Abstract
Migration through micron-size constrictions has been seen to rupture the nucleus, release nuclear-localized GFP, and cause localized accumulations of ectopic 53BP1 – a DNA repair protein. Here, constricted migration of two human cancer cell types and primary mesenchymal stem cells (MSC) increases DNA breaks throughout the nucleoplasm as assessed by endogenous damage markers and by electrophoretic ‘comet’ measurements. Migration also causes multiple DNA repair proteins to segregate away from DNA, with cytoplasmic mis-localization sustained for many hours as is relevant to delayed repair. Partial knockdown of repair factors that also regulate chromosome copy numbers is seen to increase DNA breaks in U2OS osteosarcoma cells without affecting migration and with nucleoplasmic patterns of damage similar to constricted migration. Such depletion also causes aberrant levels of DNA. Migration-induced nuclear damage is nonetheless reversible for wild-type and sub-cloned U2OS cells, except for lasting genomic differences between stable clones as revealed by DNA arrays and sequencing. Gains and losses of hundreds of megabases in many chromosomes are typical of the changes and heterogeneity in bone cancer. Phenotypic differences that arise from constricted migration of U2OS clones are further illustrated by a clone with a highly elongated and stable MSC-like shape that depends on microtubule assembly downstream of the transcription factor GATA4. Such changes are consistent with reversion to a more stem-like state upstream of cancerous osteoblastic cells. Migration-induced genomic instability can thus associate with heritable changes.
Graphical abstract
Introduction
The nucleus has long been thought to limit a cell’s ability to migrate through small, stiff pores in tissue matrix [1], but migration through constricting pores can also rupture the nuclear lamina [2]. Nuclear envelope rupture in migration through narrow channels causes GFP constructs with a nuclear localization signal (NLS) peptide to mis-localize into the cytoplasm for hours [3]. On the other hand, local enrichment within the nucleus of GFP fusions of 53BP1, one of many DNA repair factors, has suggested accumulation of DNA damage. Although consistent with initial reports of DNA damage in constricted migration [2, 4], GFP itself has a nuclear localization tendency [5], and overexpression of nuclear proteins including 53BP1 can have important functional effects [6]. Moreover, in immortalized epithelial cells (RPE-1) cells, GFP-53BP1 only appeared enriched far from the leading edge site of nuclear rupture and resolved within minutes [3], whereas in U2OS osteosarcoma cells, enrichment occurred only at the site of nuclear rupture and required hours to resolve [7]. Exposure of U2OS cells in 2D culture to DNA damage agents for 1 hr likewise causes damage that lasts for many hours [8] and that is prolonged upon depletion or mutation of chromatin binding [8] and DNA repair factors [9-11].
Here we focus first on spatiotemporal changes of endogenous DNA damage and repair factors in U2OS cells migrating through rigid micropores (of relevance to bone), and then we focus on lasting perturbations to the genome. U2OS cells are widely used for studies of genomic instability (eg. [12]) in part because osteosarcoma tumors are multi-clonal with 100’s of megabase changes in multiple chromosomes [13, 14]. Chromosomal aberrations in osteosarcomas are also characteristic of DNA repair defects [15], which motivates our scrutiny of endogenous repair factors. U2OS clones generated from single cells ultimately provide key evidence of migration-induced genotype-phenotype changes.
Results
Rupture, DNA breaks, and mis-localized repair factors after constricted migration
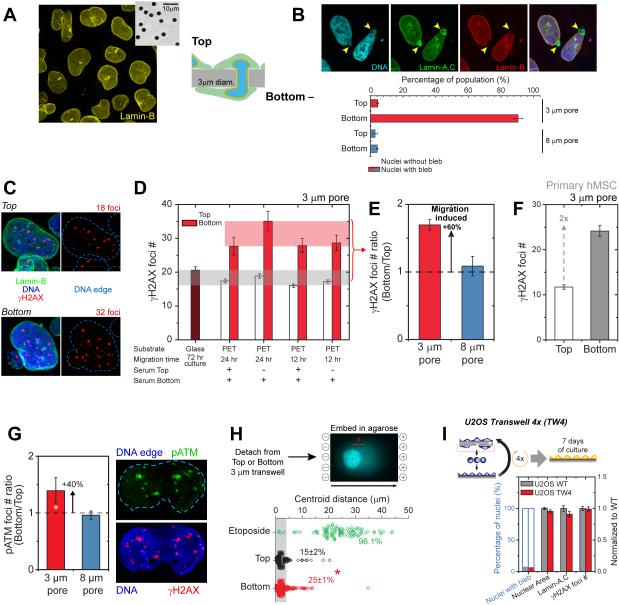
U2OS cells squeeze through transwell filters with 3-μm pores even with equal serum on both sides of the filters, and migration transforms the rounded nuclei (Figure 1A) into distorted, often elongated shapes with polar blebs on 90% of nuclei (Figure 1B). Lamin-A,C enrichment on blebs contrasts with lamin-B’s near-absence (Figure 1B, S1) as per another cell type [2]. Immunostaining for DNA damage marker γH2AX resolved 15-20 γH2AX foci in nuclei on transwell tops and in cells on glass, but γH2AX foci are greater (~60%) after constricted migration, independent of a serum gradient (Figure 1C-E). As a nucleus exits a pore, γH2AX foci concentrate near the pore (Figure S1). For 8-μm pores, top (unmigrated) and bottom (migrated) cells show no difference in γH2AX foci numbers and nuclear blebs (Figure 1B,E, S1).
Figure 1. Migration through 3-μm pores causes transient nuclear lamina rupture and DNA breaks, and repair factor mis-localization.
(A, B) U2OS nuclei on tops of transwells are rounded (inset: 3-μm pores). Migration elongates and causes blebs on poles but not with 8-μm pores (Figure S1; ≥40 nuclei per condition, n≥3 expts).
(C-F) Immunostained γH2AX foci on tops and bottoms of transwells (polyester, PET) or glass show increased damage after U2OS migration thru 3-μm but not 8-μm pores. Human mesenchymal stem cells (hMSCs) show more foci after 3-μm pore migration (Figure S1; ≥45 nuclei per condition, n=3 expts, *p<0.05).
(G) Immunostained phospho-ATM (pATM) foci show increased damage after U2OS migration thru 3-μm but not 8-μm pores (≥50 nuclei per condition, n≥3 expts, *p<0.05).
(H) Comet assay for DNA breaks in isolated U2OS nuclei show 3-μm pore migration causes more centroid shifts (threshold: 3-μm) as does Etoposide in cultures (10μM, 2hrs). (≥175 nuclei per group, n≥3 expts, *p<0.05).
(I) Post-migration recovery of lamin-A,C, DNA damage, nuclear area, and blebs. (≥130 nuclei per condition, n≥3 expts).
Damage is also evident in mesenchymal stem cells (MSCs) and lung carcinoma A549 cells. Primary MSCs from human bone marrow are osteogenic but stop growing beyond ~5 passages [16] (especially post-migration), and they express abundant lamin-A that ‘freezes’ in a highly elongated shape after pore migration [2]. However, migration increases nuclear blebs in MSCs (Figure S1) and A549s [2], and both show more γH2AX foci post-migration (Figure 1F, S1).
Foci number with activated ATM (phosphorylated ATM that can phosphorylate H2AX among other factors) is ~40% greater after U2OS cell migration through 3-μm but not 8-μm pores (Figure 1G). Electrophoresis-based ‘comet assays’ of nuclei isolated post-migration relative to nuclei detached from transwell tops (Figure 1H) also showed more of a cathode-shifted centroid of DNA (by ~60% above a threshold) with a higher mean displacement. Etoposide treatment causes abundant DNA breaks as expected [17].
