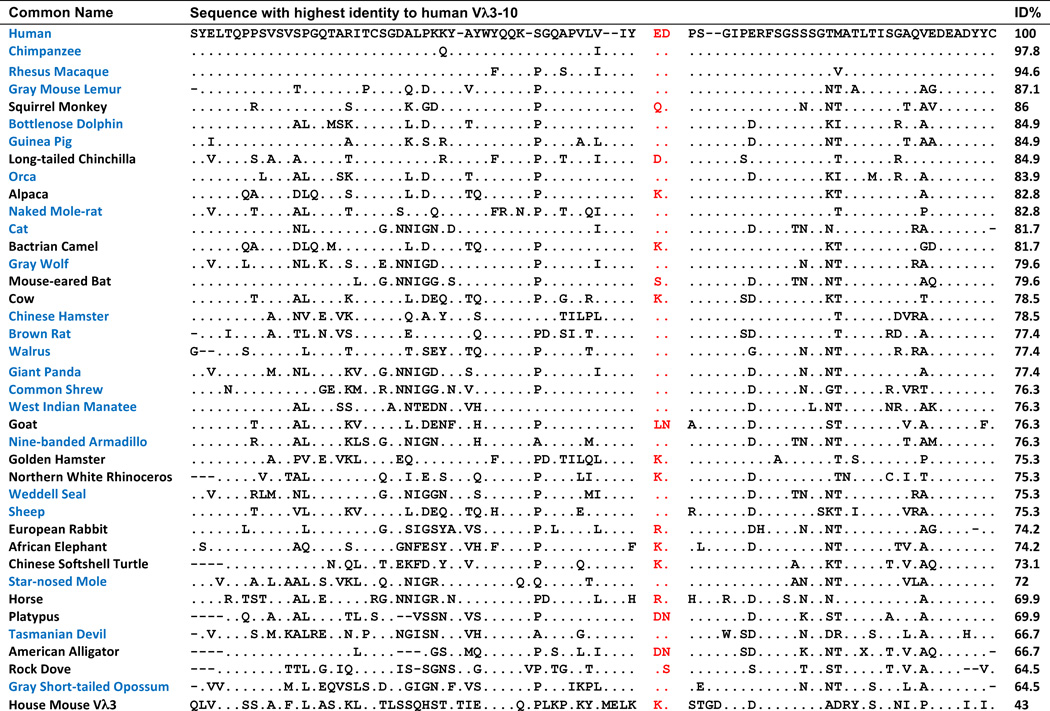
Previously we showed that the CDR L2 ED motif is highly conserved in primate phylogeny (5). However, the endogenous mouse lambda repertoire does not include a Vλ gene segment with high sequence similarity, and thus is not orthologous, to Vλ3-10 and no mouse Vλ gene segments encode for an ED in the CDR L2 (14). We therefore were interested in determining whether early common ancestors of mouse and humans had an ortholog of Vλ3-10 in its repertoire, thus implying these lambda repertoire traits were lost during the line of descent to mouse, or whether Vλ3-10 orthologs and the CDR L2 ED motif simply arose later in Mammalia evolution. We searched for potential Vλ3-10 orthologs in the RefSeq database of genomic sequences (34), and observed greater than 60% identity to the Vλ3-10 amino acid sequence in a broadly divergent group of organisms including species in the classes of mammals, reptiles and birds (Fig. 4). The observation of Vλ3-10 orthlogs containing the CDR L2 ED motif throughout the mammalian lineage including in the Tasmanian devil, a marsupial, suggests that an ED motif was present in the mammalian lineage from at least the time of speciation of placentals and marsupials which has been estimated to have occurred 162 million years ago (39). We also observed a Vλ3-10 ortholog with the CDR L2 ED motif in the brown rat, which has a recent common ancestor with mouse. The Vλ3-10 ortholog in rat has 78% identity to human whereas the highest identity mouse lambda V gene segment, VL4-1, has only 46% identity. This stands in striking contrast to the 95% identity between Vλ3-10 orthologs of human and rhesus macaques which diverged in the same order of time as mice and rats (5). Taken together, these data suggest that there was a loss of the orthologous Vλ3-10 gene segment during the line of descent in mouse and that it occurred after a relatively recent divergence from the common ancestor with rat.
Fig. 4. CDR L2 ED motif is conserved throughout mammalia lineage.
