Abstract
A requirement for understanding morphogenesis is being able to quantify expansion at the cellular scale. Here, we present new software (RootflowRT) for measuring the expansion profile of a growing root at high spatial and temporal resolution. The software implements an image processing algorithm using a novel combination of optical flow methods for deformable motion. The algorithm operates on a stack of nine images with a given time interval between each (usually 10 s) and quantifies velocity confidently at most pixels of the image. The root does not need to be marked. The software calculates components of motion parallel and perpendicular to the local tangent of the root's midline. A variation of the software has been developed that reports the overall root growth rate versus time. Using this software, we find that the growth zone of the root can be divided into two distinct regions, an apical region where the rate of motion, i.e. velocity, rises gradually with position and a subapical region where velocity rises steeply with position. In both zones, velocity increases almost linearly with position, and the transition between zones is abrupt. We observed this pattern for roots of Arabidopsis, tomato (Lycopersicon lycopersicum), lettuce (Lactuca sativa), alyssum (Aurinia saxatilis), and timothy (Phleum pratense). These velocity profiles imply that relative elongation rate is regulated in a step-wise fashion, being low but roughly uniform within the meristem and then becoming high, but again roughly uniform, within the zone of elongation. The executable code for RootflowRT is available from the corresponding author on request.
Growth underlies life. Although organisms may be distinguished from crystals by reproduction, there would be nothing to reproduce without growth. In plants, growth is important not only for development of the organism but also for physiology. An animal runs, rolls over, bites, or plays dead; instead, a plant bends away, repositions its leaves, thickens its stem, or makes thorns. All these examples, among many others, involve growth.
The first step to understanding how a plant grows is measurement. Growth overall can be measured by following the displacement of a terminus, such as the tip of a blade of grass. By attaching the tip to a position transducer, the displacement can be measured accurately (e.g. Hsiao et al., 1970; Degli Agosti et al., 1997; Frensch, 1997), and tip displacement has been measured at even greater accuracy by interferometry (Fox and Puffer, 1976; Jiang and Staude, 1989). Although such methods are useful for characterizing the overall growth output of an organ, attaching a transducer may disturb the plant, and conditions for interferometry are exacting. More fundamentally, these methods are limited because they record growth in one dimension and because they cannot be used to measure the distribution of growth within the organ. The distribution of growth reflects the growth behavior of component cells and, therefore, is linked to the underlying mechanisms powering expansion.
To study expansion locally throughout a growing organ, one begins by obtaining the velocity profile (Erickson, 1976; Silk, 1992). Velocities arise because expansion moves neighboring elements (units of cell wall, cells, or whole organs), and the velocity profile encompasses the instantaneous growth behavior. If neighboring elements have the same velocity, there is no growth between them, but if they move at different velocities, then the region between them is growing. The velocity profile is usually estimated by marking a growing organ and imaging it over time (Erickson, 1976; Silk, 1992). For marks, ink, graphite, and resin beads have been used, and even pinpricks to leaf tissue. From the images, the position of the marks is measured and velocity as a function of position is calculated. The images were originally photographs and the positions of the marks measured with a ruler, and as technology improved, the photographs were replaced by digital images and the ruler by a video cursor. Nevertheless, the basis of the approach stayed the same: The position of a particle is measured directly in a series of images and the trajectory of this particle defines its velocity. This approach is limited by the invasiveness of marking, the low density of marks that can be applied, the relatively large time that must elapse to give a measurable displacement, and by the tedious, error-prone, subjective nature of the manual measurement process. One group has improved the marking approach by developing software to recognize marks automatically (Ishikawa et al., 1991), but the approach is still limited by the potential for disturbing the plant and by the small number of marks that were applied.
An alternative means to measure the spatial profile of growth is available in principle from image processing techniques for “image sequence” analysis (Jähne, 1997; Nagel, 2000; Shapiro and Stockman, 2001). In these techniques, a stack of images is captured with a relatively short time interval between images. The stack is then treated as a three-dimensional image volume, and one or more filters are used to define spatiotemporal structures in the volume. If a defined structure is parallel to the time axis, then it was stationary; if the defined structure is at an angle to the time axis, then it was moving, and the angle defines the velocity of movement. Because the movement caused by growth is nonuniform, the filters used to define structure operate on the image volume locally. These structures are used to compute a velocity for each pixel of the image, although in practice the scene is seldom rich enough in texture for every pixel to have a well-defined velocity. Because the filter operates on a volume with pixels having neighbors in both space and time, velocity can be quantified with sub-pixel accuracy.