Micro-nuclei are satellite nuclei, often γH2AX positive, with roles in genome remodeling [18], but very few cells (2% top; 4% bottom) show micro-nuclei (Figure S1) compared to the 6- to-7-fold larger fractions of cells with significant DNA breaks in comet assays (Figure 1H). MDA-MB-231 cells show similar micro-nuclei counts [7]. Importantly, cultures of migrated U2OS cells reverse the nuclear blebs and γH2AX foci, indicating that such changes are transient (Figure 1I).
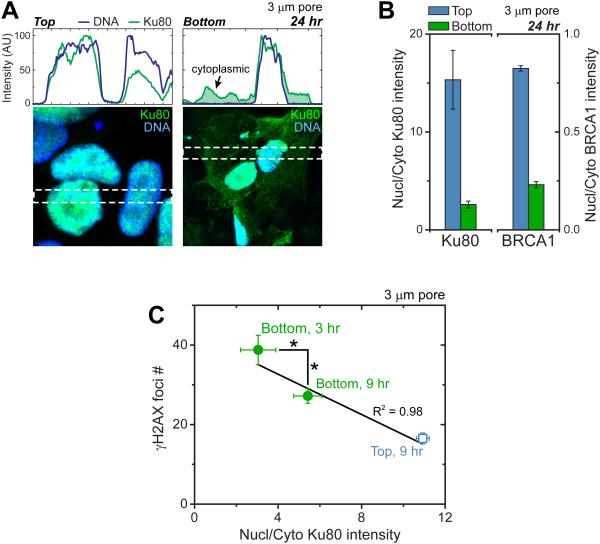
DNA repair factors Ku80 and BRCA1 are diffusible [19, 20] and mis-localize to cytoplasm after constricted migration (Figure 2A,B,S1). H2B-mCherry and GFP-NLS constructs also rupture from the fronts of nuclei in migration and re-localize within hours [3, 7, 21] while GFP-53BP1 seems delayed (Figure S1). Although over-expression of 53BP1 can rescue DNA damage in mouse embryo fibroblasts cultured on plastic from lamin-A knockout mice [6], we find over-expressed 53BP1 does not rescue migration-induced DNA damage (Figure S1). However, repair involves multiple factors. Endogenous Ku80 and BRCA1 indeed exhibit low nuclear-to-cytoplasmic ratios after migration at population (Figure 2B) and single-cell (Figure 2C) levels. Low nuclear-to-cytoplasmic ratios of Ku80 and high γH2AX foci counts decay over hours (Figure 2C), consistent with timescales post-etoposide [8]. Such deficits in repair factors could have functional effects because mouse knockouts or heterozygous mutants for BRCA1, BRCA2, Ku80, ATM, and RPA1 can alter chromosome copy numbers [22-26]. All of these DNA repair factors are expressed in U2OS cells (Figure 3A), despite reported deficits (e.g. BRCA2, p53 [15]), and variations between U2OS cultures pre- and post-migration are small. However, rupture-induced decreases in nuclear fractions of such diffusible factors can be expected to increase γH2AX foci throughout the nucleus as seen here (Figure S1), rather than being enriched near the ruptured lamina [7].
Figure 2. Constricted migration also causes repair factors mis-localization that negatively correlates with DNA damage levels.
(A-C) Immunostained intensity profiles of repair factor Ku80 highlight nuclear localization on top of 3-μm pores but cytoplasmic mis-localization on bottom (green shade). For Ku80 and BRCA1, nuclear-to-cytoplasmic intensities decrease post-migration (Figure S1; ≥10 fields of view per condition, n≥ 3expts). Ku80 re-localizes over hours on bottom as γH2AX foci count also decreases. (≥10 cells per condition, n≥2 expts, *p<0.05).
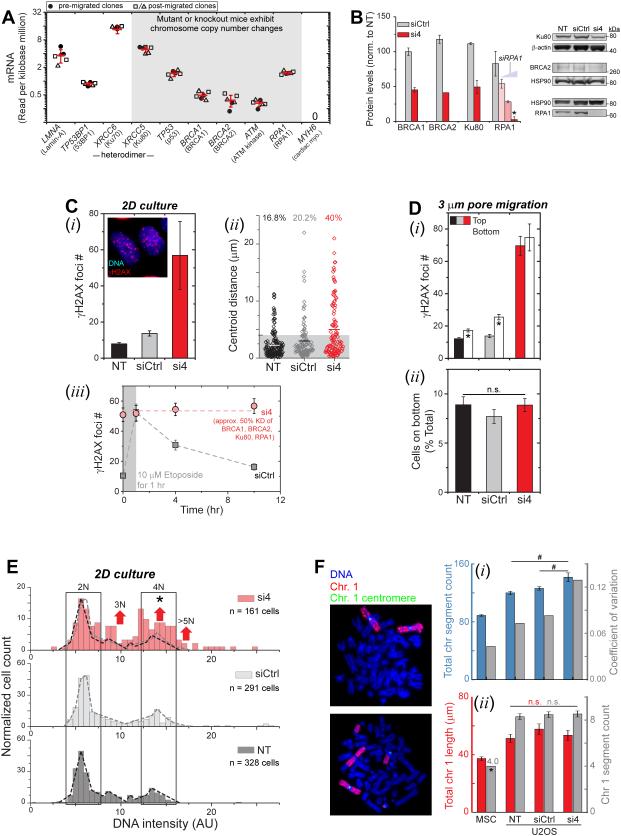
Figure 3. Partial depletion of multiple repair factors leads to DNA damage accumulation and chromosomal variation.
(A) Key nuclear or DNA repair transcripts from RNA-seq analysis of U2OS cultures, either pre- or post-migration. Mouse knockouts/mutations of key genes cause copy number variations. MYH6 (a cardiac specific gene) shows zero reads. (Normalization to Reads per kilobase million, n=8 samples).
(B) Simultaneous partial knockdown of four repair factors (si4, red) versus control (siCtrl, gray) when normalized to non-treated (NT). 25 nM of each siRNA (red) was used except for siRPA1 titration (Figure S2). Protein levels quantified by immunoblots were normalized by housekeepers (HSP90, beta-actin), with BRCA1 quantified by immunofluorescence. (n=3 blots or ≥150 nuclei, n=3 expts, *p<0.05).
(C) DNA damage increased in 2D cultures of si4 cells based on γH2AX foci (i, Figure S2) and comet assay (ii). siCtrl slightly increases damage, perhaps because of lipofectamine. DNA damage sites induced by 1 hour 10μM etoposide treatment are resolved by the siCtrl cells within hours, but γH2AX foci count of si4 cells remained at a high number (iii, ≥110 nuclei per condition, n≥3 expts, *p<0.05 vs. NT, #p<0.001 vs siCtrl).
(D) si4 increases γH2AX foci regardless of 3-μm pores migration, while damage increases in controls (i, ≥ 50 nuclei per condition, n≥3 expts, *p<0.05 between top and bottom). Migrated %-ages are similar for all conditions (ii, Figure S2; ≥3 transwell per condition, n≥3 expts).
(E, F) DNA content from Hoechst-33342 intensity shows 2N and minor 4N peaks for controls but shifts for si4 indicate poly- or aneu-ploidy (Figure S2, ≥160 nuclei per condition, n≥3 expts; *p<0.001 in two-sample Kolmogorov-Smirnov (KS) test for higher modes, with α=0.05 [50]). Metaphase spreads show higher ploidy of U2OS cells versus diploid MSCs, and more chromosome segments for si4 cells (i). Both Chr-1 length and segment counts of U2OS are higher versus MSCs, but (ii) Chr-1 counts are unaffected by si4 (≥20 spreads per condition, n≥3 expts, *p<0.05 MSC versus U2OS, #p<0.05 U2OS versus U2OS).