Amino acid sequence alignment of human Vλ3-10 to the highest similarity sequence from a representative set of animals with sequenced genomes. Dots represent amino acid residue matches. ED motif positions are shown in red. Mammals with sequences that include the CDR L2 ED motif are shown in blue. Amino acid sequence identity to human Vλ3-10 is shown in rightmost column. Mouse Vλ3-1 is included for reference.
DISCUSSION
A V2-directed response to RV144 immuogens is conserved between humans and rhesus macaques due to the common utilization of a recognition component, the CDR L2 ED motif, and it is this component that interacts with a critical residue, K169, that is correlated with reduced transmission risk in the RV144 trial (5). In this study, we showed that immunization with the same RV144 vaccine immunogens in a humanized mouse model that lacks the human Vλ repertoire and consequently the CDR L2 ED motif, nevertheless generates a robust V2-directed response. The V2-targeting antibodies from this mouse model that were elicited had a remarkable similarity to those induced in the RV144 vaccine trial, utilizing the same human VH gene segment and a close alternative endogenous mouse Vλ gene segment as human RV144 antibody CH59. We showed that the V2 epitope response in the humanized mice was shifted away from the K169 residue. Structural determination revealed a mechanism by which the mouse antibody could recognize the V2 without a principal interaction between the ED motif and K169. Thus the absence of the ED motif does not prohibit a similar V2 response as that observed in humans, but it altered the precise footprint of that antibody response away from a critical residue identified to be important for transmission risk.
The V2 response to RV144 immunogens in the VHDJH, VκJκ VelocImmune® humanized mouse is essentially the closest alternative to CH59 in humans that is available in the VH/Vκ humanized mouse repertoire. This remarkably conserved response not only demonstrates the viability of humanized mouse models to recapitulate human antibody responses in a vaccination setting, but also demonstrates the extraordinary selective pressure for the immune system to respond to the same immunogen in a strikingly similar fashion.
These observations have implications for HIV vaccine design. First, if a strain-specific effector function-mediating response can be protective, it stands that broadening the diversity of such a response within the V2 epitope could increase vaccine efficacy. Immunizing with a diverse panel of V2 sequences could leverage the immunodominance of the V2 response to elicit several different V2-targeting antibodies that individually may be narrowly specific, but in combination are broad enough to be protective against multiple strains of infecting virus. In this regard, we have recently shown that a pentavalent B/E/E/E/E gp120 vaccine boost of the ALVAC used in the RV144 trial protected against a mucosal R5 SHIV challenge (Bradley T, Haynes B et al. unpublished data). Second, if an easy-to-induce strain-specific dominant V2 polypeptide response outcompetes early ancestors of V1V2 glycan bnAb lineage development of HIV Env antibody types such as CH01, PG9, VRC-26, one strategy that has been proposed is termed B cell lineage design, in which sequential Envs targeted at lineage members are administered to selectively drive bnAb development (40). Alternatively, strategies to select against a CH59-like response might be necessary for bnAb induction. Strategies such as antigen induced germinal center B cell apoptosis(41, 42) could be used to negatively select CH59-like antibodies, thus in theory providing a survival advantage for V1V2-glycan bnAb lineage members.
Supplementary Material
Acknowledgments
Financial Support:
This work was supported by the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID; UMI-AI100645) grant from NIH/NIAID/DAIDS.
We thank Shi-Mao Xia, Melissa Cooper for performing neutralization assays. Crystallography was performed in the Duke University X-ray Crystallography Core Lab. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. SER-CAT supporting institutions may be found at www.ser-cat.org/members.html.
C.A.K., L.E.M and A.J.M are employed by Regeneron Pharmaceuticals, Inc. B.F.H. holds a patent with Regeneron Pharmaceuticals, Inc. related to vaccine development in VelocImmune mice.
Footnotes
Disclosures:
The authors declare competing financial interests: The authors have no additional financial interests.