Algorithms based on image sequence analysis work at short time intervals, do not require marking the plant, and remove the need for tedious and subjective manual measurement. Although the mathematical principles behind image sequence analysis were delineated years ago (Fennema and Thompson, 1979; Horn and Schunck, 1981), it has only been recently that improved algorithms and, in particular, enhanced processing power have brought these methods into the realm of practicality. Recently, two groups have used image sequence analysis to quantify growth in plants, with the first group applying it to the coleoptile (Barron and Liptay, 1994, 1997; Liptay et al., 1995) and the second group applying it to both roots (Walter et al., 2002) and leaves (Schmundt et al., 1998), with the latter paper reporting fully two-dimensional velocity fields.
Although these papers make an important start, they have limitations. The method for the coleoptile treated the growing organ as a rigid body and found the velocity of tip movement only, thus functioning in essence like a position transducer, except that velocity in any direction could be measured (Barron and Liptay, 1994). In the method of the second group, values of velocity were confident at relatively few pixels, thus requiring extensive interpolation (Schmundt et al., 1998). An objective of the present work was to develop an algorithm for estimating the velocity field of a deformable object, such as a plant root, returning confident velocity values densely across an image, and handling intervals between frames on the order of seconds.
Furthermore, we developed the algorithm reported here for our studies of growth in the root, in which relative elongation rates differ markedly in size between elongation zone and meristem. The meristem is awkward for marking methods because it is difficult to apply more than one or two marks within it, and the displacements are usually too small to measure manually within the customary time intervals. The previously cited paper using image sequence analysis on the root (Walter et al., 2002) does not have data for the meristem. By any technique, growth in the meristem remains poorly characterized. An additional objective of the present work was to develop an algorithm for image sequence analysis with sufficient versatility to quantify velocity confidently within the elongation zone and meristem from the same sequence of images.
Here, we report an algorithm that quantifies velocity confidently at more than 50% of the root pixels, with time steps between images as low as 2 s and an accuracy of better than 1 pixel per nine frames. The velocity profiles we have obtained with the algorithm for the root show velocity increasing more or less linearly through, and perhaps beyond, the meristem, and, surprisingly, also increasing quite linearly, albeit more steeply, through the zone of elongation.
RESULTS
Overview of the Software
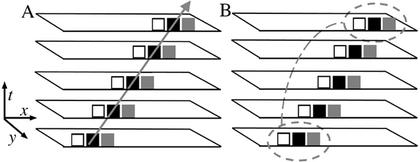
Here, we overview the main features of the software (RootflowRT); computational details are presented elsewhere (Jiang et al., 2003). A set of nine images are captured, with the same time interval between each image. The nine images, called a “stack,” are treated as a single image volume. A motionless feature will parallel the time axis, whereas a moving feature will be at an angle, an angle that defines its velocity. The image processing task is to find those angles for as many pixels as possible. The novelty of our algorithm is that it combines two complementary procedures for doing this, in essence as follows. The first uses the so-called “structure tensor” (Jähne, 1997; Farnebäck, 2000) and can be thought of as finding a line through the stack, from a pixel in the starting image, that minimizes changes in intensity, as expected for the same feature through time (Fig. 1A). The virtues of the tensor method are that it is fast to compute and it reports the statistical confidence of values; the drawbacks are that it is noisy and often gives confident data for few pixels. The second method is called “robust matching” (Black and Anandan, 1996; Black and Rangarajan, 1996; Zhuang et al., 1999), a brute force search to match a target neighborhood between first and last images (Fig. 1B). Matching is accurate and also includes statistical confidence but is computationally intensive and, hence, slow to run.
Figure 1.
Schematic representation of the two major processing steps in the algorithm for quantifying velocity. The drawing shows stacks with five images, but nine are actually used. A, Tensor method finds the line that minimizes intensity changes along its length, in this case going through all black pixels. B, “Robust matching” method matches a local neighborhood in first and last images.