Depletion of a subset of repair factors favors DNA damage accumulation but not death
To begin to assess possible effects of partial loss of repair factors from U2OS nuclei, we partially knocked down BRCA1, BRCA2, Ku80, and RPA1 (Figure 3B, S2). DNA damage increases with knockdown of individual factors or the combination (denoted si4) as shown by both γH2AX foci and by comet assay (Figure 3Ci,ii, S2), with foci again seen throughout the nucleus (Figure 3C-i). Knockdown of BRCA2 notably increases γH2AX foci (Figure S2), despite BRCA2 being functionally low in U2OS cells [15]. The si4 is intended to approximate nuclear depletion of multiple factors after constricted migration (Figure 2A,B,S1), and the increased DNA damage is consistent with past studies of individual factors in other cells [27-30]. Control transfections with siCtrl/lipofectamine also increase DNA damage (by ~40% when averaged across all control assays) and might reflect cell stress in transfection [31]. Etoposide-induced γH2AX foci in U2OS cultures require many hours to resolve [8] and remain high with si4 (Figure 3Ciii). Migration-induced mis-localization of multiple repair factors for many hours (Figure 2A-C, S1) could thus delay repair so that damage accumulates (Figure 1C-F) – as seen again with siCtrl cells (bottom vs. top, Figure 3Di). Additivity and linearity of the various knockdowns is suggested (Figure S2) but requires further study. Regardless, si4 knockdown cells on bottom show the same high number of γH2AX foci as non-migrated cells on top (Figure 3Di), and because cell death is also elevated with si4 (unlike control cells, Figure S2), the high number of foci might reflect maximum levels before γH2AX fills the nucleus and precipitates cell death.
A high level of DNA damage on top with si4 (or etoposide) does not affect the fraction of cells that migrate through a pore (Figure 3Dii or Figure S2), and the fraction of cells that survive such migration can also be reduced pharmacologically without affecting DNA damage (Figure S2). This includes careful titrations with ATMi, an inihibitor of ATM kinase that exerts effects independent of DNA repair [32]. However, DNA staining analyses in interphase cells does show that si4 causes an increase in DNA levels per cell (Figure 3E, S2), and DNA content in some si4 cells also exhibits odd ploidy and can exceed 4N. Such differences begin to suggest changes in chromosome copy numbers that could be imbalanced (as aneuploidy).
For high resolution microscopy of chromosome ploidy in a population of U2OS cells that were genomically homogeneous to start with, U2OS clones were generated by expansion of single cells pipetted into 96-well plates, and one clone was chosen (Ctrl_clone-1, Table S1) for si4 knockdown. Genomic homogeneity of 95.2% for this clone was determined from comparative genome hybridization arrays (aCGH, performed by Cell Line Genetics, Madison, WI) which remains a ‘gold standard’ for chromosome copy number variations [33]. Imaging of metaphase chromosomes shows si4 treatment gives more chromosomes (15-20 more) versus siCtrl and non-treated cells (Figure 3F-i). The highest variation is with si4 treatment and is 3-fold greater than normal diploid MSCs, suggesting high genome variation and diverse aneuploidy within si4 cultures. All U2OS cells also exhibit much higher ploidy than MSCs, which is likewise evident in genomic analyses by aCGH (i.e. 41% more; Table S1). Fluorescence in situ hybridization (FISH) applied to Chr-1 (the longest is best for microscopy) shows the expected gains in chromosome copy number: comparing U2OS to MSCs (Figure 3F-ii), Chr-1 segments are shorter and/or fused to other chromosomes but greater in total number (~8 vs 4) and length (46% more). However, the lack of difference for Chr-1 between U2OS samples suggests that other chromosomes confer the higher ploidy of si4 cells, consistent with aneupoloidy caused by depletion of repair factors.
Changes in chromosome copy number after constricted migration
Chromosome copy number changes after constricted migration of U2OS cells seemed possible because: (i) nuclear-to-cytoplasmic ratios of DNA repair factors are greatly decreased by migration (Figure 2A-C), and (ii) some of these factors when knocked out or mutated in mice cause changes in chromosome copy number [22-26]. In addition, we find (iii) partial, combined knockdown of multiple repair factors affects ploidy (Figure 3E,F) and increases DNA damage (Figure 3C,D). The genomes of U2OS cells were therefore analyzed before and after constricted migration, and we again used comparative genome hybridization arrays, aCGH (with clonality per Table S1). Single nucleotide polymorphism arrays (SNPa) and whole exome sequencing (WES) were also used and can further reveal chromosome copy number changes that produce loss of heterozygosity (LOH). RNA-sequencing (RNA-Seq) was used to correlate transcript levels and genomic changes.
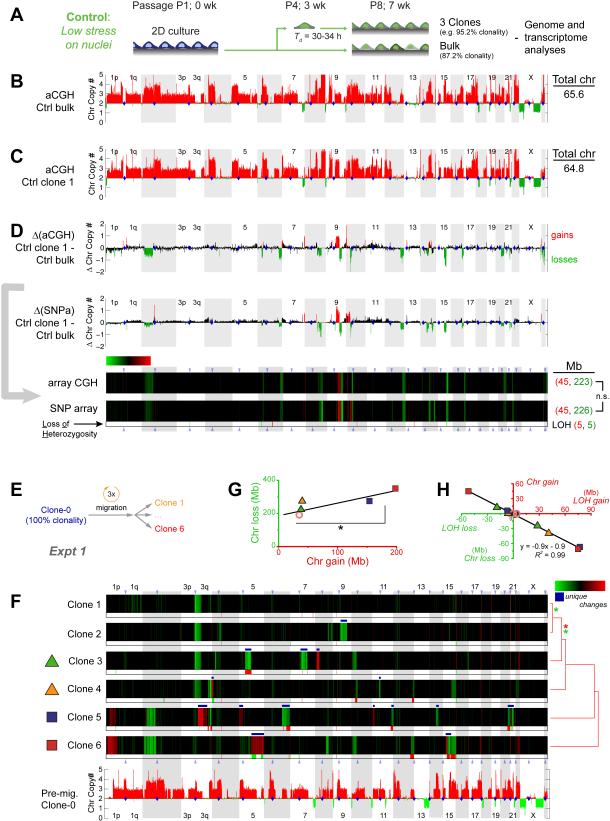
U2OS clones generated in standard 2D cultures done in parallel with a typical long-term, bulk culture of U2OS cells (Figure 4A) show heterogeneity in the latter (i.e. 87% clonality, Table S1). This is consistent with minimal genomic drift in long-term culture and migration on plastic [34]. Clonal expansion not only produces sufficient DNA for accurate genomic analyses but also provides evidence of viable, proliferating cancer cells—typical of malignancy—rather than non-viable or senescent cells with excessive DNA damage that might be analyzed by single cell methods. Importantly, expansion from single U2OS cells can maintain high clonality for months in standard 2D cultures (>95%; Table S1). Furthermore, as expected from the high ploidy of U2OS cells (Figure 3F), aCGH and SNPa as well as whole exome sequencing (WES) consistently yield similar chromosome copy number patterns across the genome of a given clone (Figure 4B,C,S3), revealing 3-4 copies of many chromosomes (or parts of chromosomes).
Figure 4. Constricted migration introduces genomic variation.
(A) U2OS culture with 87.2% clonality (Table S1,S6) diluted to single cells in 96-well plates to generate control clones. Genomic analyses included comparative genome hybridization arrays (aCGH) and single nucleotide polymorphism arrays (SNPa).
(B, C) Chromosome copy numbers from aCGH for bulk U2OS (Ctrl_bulk) and one clone (Ctrl_clone-1, Figure S3). Total chromosome numbers are calculated per Table S2.
(D) Differences in chromosome copy numbers between Ctrl_bulk and Ctrl_clone-1 are heat-mapped as changes in chromosome copy numbers (ΔChr copy#). Gain (red) and loss (green) calls in aCGH data were thresholded at ±0.48 based on ΔChr copy# distributions from all pairwise comparisons in Table S1. Ctrl clone 1 shows more net losses and fewer chromosomes. Gain and loss calls for this SNPa comparison were thresholded at +0.6 and -0.2 to match gains and losses of aCG, and KS tests (Figure 3E) show no difference between gain distributions and between loss distributions. SNPa also shows few LOH’s (below heatmap).