References and Notes
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH M.-T. Investigators. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiehe K, Easterhoff D, Luo K, Nicely NI, Bradley T, Jaeger FH, Dennison SM, Zhang R, Lloyd KE, Stolarchuk C, Parks R, Sutherland LL, Scearce RM, Morris L, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Sinangil F, Phogat S, Michael NL, Kim JH, Kelsoe G, Montefiori DC, Tomaras GD, Bonsignori M, Santra S, Kepler TB, Alam SM, Moody MA, Liao HX, Haynes BF. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41:909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Program NCS, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, Mankoff Z, Schmidt SD, Lifson JD, Mascola JR, Nussenzweig MC, Martin MA. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macdonald LE, Karow M, Stevens S, Auerbach W, Poueymirou WT, Yasenchak J, Frendewey D, Valenzuela DM, Giallourakis CC, Alt FW, Yancopoulos GD, Murphy AJ. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc Natl Acad Sci U S A. 2014;111:5147–5152. doi: 10.1073/pnas.1323896111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy AJ, Macdonald LE, Stevens S, Karow M, Dore AT, Pobursky K, Huang TT, Poueymirou WT, Esau L, Meola M, Mikulka W, Krueger P, Fairhurst J, Valenzuela DM, Papadopoulos N, Yancopoulos GD. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci U S A. 2014;111:5153–5158. doi: 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam SM, Liao HX, Tomaras GD, Bonsignori M, Tsao CY, Hwang KK, Chen H, Lloyd KE, Bowman C, Sutherland L, Jeffries TL, Jr, Kozink DM, Stewart S, Anasti K, Jaeger FH, Parks R, Yates NL, Overman RG, Sinangil F, Berman PW, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Karasavva N, Rerks-Ngarm S, Kim JH, Michael NL, Zolla-Pazner S, Santra S, Letvin NL, Harrison SC, Haynes BF. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J Virol. 2013;87:1554–1568. doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody MA, Santra S, Vandergrift NA, Sutherland LL, Gurley TC, Drinker MS, Allen AA, Xia SM, Meyerhoff RR, Parks R, Lloyd KE, Easterhoff D, Alam SM, Liao HX, Ward BM, Ferrari G, Montefiori DC, Tomaras GD, Seder RA, Letvin NL, Haynes BF. Toll-like receptor 7/8 (TLR7/8) and TLR9 agonists cooperate to enhance HIV-1 envelope antibody responses in rhesus macaques. J Virol. 2014;88:3329–3339. doi: 10.1128/JVI.03309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scearce RM, Eisenbarth GS. Production of monoclonal antibodies reacting with the cytoplasm and surface of differentiated cells. Methods Enzymol. 1983;103:459–469. doi: 10.1016/s0076-6879(83)03032-3. [DOI] [PubMed] [Google Scholar]
- 18.Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X, Duffy R, Howington R, Cope A, Sadagopal S, Park H, Pal R, Kwa S, Ding S, Yang OO, Fouda GG, Le Grand R, Bolton D, Esteban M, Phogat S, Roederer M, Amara RR, Picker LJ, Seder RA, McElrath MJ, Barnett S, Permar SR, Shattock R, DeVico AL, Felber BK, Pavlakis GN, Pantaleo G, Korber BT, Montefiori DC, Tomaras GD. Vaccine-Induced Linear Epitope-Specific Antibodies to Simian Immunodeficiency Virus SIVmac239 Envelope Are Distinct from Those Induced to the Human Immunodeficiency Virus Type 1 Envelope in Nonhuman Primates. J Virol. 2015;89:8643–8650. doi: 10.1128/JVI.03635-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicely NI, Dennison SM, Spicer L, Scearce RM, Kelsoe G, Ueda Y, Chen H, Liao HX, Alam SM, Haynes BF. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat Struct Mol Biol. 2010;17:1492–1494. doi: 10.1038/nsmb.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicely NI, Wiehe K, Kepler TB, Jaeger FH, Dennison SM, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Robb ML, O'Connell RJ, Michael NL, Kim JH, Liao HX, Munir Alam S, Hwang KK, Bonsignori M, Haynes BF. Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee. EBioMedicine. 2015;2:713–722. doi: 10.1016/j.ebiom.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, Ma BJ, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Matthews BW. Solvent content of protein crystals. Journal of molecular biology. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta crystallographica. Section D, Biological crystallography. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinstry WJ, Polekhina G, Diefenbach-Jagger H, Ho PW, Sato K, Onuma E, Gillespie MT, Martin TJ, Parker MW. Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor. J Biol Chem. 2009;284:15557–15563. doi: 10.1074/jbc.M900044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGuire KL, Vitetta ES. kappa/lambda Shifts do not occur during maturation of murine B cells. J Immunol. 1981;127:1670–1673. [PubMed] [Google Scholar]
- 39.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 40.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han S, Zheng B, Dal Porto J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. IV. Affinity-dependent, antigen-driven B cell apoptosis in germinal centers as a mechanism for maintaining self-tolerance. J Exp Med. 1995;182:1635–1644. doi: 10.1084/jem.182.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shokat KM, Goodnow CC. Antigen-induced B-cell death and elimination during germinal-centre immune responses. Nature. 1995;375:334–338. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.