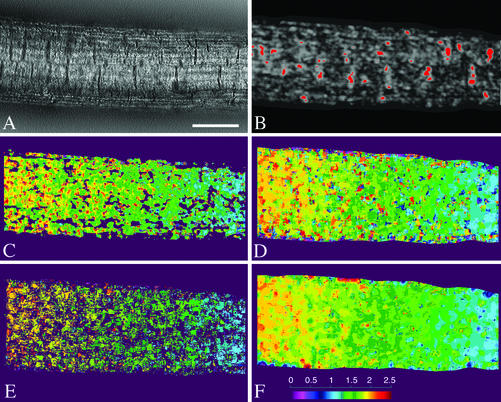
The algorithm in RootflowRT first calculates a velocity field with the tensor method (Fig. 2, A–C). The tensor step is also used to define a mask, segmenting the image into moving and nonmoving components (Fig. 2B). The confident tensor values are then used to constrain and, hence, accelerate a second round where the velocity field is calculated by matching (Fig. 2, D and E). Although confident tensor pixels are few, they tend to be dispersed over the root image (Fig. 2B). The implemented robust matching procedure includes criteria to ensure valid matches, including consistent matching forward and backward in time. The final velocity field is made by combining the confident pixels from both approaches; typically, 5% of the pixels are estimated by the tensor and 50% to 60% estimated by the robust matching. Because confidently estimated pixels appear well spread across the root, the remaining pixels are assigned velocity values based on interpolation (Fig. 2F).
Figure 2.
Steps in the robust tensor velocity estimation algorithm. A, One input frame from a nine-frame image stack in the middle of the root's growth zone. Scale bar = 50 μm. B, Result of the first stage of motion estimation, showing object segmentation based on a two-step motion mask with high confidence tensor-based velocity regions in red. Velocities are computed parallel and perpendicular to the medial axis of the root, and the velocities shown here are the parallel component. C, Pure, tensor-based velocity field, regardless of confidence. D, Velocity field from one step of robust estimation showing the displacement field between the first and ninth frames (forward matching). E, Velocity field after eliminating unreliable data with the forward-backward motion consistency test. F, Final robust tensor velocity field after motion interpolation. Images C to F are pseudo-colored, as indicated in the color bar in F, with velocities ranging from 0 to 2.5 pixels per frame.
The output thus far is a two-dimensional velocity field. For the purpose of studying elongation, the mask is used to generate a root midline and velocities are computed parallel and perpendicular to this curve, with the parallel component taken to represent elongation. Because the determination of the midline algorithmically is sometimes confounded by root hairs or image irregularities, the software instead can accept midline coordinates entered manually. The velocity profile along the x axis is then determined by averaging the velocities perpendicular to each pixel along the midline.
To obtain the velocity profile for the total growth zone, the root is imaged in a series of overlapping stacks and profiles are determined for each stack. Subsequently, the stack profiles are concatenated by the software into a single profile based on distance from the quiescent center and the movement of the stage between image stacks, taking into account the movement of the tip, found by estimating the velocity in the region of the quiescent center. The approximate quiescent center coordinates are input by the user to improve precision. The movement of the stage was determined originally by marking the surface of the agar with graphite particles and capturing a background image at the same position as each stack (van der Weele, 2001) but more recently by using a position transducer to move the stage on command. RootflowRT accepts either kind of input to produce a single profile from those of each stack.
For Arabidopsis roots growing about 500 μm h–1, acceptable velocity profiles were computed with time intervals between images ranging between 2 and 20 s (Jiang, 2001), and we selected a 10-s interval as standard. This results in 80 s used per stack and a total imaging time of 5 to 10 min depending on the size of the growth zone. Various magnifications were used, depending on the size of the root, but all in the range of 1 μm pixel–1, which provides an upper limit for spatial resolution. Velocities were measurable from a high of about 0.3 pixels s–1 to a low of approximately 0.01 pixels s–1, defining the temporal resolution.
Spatial Profile of Velocity in the Root Growth Zone
As a root grows, its tip is propelled through the soil by the cumulative elongation of all of the cells in the growth zone. An element at the very tip moves at maximal velocity; elements located at positions progressively distant from the tip move at progressively lower velocities, until the end of the growth zone is reached, where the velocity becomes zero. For mathematical convenience, it is useful to work in a coordinate system where the tip of the root defines the origin and growth displaces elements away from the tip toward the base (Erickson, 1976). In this frame, an element at the very tip has a velocity of zero, and as the location of the element moves basally, its velocity increases until reaching a maximum where growth ceases. All velocity profiles reported here are in this frame, with the quiescent center used as the origin rather than the root tip because velocity values at the very apex and within the root cap can be distorted by the film of water surrounding the tip and by the splaying out of root cap cells as they separate from the root body. Note that in Arabidopsis, the quiescent center amounts to four cells and acts as the biological origin for all cells in the root.