(E, F) 100% clonal U2OS after 3 migrations through 3-μm pores was used to make six single cell clones expanded and analyzed by SNPa’s. Compared to pre-migration clone-0, migration causes unique ΔChr copy# (white *) and LOH’s. Clones are listed per hierarchical clustering (city-block metric) of their ΔChr copy#, and asterisks indicate statistical significance (p<0.05 in KS tests) between the distributions of gains (red *) or distributions of losses (green *). Gain and loss calls were thresholded at ±0.42 based on aCGH versus SNPa (see Methods).
(G, H) Chromosome gains and losses reach 100’s of Mb, and two clones (5, 6) show the highest gains (*p<0.05). LOH’s anti-correlate with chromosome copy number changes.
Changes in chromosome copy number between any two samples were calculated as illustrated for Ctrl bulk and Ctrl clone 1 (Figure 4D). Heatmaps for easier visualization of the genome show gains in red, losses in green, and no change in black. Two different arrays (aCGH and SNPa) from different manufacturers have different probes, different standards, and different genome coverage, but the heatmap from the ‘gold standard’ aCGH [33] is largely the same as that from SNPa’s (Figure 4D). SNPa’s yield additional, highly accurate information on loss of heterozygosity (LOH: which indicates complete loss of either ‘mother’ or ‘father’ derived alleles), and the relatively rare LOHs are summarized by red or green tick marks against a light background beneath chromosomal heatmaps (Figure 4D). LOH differences are low (~10 megabases, Mb) for control cells that proliferate on plastic for weeks.
A clone that is genomically 100% homogenous based on aCGH was subjected to three rounds of constricted migration through transwells (TW3), and for genomic analyses single cell clones were once again isolated and expanded from the migrated population by 96-well serial dilution (Figure 4E). Three rounds of 3-μ m migration were chosen because a single round generated minimal genome variation (Figure S3). Subtracting the starting clone’s genome from genomes of the triply migrated TW3 clones reveals unique gains and/or losses in parts of many chromosomes (Figure 4F white *’s and blue bars). Changes in chromosome copy number can total 100’s of Mb’s (clones 5 and 6), which differ significantly from less affected clones (Figure 4G).
A subset of the chromosome changes in clones 3 to 6 also involved the gain and/or loss of LOH regions (Figure 4F). The significant anti-correlation of LOH with changes in chromosome copy number (Figure 4H) indeed confirms that a gain/loss of LOH associates tightly with a loss/gain within a chromosome. LOH gains exceed LOH losses for a given sample (also in data of independent experiments below), and the low LOH losses could reflect either a low frequency process of DNA mis-repair or else measurement uncertainties (see below). Constricted migration of a 100% clonal population can thus give rise to genomic variation even though mechanisms require further study.
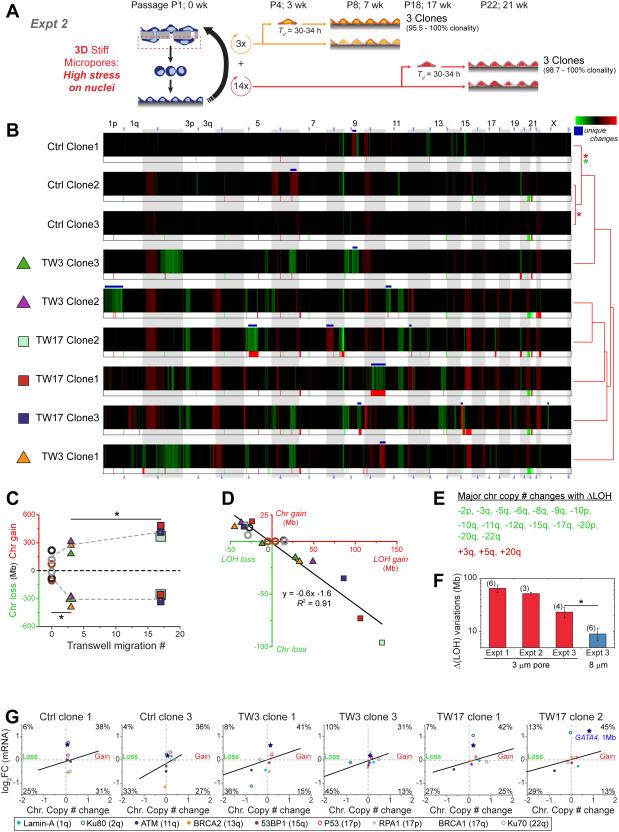
A standard 2D bulk U2OS culture (from Figure 4A) was also subjected to pore migration, with distinct single cell clones isolated after three rounds of migration (TW3) and seventeen rounds (TW17) (Figure 5A). Starting with a non-clonal ‘bulk’ culture (87% clonal) is important to assessing whether statistically significant genome heterogeneity can be added to a standard cell culture, which seems typical of tumors that become multi-clonal (e.g. osteosarcoma) [15]. In order to control for passage number, transwell migrations were done in parallel to the 2D culture: clonal selection (in 96-well plates) and DNA isolation for TW3 and non-migrated controls were thus done at the same time. Subtraction of pre-migration ‘bulk’ (Ctrl_bulk of Figure 4B) to generate heatmaps (per Figure 4D) reveals partial gains and losses in chromosome copy numbers (Figure 5B), and while the control clones differ the least in clustering together, all clones exhibit uniqueness. TW3 clone 2 shows a unique loss of 1 copy of Chr-1p from 3-4 copies of this arm in Ctrl_bulk (or any control clones, Figure 4B). TW17_clone-1 shows a unique loss of Chr-10p plus half of -10q from 3-4 copies in control clones. Whole exome sequencing (WES) of key samples shows the same trends (Figure S4).
Figure 5. More migration causes more genomic variation, with changes evident in expression.
(A) Migration of ‘bulk’ U2OS for three (TW3) or seventeen (TW17) rounds was followed by clonal expansion (Table S1), SNPa, and RNA-seq analyses. To control for genomic variation in culture, TW3 and Ctrl clones were passaged in parallel.
(B) Relative to pre-migration ‘bulk’, ΔChr copy# shows Ctrl clones cluster together versus migrated clones, which all show many more ΔChr copy#, with many unique changes (white *). ΔLOH regions were greatest in TW17 clones. Clones are listed per hierarchical clustering (city-block metric) of their ΔChr copy#, and asterisks indicate significance (p<0.05 in KS tests) between the distributions of gains (red *) or distributions of losses (green *). Gain and loss calls for these samples were thresholded per Figure 4F. Table S3 compares samples to Control_clone-1 and leads to similar conclusions of genome variation.
(C, D) ΔChr copy# reached 100’s Mb and increased with rounds of migration (*p<0.05, student t-test), and LOH’s and ΔChr copy# anti-correlate per Figure 4H.
(E) Major ΔChr copy# (loss green; gains red) that show changes in LOH.
(F) ΔLOH variations within samples of independent TW3 experiments were derived from pairwise comparisons of changes in SNV-confirmed LOH’s, with means calculated for all pairs. Experiment-1 is from Figure 4E-H migration of a clone; experiment-2 s from Figure 5A-D migration of bulk; and experiment-3 involves four more 3-μm TW3 clones and six 8-μm TW3 clones migrating from a bulk culture (Figure S3). Even though ΔLOH variations were observed within all groups, consistent with heterogeneity, variation for 3-μm pores always exceeds that for 8-μm pores (*p<0.05).
(G) Transcript changes (RNA-Seq) correlate with ΔChr copy# from SNP-arrays (1 Mb averaging for both datasets, Table S3). Data within the white oval have log2(RNA_ratio) and ΔChr copy#<0.5. For significance tests, the thresholded percentage of data in (upper-right + lower-left) versus (lower-right + upper-left) was calculated for all plots: p=5×10-6 (Table. S4,S5). Genomic heatmaps of RNA-seq data, aligned with the ΔChr copy# (Figure S4). Blue star indicates 1 Mb window containing GATA4, and colored circles indicate 1Mb window containing nuclear factors of Figure 3A.