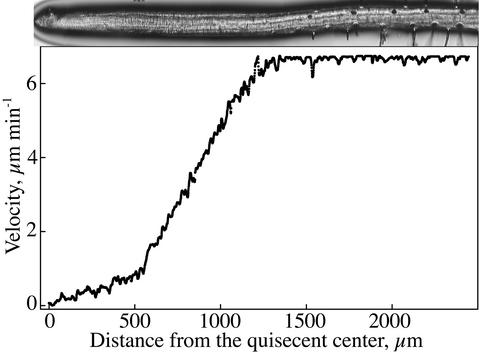
A representative example of a velocity profile obtained by the algorithm is shown, with a photograph of the root approximately on the same scale included for reference (Fig. 3). The profile has three distinct regions: starting at the quiescent center (x = 0), velocity increases first gradually with position, then steeply, and finally it becomes constant, indicating the end of the growth zone. The regions are separated by fairly abrupt transitions, with the first transition invariably more abrupt than the second. The increase in velocity in both the first and second regions appears to have a considerable linear character. The first region spans the meristem and may extend beyond it (van der Weele, 2001), whereas the second region is the elongation zone. The third region (constant velocity and no growth) begins basal of the position where root hairs initiate (Ma et al., 2003). For convenience, we will refer to the first region as the “meristem,” although cell division remains to be delineated.
Figure 3.
Velocity profile for Arabidopsis obtained with the new software. The root from which the profile was obtained is reproduced above, roughly to scale. Data points are every pixel (=1.1 μm) and do not show up as single symbols.
The profiles have high-frequency spatial fluctuations, probably indicating computational noise. Profiles occasionally have larger, irregular, spatial fluctuations and less pronounced transitions. In approximately 15% of the profiles, velocity is constant (i.e. has a slope of 0) over an appreciable region of the meristem, suggesting either that expansion stops transiently in the meristem or that the algorithm is misled, for example by anomalous behavior of lateral root cap cells. Similar regions of constant velocity are present in the maize (Zea mays) meristem in the profiles published by Erickson and Sax (1956).
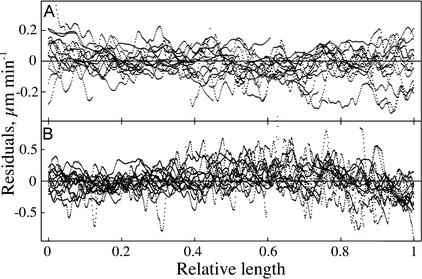
In view of approximately linear regimes in the velocity profile, the data were fitted to a single model comprising three linear equations joined at two breakpoints. The goodness of fit is evident (Fig. 4A). This impression was confirmed by analysis of residuals for 15 different roots (Fig. 5). For the first line (through the meristem; Fig. 5A), the residuals are small and evenly distributed around the mean, providing no evidence for a systematic departure from linearity. For the second line (through the elongation zone; Fig. 5B), the residuals are again small and randomly distributed, except near the end where they cluster beneath the mean, indicating that the transition to constant (zero) velocity is more gradual than the intersection of two lines.
Figure 4.
Comparison of the velocity profile and its derivative, the profile of relative elongation rate. A and B, Velocity profiles of two different roots fitted to either a three-piece linear model (A) or overlapping polynomials (B; Beemster and Baskin, 1998). Gray wiggly lines are raw data, and solid lines are the fits. C and D, Derivatives obtained analytically from the fitted velocity profiles. The derivative ordinate was converted from percentage per minute to percentage per hour for easier comparison with data in the literature.
Figure 5.
Residual analysis of the linear regression fits to the data from 15 individual Arabidopsis roots. A, Data for the meristem. B, Data for the zone of elongation. The distance coordinate (x axis) is expressed as a relative length, namely the ratio of the position of a point within a zone to the total length of the zone.
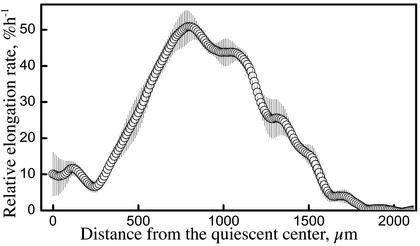
Although the velocity profiles are well fitted by lines, linearity is an oversimplification. Instead of lines, one may fit a set of overlapping polynomials (Beemster and Baskin, 1998), which can conform closely to the raw data (Fig. 4B). The implications of the different fits are best grasped by considering the derivative of the velocity plot, which is the profile of relative elongation rate (Fig. 4, C and D). For a velocity profile of two lines, the derivative is a “step stool” (Heaviside step function), indicating a constant rate of relative elongation for the meristem and a constant, albeit higher, rate for the elongation zone, with a rapid transition between states. In contrast, the velocity profile fit locally to polynomials gives rise to a more complex derivative, which may be considered to represent an overall step function shape convolved with higher spatial frequency fluctuations. These higher frequency fluctuations appear to be partly synchronized among roots, insofar as they are retained as “wobbles” in a plot averaging the polynomial-based derivatives of the velocity profiles of five roots (Fig. 6). Note that the averaging deemphasizes the step stool-like appearance of the derivative not only because the location of the abrupt transitions differed among roots but also because the fitted polynomial is 300 μm long and when fit to an abrupt transition gives rise to a derivative that changes smoothly over much of the 300-μm interval (van der Weele, 2001).
Figure 6.
Spatial profile of relative elongation rate (strain rate) for 9-d-old Arabidopsis roots. The raw velocity data for each root was fitted with overlapping polynomials (Beemster and Baskin, 1998), interpolated to 1-μm intervals, and differentiated analytically. The symbols show the average for five roots, ±se The ordinate was converted to percentage per hour for easier comparison with data in the literature.
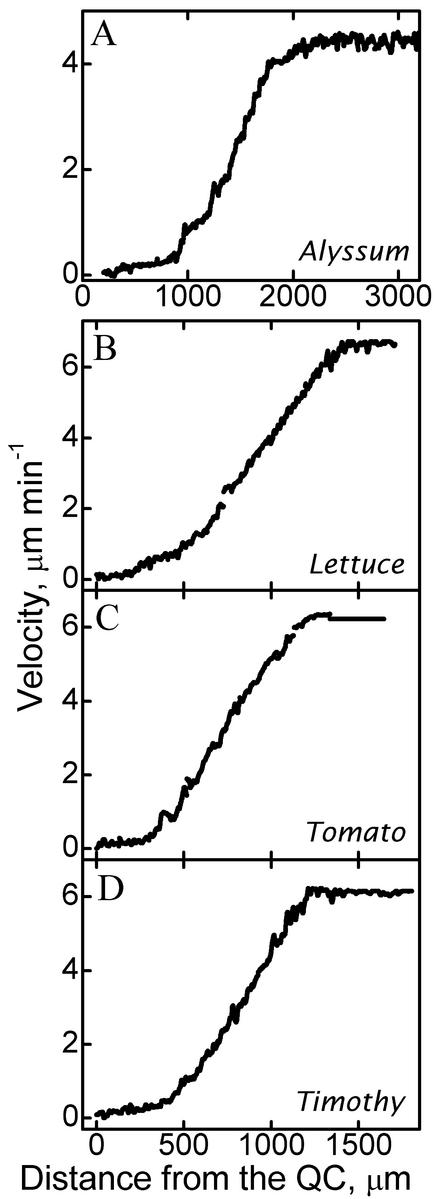
To determine whether the biphasic profile of velocity found for Arabidopsis typifies other species, we obtained data for alyssum (Aurinia saxatilis), lettuce (Lactuca sativa), tomato (Lycopersicon lycopersicum), and timothy (Phleum pratense). These species have roots of different optical texture. As in Arabidopsis, the velocity profiles have two regions of roughly linear increase separated by an abrupt transition (Fig. 7). As for Arabidopsis, linear regressions fitted to the data from these species conform closely to the curves (data not shown).
Figure 7.
Velocity profiles for four species: A, alyssum; B, lettuce; C, tomato; and D, timothy. The location of the quiescent center (QC) was approximated from the image of the tip.