Quantitation of chromosome copy number from the heatmaps indicates statistically greater loss of 100’s of Mb’s for TW3s on average compared to control clones (Figure 5C). TW17s show more gains on average relative to TW3s and control clones (Figure 5C). LOH changes are again a small subset of anti-correlated changes in chromosome copy number (Figure 5D), and arise again from losses in many more chromosomes compared to gains (Figure 5E), suggestive of typical LOH mechanisms [35]. Changes in single nucleotide variations (SNVs) largely overlap with changes in LOH (Figure S4), and compared to control clones the TW17 clones show many more SNVs (Figure S3). Pairwise analyses of SNVs in additional control clones further show minimal variation, as expected of clonality (Figure S3), while variation increases with rounds of migration to the highest levels between TW17 clones. Clones thus diverge genomically with migration.
Changes in LOHs of each chromosome were also checked as SNVs (Figure S3): LOH gains mostly coincide with SNV calls, but LOH losses do not. For a high confidence measure of heterogeneity of TW3s generated in the two experiments (i.e. six clones in Figure 4E-H from one clone, and three clones in Figure 5A-E from bulk), pairwise differences were determined for SNV-confirmed LOHs (as total Mb’s = mostly gains + a few losses), and the mean ΔLOH was calculated across all pairs. The two experiments above plus a third quantify ΔLOH variations (Figure 5F), which reflect both chromosome copy number changes and SNV coincidence. The third experiment also included TW3 clones generated by migration through larger 8-μ m pores. Although large pores cause minimal nuclear blebbing and DNA damage (Figure 1), ATMi toxicity in migration clearly suggests some pore-induced stresses on the cells (Figure S2). LOH heterogeneity is nonetheless greatest for 3-μ m pores.
RNA-Seq analyses of many samples above was once again compared to Ctrl_clone_1. Zero reads of mRNA from the Y chromosome (Table S2), in addition to zero LOH and SNV calls, are consistent with derivation of the U2OS line from a female patient; in contrast, 40-57% of genes from all other chromosomes are expressed. Fold changes in transcript (1 Mb averaged) from this fourth ‘omic method were mapped to changes in chromosome copy number (Figure 5G,S4). Most transcript changes correlated with changes in gene copy number, with high statistical significance for the dataset (p<<0.01) (Figure 5G, Table S3), per expected “gene dosage” effects [36].
For DNA repair factors that can cause variations when mutated (Figure 3A), TW3_clone-3 shows no mRNA deficiencies, and so its genome variations are not explained by defects in these repair factors. ATM mRNA is low in three migrated clones, but ATM does not affect DNA damage in U2OS cells (Figure S2). However, TW3_clone-1 shows a 2-fold decrease in Ku80 that could suffice to increase DNA damage in this one clone (Figure S2). A 2-fold decrease in BRCA2 in Ctrl clone 3 is perhaps consistent with past reports for U2OS [15], and a lack of correlation with chromosome copy number merely illustrates alternative, epigenetic regulation [37]. Because low expression of key repair factors is evidently rare, transient depletion of protein after nuclear rupture remains a better explanation for DNA damage in constricted migration (Figure 1).
Phenotypic changes reflect genomic changes
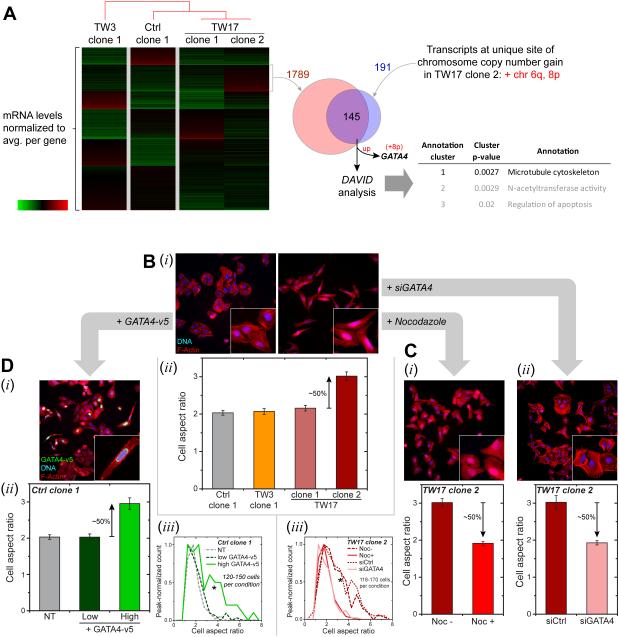
Proliferation rates of all clones generated after migration appeared similar (Figure S5), which is sensible because constricted migration seems unlikely to select for proliferation. However, while imaging cells for cell counts of various clones, TW17_clone-2 appeared the most spindle shaped. A k-means clustering analysis of mRNA levels showed 1789 genes were uniquely up in this clone (Figure 6A). Chromosome maps (Figure 5B) also show this clone has unique gains in Chr-6q and -8p, corresponding to 191 genes for which we have RNA-seq results: 76% of these transcripts (145 genes) are up per gene dosage effects (Figure 5G). Functional analyses of the many transcript changes (by DAVID Bioinformatics 6.7 [38]) reveals enrichment for the microtubule (MT) cytoskeletal system only in TW17_clone-2.
Figure 6. Phenotypic changes driven by migration-induced genome variations.
(A) mRNA from four clones (Ctrl_clone-1, TW3_clone-1, TW17_clone-1 and -2) were normalized by the average value for each gene. K-means clustering applied to the transcriptome data revealed a transcript cluster uniquely up-regulated in TW17_clone-2 (1789 genes). Clones are listed per hierarchical clustering (city-block metric) of transcript levels. Only two chromosome gain regions are unique to TW17_clone-2: Chr 6q and 8p. These unique regions correspond to 191 transcripts in RNA-seq data, with 145 genes upregulated. Functional annotation analysis (DAVID Bioinformatics v.6.7) reveals enrichment of MT cytoskeleton, suggestive of MT upregulation in TW17_clone-2. GATA4 also resides in Chr-8p and is in the overlap list. Similar comparisons with all samples in Figure 5 yield Chr-8p as the only unique chromosome gain region and gives 107 overlapping genes, with MT cytoskeleton statistically enriched and GATA4 listed.
(B) Cell aspect ratio from F-actin staining by phalloidin (i) calculated from major over minor axes (ii). Higher aspect ratios were observed only for TW17_clone-2, indicating more elongated cells (Figure S5, ≥150 cells per condition, n≥ 3expts, *p<0.05).
(C) De-polymerization of MT’s with 10 μM Nocodazole and GATA4 depletion by siGATA4 (Figure S5) on TW17_clone-2 leads to more rounded cells with decreased aspect ratio (i,ii) and shifts in aspect ratio distributions (iii) (≥140 cells per condition, n≥3 expts, KS tests: *p<0.05).
(D) Overexpression of GATA4-v5 in Ctrl_clone-1 elongates cells (i) per higher aspect ratio (ii, Figure S5), shifting the aspect ratio distribution (iii, ≥120 cells per condition, n≥3 expts, KS tests: *p<0.05). Anti-v5 identifies expressing cells.
MT organization has long been associated with cell morphology and polarity [39], and MTs span the long axis of prototypical spindle shaped MSCs (Figure S5). The elongated aspect ratio of most TW17_clone-2 cells propagates with further cloning (Figure 6B, S5). Elongated cells with aspect ratio >5 are extremely rare in cultures of the other clones (1 ± 1%), and given that we randomly chose only three TW17 clones, selection of such rare elongated clones is unlikely (p<0.05). High clonality (Table S1) also makes selection unlikely. An elongated shape is thus a likely consequence of genomic changes caused by migration. Importantly, Nocodazole induced depolymerization of MTs in TW17_clone-2 decreases cell aspect ratios to a level similar to rounded clones (Figure 6Ci), confirming a key role of MT assembly in an elongated shape.
In scanning the 145 genes on Chr-6 or -8 that are uniquely up-regulated in TW17_clone-2, we noted a ~2-fold upregulation of GATA4 (Figure 6A), which is a transcription factor that drives an endothelial-to-mesenchymal transition (EMT) process in cardiogenesis where – more specifically –GATA4 repression decreases MT-associated proteins [40]. Cardiac genes such as cardiac myosin are not expressed in U2OS cells (Figure 3A), but GATA4 maintains osteoblast progenitors [41], which often originate from MSCs [42]. GATA4 inhibits osteogenic differentiation [43] and is relevant to the osteosarcoma origin of U2OS cells.
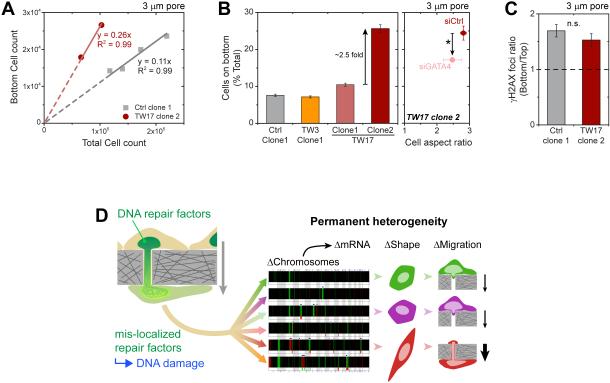
Averaging all genes within 1 Mb of GATA4 confirmed its significant upregulation only in TW17_clone-2 (Figure 5G). If high levels of GATA4 in this clone (Figure 6A) suffice to drive the spindle shape, then (i) GATA4 knockdown in TW17_clone-2 (Figure S5) should decrease the aspect ratio, and (ii) GATA4 overexpression in a rounded clone (Figure S5) should drive elongation. These predictions hold true (Figure 6Cii, iii) and even yield the expected shape distributions (Figure 6D). Spindle shaped U2OS cells show MT’s from end-to-end, consistent with MSCs and with morphological roles (Figure S5). The elongated clone also migrates more efficiently through pores unless treated with siGATA4 (Figure 7A,B), although migration-induced DNA damage still occurs (Figure 7C).
Figure 7. Clone with elongated morphology migrates more rapidly, but migration still increases DNA damage.
(A-B) TW17_clone-2 squeeze through 3-μm pores in greater numbers than other clones, with a greater proportion of cells on the bottom (≥3 transwells per condition, n≥3 expts, *p<0.05). siGATA4 treatment leads to lower number of migrated cells. Cell aspect ratio of the migrated siGATA4 cells tend to be lower, with a wider variance (≥125 cells per conditions, n≥3 expts, *p<0.05).
(C) TW17_clone-2 is still prone to migration-induced DNA damage (≥50 cells per condition, n≥3 expts).
(D) Overall, cell migration through micron-size constrictions causes transient mis-localization of DNA repair factors and thereby causes DNA damage, which leads to permanent heterogeneity in chromosome copy numbers, expression levels, cell shape, and migration capability.
Discussion
Constricted migration increases the number of DNA damage foci (based on three independent approaches) and depletes DNA repair factors for hours (Figure 1,2). Partial knockdown of such factors in 2D cultures delays DNA repair by at least hours and increases both damage and chromosome aberrations (Figure 3). Damage foci are nucleoplasmic (Figure S1), consistent with repair factor loss but contrasting with GFP-53BP1 foci at the distal end of a migrating nucleus [3] and with DNA damage just at the leading edge of the nucleus [7]. Chromatin fragmentation as a nucleus enters and elongates in a small pore was also considered but seems unlikely because stretched chromatin maintains its integrity even if cleaved by a nuclease [44].
Constricted migration of U2OS clones additionally causes many unique gains and losses in large segments of chromosomes in diverse clones (Figure 4,5), consistent with copy number variations in mouse mutants of some DNA repair factors [22-26]. While clonality of U2OS cells can be reasonably maintained in 2D culture (Table S1), constricted migration could simply be amplifying deficient repair pathway(s) that lead to the multi-clonality in osteosarcoma tumors [13, 14]. Nuclear entry of DNA damage factors has also been speculated [3, 7], but evidence in osteosarcomas is lacking. Moreover, the pan-nucleoplasmic distribution of DNA damage foci (Figure 1) would require entry of loci-specific, genome-spanning nucleases and simultaneous exclusion of non-specific nucleases, which would tend to damage DNA primarily near the site of envelope rupture.
Mechanisms of migration-induced increases in DNA damage based on loss of repair factor(s) are both similar to and different from mechanisms elaborated for lamin-A null fibroblasts in standard cultures [6]. Repetitive rupture of the nuclear envelope in such cultures on rigid substrates causes cytoplasmic mis-localization of multiple mobile nuclear factors [45] (which is mimimized on soft, tissue-like substrates that reduce nuclear stress [46]); 53BP1 protein is thus expected to be more cytoplasmic, which could explain its rapid degradation in lamin-A null fibroblasts in the absence of any transcript change [6]. Importantly, overexpression of 53BP1 has proven sufficient to rescue DNA damage in lamin-A low cells [6]. Loss of 53BP1 likewise occurs early across many human cancers of different tissue and cell types, but it occurs more consistently than appearance of γH2AX [47], which suggests decoupling from 53BP1. Indeed, after constricted migration of U2OS cancer cells that have abundant lamin-A (Figure 1) and pre-existing cancerous changes (Figure 3), overexpression of 53BP1 is not sufficient to reproducibly provide a significant rescue (Figure S1). Mis-localization after migration is, however, transient for repair factors (Figure 1,2) that are known to regulate chromosome copy number variations – including BRCA's that explain genomic aberrations in osteosarcoma [15]. The fact that lamin-A deficiencies and mutations do not associate closely with carcinogenesis further implicates other important mechanisms unrelated to rupture, such as squeezing-dependent segregation of repair factors away from chromatin within an intact nucleus [48]. Lastly, while mechanistic links between our main observations of DNA damage, repair factor depletion, and genome variation might benefit from more direct comparisons with the effects of lamin-A depletion, the EMT-like change in cancer cell phenotype after migration (Figures 6,7) illustrates invasion-mutation mechanisms pertinent to metastasis and intra/inter-tumor heterogeneity [49].
Experimental Procedures
Cell culture
U2OS, osteosarcoma cell line, and A549, human lung adenocarcinoma cell line, were cultured in DMEM high glucose media and Ham’s F12 nutrient mixture (Gibco, Life Technologies), respectively, supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma-Aldrich). MSCs were cultured as described previously [2].
Transwell migration
For migration through transwells (Corning Inc.), cells were seeded at 300,000 cells/cm2 onto the top side of the filter membrane and left to migrate in normal culture condition for 24 hours. The number of migrated cells on bottom are proportional to the cells added on top in a given set of experiments, and so different experiments are readily compared by normalizing to a control sample such as non-treated. In order to isolate the cells from the transwell, cells were detached from the transwell by using 0.05% Trypsin-EDTA (Gibco, Life Technologies). If the isolated cells were to undergo another transwell migration, they were expanded for a week to reach the required number of cells. Alkaline comet assays of the migrated cells were carried out per manufacturer’s instruction (Cell Biolabs). Image processing to determine the centroid of main nuclear body and its comet tail was done in MATLAB. Intensity thresholding was used to locate the comet area, while distribution of the intensity derivatives was used locate the main nuclear body area. The centroids were calculated from the area locations.