Tip-Tracking Algorithm
Although determining the spatial distribution of growth was the major motivation behind developing the algorithm, the spatial algorithm was modified to measure the velocity of the root tip versus time. In tip-tracking mode, images of the growing root tip are collected at a given frequency for up to 300 frames. Because the root cap may contain tissue fragments moving irregularly as they separate from the root body, fully automatic segmentation of the extreme tip is difficult; therefore, the user initializes the routine by entering the approximate coordinates of the quiescent center. The algorithm then defines a region of interest, with the quiescent center at one edge. The size of the region is defined by the user, and we generally used 200 × 100 pixels. A tensor method alone is used to calculate velocities for the pixels in this box, starting with the first nine frames and moving through the entire sequence by adding the next frame and dropping the last one. All confident velocities are averaged, and this value is reported as tip velocity and used to update quiescent center coordinates.
Results of tip tracking have found that roots grow more or less steadily during imaging for up to 1 h (data not shown) but that small fluctuations in tip velocity are usually present (Fig. 8). The fluctuations are irregular, with magnitudes typically less than 10% of the mean velocity and apparent period ranging from 5 to 20 min. Satisfactory records could be obtained with 5- or 10-s intervals between frames. To see if fluctuations could be reduced, we tracked roots illuminated with yellow light (as used by Beemster and Baskin, 1998) or with infrared; in addition, we illuminated the shoot with yellow light either at the intensity present in the growth chamber (200 μmol m–2 s–1) or less (70 μmol m–2 s–1). Fluctuations were assessed by analysis of variance and residuals. On average, fluctuations were slightly reduced by infrared, but they were increased significantly by the higher of the two light intensities on the shoot (not shown). Based on these results, we have standardized infrared light to illuminate the root and one-half-strength light to the shoot.
Figure 8.
Time courses of overall root growth rate (tip velocity) obtained with the tip-tracking algorithm. Records of two representative roots are shown. Different mean velocities result in part from the roots being of different ages.
DISCUSSION
A New Algorithm for Image Sequence Analysis Applicable to the Plant Root
To measure the velocity field embodying root growth accurately and easily, we turned to image processing techniques for estimating deformable motion. Previously, we measured the velocity field manually by marking the plant and measuring the displacement of the marks (Beemster and Baskin, 1998), but the subjective and tedious nature of the method spurred us to find an alternative. Quantifying deformable motion, as in a growing root, is demanding because the object changes while it moves. This kind of motion analysis has been worked on extensively (Nagel, 2000), but algorithms typically invoke assumptions specific to a given application, such as robot vision, graphics, or meteorology (e.g. Metaxas and Terzopoulos, 1993; Vedula et al., 2000; Zhou et al., 2001). For quantifying deformable motion in biology, there are unique challenges, including the lack of explicit motion models, low-contrast images, high variability in local intensity, nonuniform background, and multifaceted motion (simultaneously observable fluid-like, appendage-like, thread-like motions). Moreover, biologists typically wish to represent the velocity field quantitatively, a constraint that is often absent from other kinds of application.
In the most common class of such methods, called “optical flow,” the intensity of a small, moving region of the image is assumed to be conserved (Fennema and Thompson, 1979; Horn and Schunck, 1981; Beauchemin and Barron, 1995). The concept arose out of studies on the human visual system (Gibson, 1966), hence the word “optical,” but the concept is independent of the imaging modality. The optical flow-based algorithms that have been applied to growth and motility can be divided roughly into three classes: parametric, tensor, and matching. Parametric methods fit an affine transformation to the motion within a region of interest (Odobez and Bouthemy, 1995; Black and Jepson, 1996); this is an approximate solution because all pixels in the region are constrained to undergo the same transformation. The tensor methods are differential methods based on intensity gradients (Jähne, 1997); they are computationally fast but error prone. Finally, matching methods find corresponding regions in a pair of images by maximizing a similarity criterion or minimizing an error criterion (Beauchemin and Barron, 1995; Black and Anandan, 1996). Matching methods tend to be slow, sensitive to neighborhood size, local image content, and outliers; however, they can be highly accurate, particularly when the search process incorporates robust statistics.