Genome and Transcriptome analysis
DNA isolation used the Blood & Cell Culture DNA Mini Kit (Qiagen) per manufacturer’s instruction. Chromosome copy number was measured using array comparative genomic hybridization (aCGH) SurePrint G3 Human Genome CGH+SNP Microarray 4x180k (Agilent), which involves ~110,000 probes for CGH and ~60,000 probes for SNP detection. Isolated DNA samples were shipped to Cell Line Genetics, WI, for aCGH measurements. Cell Line Genetics used standard CytoGenetics (Agilent) to provide a summary analysis and raw data for each sample: see Table S4 for a representative summary report and raw data table from aCGH (data files are large but all raw data is available for review at any time). In addition to the sample clonality (as indicated by “clonal fraction” in Table S4), raw data indicates regions of both chromosome copy number and LOH variation. Further analyses were done with custom algorithms written in MATLAB (MathWorks). In order to validate aCGH results, the same DNA samples were also sent to The Center for Applied Genomics Core in The Children’s Hospital of Philadelphia, PA, for Single Nucleotide Polymorphism array (SNPa) HumanOmniExpress-24 BeadChip Kit (Illumina). For this array, >700,000 probes have an average inter-probe distance of ~4kb along the entire genome. For each sample, the Genomics Core provided the data in the form of GenomeStudio files (Illumina). Chromosome copy number and LOH regions were analyzed in GenomeStudio by using cnvPartition plug-in (Illumina). Regions with one chromosome copy number are not associated with LOH by the Illumina’s algorithm. Hence, regions with one chromosome copy number as given by the GenomeStudio are added to the LOH region lists. Comparison analyses between SNPa and aCGH were again done in MATLAB. SNP array experiments also provide genotype data, which was used to give Single Nucleotide Variation (SNV) data. Genotyping in this Illumina system rely on the correlation between total intensity and intensity ratio of the two probes, one for CG and another for AT. These correlations were mapped to a standard clustering file (Illumina) to give the SNP calls. In order to compare different samples, probes with ‘no-call’ (either due to low read intensity or located outside the ‘call’ cluster) were removed from further analysis. In order to increase the confidence of LOH data given by the GenomeStudio, the changes in LOH of each chromosome from each sample were cross referenced to their corresponding SNV data.
The isolated DNA samples were sent to the Next-Generation Sequencing Core at the Perelman School of Medicine, University of Pennsylvania, PA, for exons capture by using SureSelect Clinical Research Exome kit (Agilent), per manufacturer’s standard protocol. 3 samples were pooled together and submitted to HiSeq 2500 (Illumina) for 100 bp paired-end sequencing, resulting in ~80,000,000 reads for each sample. Chromosome copy number analysis was done by using CNVkit software package (https://media.readthedocs.org/pdf/cnvkit/latest/cnvkit.pdf).
RNA isolation used RNeasy plus Mini Kit (Qiagen). For RNA-seq analyses, RNA samples were also sent to the Next-Generation Sequencing Core. Libraries for RNA-seq were made by using TruSeq Stranded mRNA Library Prep kit (Illumina) per manufacturer’s instruction, followed by 100 bp paired-end sequencing with HiSeq 2500. 10 cDNA libraries were pooled together, resulting in ~16,000,000 reads for each sample. Reads per kilobase million for each gene were calculated by normalizing the read of each gene by the sample’s total read count (in million) and by the gene length (in kilobase). Data processing and clustering were done in MATLAB and function annotation analysis were done with DAVID Bioinformatics 6.7 [38].
Immunostaining and imaging
Transwell membrane was fixed in 4% formaldehyde (Sigma) for 15 minutes, followed by permeabilization by 0.25% Triton-X (Sigma) for 10 minutes, blocked by 5% BSA (Sigma) and overnight incubation in various primary antibodies: lamin-A/C (Santa Cruz and Cell Signaling), Lamin-B (Santa Cruz), γH2AX (Millipore), Ku80 (Cell Signaling), BRCA1 (Santa Cruz), BRCA2 (Milipore), RPA1 (Santa Cruz), phosphorylated S1981 ATM (Abcam), α/β tubulin (Cell Signaling) and v5 (Abcam). Finally, the primary antibodies were tagged with the corresponding secondary antibodies for 1.5 hours (ThermoFisher). For F-actin staining, 100 ng/mL TRITC-phalloidin (Sigma-Aldrich) was also added to the secondary antibody solution. DNA was stained with 8μM Hoechst 33342 (ThermoFisher) for 15 minutes. When mounting is involved, Prolong Gold antifade reagent was used (Invitrogen, Life Technologies). Epifluorescence imaging was performed using Olympus IX71, with a digital EMCCD camera (Cascade 512B, Photometrics) and a 40x/0.6 NA objective. Confocal imaging was done in Leica TCS SP8 system, by either 63x/1.4 NA oil-immersion or 40x/1.2 NA water-immersion objectives. Various image quantification and processing were done with either ImageJ or MATLAB.
Protein modulation in U2OS cells
siRNAs used in the main study were purchased from Dharmacon (ON-TARGET plus Smart pool siBRCA1, L-003461-00, siBRCA2, L-003462-00, siKu80, L-010491-00, siRPA1, L-015749-01 and non-targeting siRNA, D-001810-10). For specificity validation (Figure S2), additional siRNAs were purchased from ThermoFisher (Silencer Select siBRCA2 s2085, siKu80 s14953, siKu80 s14954 and siRPA1 s12127), and siBRCA1 (5'-AGAUAGUUCUACCAGUAAA-3’). U2OS cells were passaged 24 hours prior to transfection. Pooled siRNA oligos (25nM; 4 siRNAs/target) and 1 μ g/mL Lipofectamine 2000 were prepared according to the manufacturer’s instructions and incubated for 24 hours in high glucose DMEM with 10% FBS. Knockdown efficiency was determined by Western blot following standard methods. After 3 days of treatment with the various siRNA’s, all showed pan-nucleoplasmic foci of γH2AX and only siRPA1 also caused global foci of γH2AX, but this was evident in only a minor fraction of cells (<20%) even after ~50% knockdown.
In order to establish an inducible GATA4-v5 line, U2OS Ctrl clone 1 cells were plated onto 24 well plates and cultured for 24 hours. On the following day, cells were incubated with 100 μ L (≥2.5×105 IFU) of rtTA lentivirus and 100 μL (≥2.5×105 IFU) of tetO-GATA4-V5 lentivirus (provided with methods by the Dr. John D. Gearhart, University of Pennsylvania), 8 μg/mL polybrene (Milipore) and 200 μ L of full culture media for 2 days. After viral transduction, cells were re-plated and on the next day, GATA4-v5 expression was induced by culturing the cells in 2 μ g/mL doxycycline for at least 5 days. In order to de-polymerize the microtubule, TW17 clone 2 cells were treated with 100 ng/mL Nocodazole (Sigma) for 12 hours.
Fluorescence in situ hybridization (FISH)
U2OS and iPS-MSC cells were fixed by methanol and mounted onto glass slides, chromosome 1 was labeled with an orange whole chromosome painting probe (Creative Bioarray); its centromere was simultaneously labeled with a green VividFISH chromosome enumeration probe (GeneCopoeia). Cell fixation, slide preparation, and probe hybridization were performed according to manufacturer protocol. Post-hybridization, the slides were mounted with AntiFade-DAPI (GeneCopoeia). The labeled, DAPI-stained cells were then imaged using a 150x/1.5 NA oil-immersion objective.
Supplementary Material
Highlights.
Constricted migration causes mis-localization of DNA repair proteins and DNA breaks
Depletion of repair factors leads to DNA damage and chromosomal aberrations
Migration of cancer clones through small pores causes lasting genomic heterogeneity
Gene dosage effects in the transcriptome can perturb cell shape and motility
Acknowledgment
The authors thank John D. Gearhart (University of Pennsylvania) for the tetO-GATA4-v5 lentiviral system. The authors in this study were supported by the National Cancer Institute of the National Institutes of Health under PSOC Award Number U54 CA193417. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
eTOC blurb
Irianto et al. demonstrate that cell migration through micron-size constrictions leads to transient DNA damage and cytoplasmic mis-localization of multiple DNA repair factors, with lasting genomic heterogeneity that translate to phenotypic changes. Migration-induced genomic instability can thus associate with heritable changes.
Author Contributions
J.I., Y.X, C.P., A.A. designed and performed experiments. J.J., C.A., M.T. performed experiments. S.H., R.B., A.L., R.G., D.D. analyzed results. J.I. and D.D. wrote the manuscript.
References
- 1.Chamberlain JK, Lichtman MA. Marrow cell egress: specificity of the site of penetration into the sinus. Blood. 1978;52:959–968. [PubMed] [Google Scholar]
- 2.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 4.Irianto J, Pfeifer CR, Xia Y, Athirasala A, Ivanovska IL, Greenberg RE, Discher DE. Constricted cell migration causes nuclear lamina damage, DNA breaks, and squeeze-out of repair factors. bioRxiv. 2015 [Google Scholar]
- 5.Ogawa H, Inouye S, Tsuji FI, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalo S. DNA damage and lamins. Adv. Exp. Med. Biol. 2014;773:377–399. doi: 10.1007/978-1-4899-8032-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene. 2010;29:2281–2291. doi: 10.1038/onc.2009.501. [DOI] [PubMed] [Google Scholar]
- 9.Harding SM, Bristow RG. Discordance between phosphorylation and recruitment of 53BP1 in response to DNA double-strand breaks. Cell Cycle. 2012;11:1432–1444. doi: 10.4161/cc.19824. [DOI] [PubMed] [Google Scholar]
- 10.Alagoz M, Katsuki Y, Ogiwara H, Ogi T, Shibata A, Kakarougkas A, Jeggo P. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015;43:7931–7944. doi: 10.1093/nar/gkv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo C, Nakazawa Y, Woodbine L, Bjorkman A, Shimada M, Fawcett H, Jia N, Ohyama K, Li TS, Nagayama Y, et al. XRCC4 deficiency in human subjects causes a marked neurological phenotype but no overt immunodeficiency. J. Allergy Clin. Immunol. 2015;136:1007–1017. doi: 10.1016/j.jaci.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irianto J, Pfeifer CR, Xia Y, Discher DE. SnapShot: Mechanosensing Matrix. Cell. 2016;165:1820–1820. doi: 10.1016/j.cell.2016.06.002. e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F, Castro-Giner F, Weischenfeldt J, Kovacova M, Krieg A, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun. 2015;6:8940. doi: 10.1038/ncomms9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olive PL, Banath JP, Durand RE. Detection of etoposide resistance by measuring DNA damage in individual Chinese hamster cells. J. Natl. Cancer Inst. 1990;82:779–783. doi: 10.1093/jnci/82.9.779. [DOI] [PubMed] [Google Scholar]
- 18.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Lu LY, Yu X. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1:117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, Liu AJ, Greenberg RA, Discher DE. Nuclear constriction segregates mobile nuclear proteins away from chromatin. MBoC. 2016 doi: 10.1091/mbc.E16-06-0428. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege H, van Beers E, Velds A, Liu X, Joosse SA, Klarenbeek S, Schut E, Kerkhoven R, Klijn CN, Wessels LF, et al. Cross-species comparison of aCGH data from mouse and human BRCA1- and BRCA2-mutated breast cancers. BMC Cancer. 2010;10:455. doi: 10.1186/1471-2407-10-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 25.Dudgeon C, Chan C, Kang W, Sun Y, Emerson R, Robins H, Levine AJ. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 2014;28:2613–2620. doi: 10.1101/gad.252148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 27.Hass CS, Gakhar L, Wold MS. Functional characterization of a cancer causing mutation in human replication protein A. Mol. Cancer Res. 2010;8:1017–1026. doi: 10.1158/1541-7786.MCR-10-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon GS, Kim KY, Hwang YJ, Jung MK, An S, Ouchi M, Ouchi T, Kowall N, Lee J, Ryu H. Deregulation of BRCA1 leads to impaired spatiotemporal dynamics of gamma-H2AX and DNA damage responses in Huntington's disease. Mol. Neurobiol. 2012;45:550–563. doi: 10.1007/s12035-012-8274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen RB, Ozes A, Kim T, Estep A. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair (Amst) 2013;12:306–311. doi: 10.1016/j.dnarep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandersickel V, Depuydt J, Van Bockstaele B, Perletti G, Philippe J, Thierens H, Vral A. Early increase of radiation-induced gammaH2AX foci in a human Ku70/80 knockdown cell line characterized by an enhanced radiosensitivity. J Radiat Res. 2010;51:633–641. doi: 10.1269/jrr.10033. [DOI] [PubMed] [Google Scholar]
- 31.Fiszer-Kierzkowska A, Vydra N, Wysocka-Wycisk A, Kronekova Z, Jarzab M, Lisowska KM, Krawczyk Z. Liposome-based DNA carriers may induce cellular stress response and change gene expression pattern in transfected cells. BMC Mol. Biol. 2011;12:27. doi: 10.1186/1471-2199-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen WT, Ebelt ND, Stracker TH, Xhemalce B, Van Den Berg CL, Miller KM. ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion. Elife. 2015;4 doi: 10.7554/eLife.07270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39:S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Z, Zou K, Zhuang L, Qin J, Li H, Li C, Zhang Z, Chen X, Cen J, Meng Z, et al. Hepatocellular carcinoma cell lines retain the genomic and transcriptomic landscapes of primary human cancers. Sci. Rep. 2016;6:27411. doi: 10.1038/srep27411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiagalingam S, Laken S, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B, Lengauer C. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2698–2702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sankaranarayanan P, Schomay TE, Aiello KA, Alter O. Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival. PLoS One. 2015;10:e0121396. doi: 10.1371/journal.pone.0121396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Tilney LG, Gibbins JR. Microtubules in the formation and development of the primary mesenchyme in Arbacia punctulata. II. An experimental analysis of their role in development and maintenance of cell shape. J. Cell Biol. 1969;41:227–250. doi: 10.1083/jcb.41.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guemes M, Garcia AJ, Rigueur D, Runke S, Wang W, Zhao G, Mayorga VH, Atti E, Tetradis S, Peault B, et al. GATA4 is essential for bone mineralization via ERalpha and TGFbeta/BMP pathways. J. Bone Miner. Res. 2014;29:2676–2687. doi: 10.1002/jbmr.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Song I, Kim K, Kim JH, Lee YK, Jung HJ, Byun HO, Yoon G, Kim N. GATA4 negatively regulates osteoblast differentiation by downregulation of Runx2. BMB Rep. 2014;47:463–468. doi: 10.5483/BMBRep.2014.47.8.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irianto J, Xia Y, Pfeifer CR, Greenberg RA, Discher DE. As a nucleus enters a small pore, chromatin stretches and maintains integrity even with DNA breaks. Biophys. J. 2016 doi: 10.1016/j.bpj.2016.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 2011;20:4175–4186. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 46.Tamiello C, Kamps MA, van den Wijngaard A, Verstraeten VL, Baaijens FP, Broers JL, Bouten CC. Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus. 2013;4:61–73. doi: 10.4161/nucl.23388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuciforo PG, Luise C, Capra M, Pelosi G, d'Adda di Fagagna F. Complex engagement of DNA damage response pathways in human cancer and in lung tumor progression. Carcinogenesis. 2007;28:2082–2088. doi: 10.1093/carcin/bgm108. [DOI] [PubMed] [Google Scholar]
- 48.Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, Liu AJ, Greenberg RA, Discher DE. Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell. 2016 doi: 10.1091/mbc.E16-06-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dingal PC, Bradshaw AM, Cho S, Raab M, Buxboim A, Swift J, Discher DE. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat Mater. 2015;14:951–960. doi: 10.1038/nmat4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.