In studies of growth and cell motility in biology, there are a few examples of the use of each type of algorithm. Tracqui and coworkers have estimated motion parametrically in crawling and dividing cultured animal cells as well in as monolayers involved in wound healing (Germain et al., 1999; Ronot et al., 2000). In their approach, the motion of an entire cell was reduced to a single transformation and, thus, was appreciably simplified. Matching methods have been applied to muscle contraction, taking advantage of the repetitive structure of the sarcomere (Zoccolan et al., 2001). Tensor methods are the most popular and have been applied to cells moving in slime molds (Siegert et al., 1994; Dormann et al., 1996, 1997), although without evaluating the confidence of the recovered velocities, and to the growth of plant leaves (Hauβecker and Jähne, 1997; Schmundt et al., 1998) and roots (Walter et al., 2002), where the relatively sparse frequency of confident pixels obtained led to extensive interpolation. In their analysis of the maize root, Walter et al. (2002) took advantage of the fast computation of their tensor-based algorithm and followed the features of the growth zone for a whole day, capturing images once per minute, with approximately 20 μm pixel–1. In contrast, our method emphasizes resolution, capturing images once per 10 s, with approximately 1 μm pixel–1, and computing a dense velocity field, but only for a single time.
We have applied RootflowRT to analyze root elongation under water stress (van der Weele, 2001) and phosphorus stress (Ma et al., 2003). The algorithm combines tensor analysis with robust matching procedures, using the speed of the tensor method to accelerate matching while retaining the reliability of matching. The root does not require marking, and the images are relatively low contrast and separated by short time intervals (10 s). The algorithm is automated, estimates velocity with sub-pixel accuracy, applies statistical tests to exclude values below a given level of confidence, and returns confident velocities for the majority of pixels in the root. Along with velocity data, the software also provides an output file tabulating various parameters. Because the root is essentially cylindrical, the software calculates velocity parallel and perpendicular to the local tangent of the root's midline.
RootflowRT also concatenates the velocity fields from overlapping stacks spanning the root growth zone, which cannot be imaged at sufficiently high resolution in a single field of view. Lowering the magnification results in insufficient gray level texture for reliable feature extraction over the short time intervals used. Combining the profiles from individual stacks into a single profile requires knowing the displacement of the stage between stacks and also the displacement of the tip between successive stacks because the quiescent center has an x axis coordinate of zero but is moving in the laboratory frame. To combine the profiles, we assume that the velocity field is time invariant (i.e. for all x, dV(x)/dt = 0), an assumption that is also made in the traditional marking methods (Silk, 1992). At the highest level of resolution, this assumption is wrong; fluctuations in overall root elongation rate occur (Fig. 8) and for some stacks values of velocity do not match exactly in the region of overlap. For these cases, the software lifts one profile in the y axis (velocity) direction by an amount that minimizes the least square difference between points within the overlap. The values of the lifts (if any) are tabulated for the user; therefore, roots with large discrepancies can be excluded.
Fluctuations can distort the velocity profile in three ways. First, changes in velocity that occur within the 80-s interval of the image stack will lead to an average velocity determination; few fluctuations occur on this time scale. Second, as described above, fluctuations cause a mismatch in the velocity where neighboring stacks overlap. To date, the mismatch amounts to no more than 10% of the velocity itself and is usually much less. Note that the derivative of the velocity profile, which is of chief interest, is not affected by lifting one part of the profile relative to another. Third, the fluctuations in tip velocity mean that the succeeding stacks cannot be placed exactly with respect to the quiescent center. This uncertainty amounts to approximately 1 pixel on the x axis and, thus, is unlikely to distort the final profile appreciably. Given the time required to image the entire growth zone, our algorithmic procedure represents an increase in temporal resolution over previous manual methods by more than an order of magnitude.
A New “Old” View of Growth in the Root
For many years, the profile of velocity in the root growth zone has been estimated by marking experiments and widely accepted to be a smooth curve, resembling a sigmoid (Goodwin and Stepka, 1945; Erickson and Sax, 1956; Sharp et al., 1988; Mullen et al., 1998). In contrast, the results of our algorithmic determination show a velocity profile that has a considerable linear character, more closely resembling three lines than a sigmoid. The difference in the shape of the velocity profile is unlikely to be explained by different environments because previous marking experiments recovered a sigmoid velocity curve for Arabidopsis roots grown under the same conditions as used here (Beemster and Baskin, 1998), and RootflowRT found abrupt velocity profiles for Arabidopsis roots growing inside the agar medium (Ma et al., 2003). We suggest that the linear velocity profile reflects a nearly instantaneous picture of growth, whereas the sigmoid presents an average picture, possibly compromised by measurement error or undersampling.
Marking experiments are intrinsically error prone because they rely on subjective and manual measurements, and they are limited by the number of marks that can be applied to the root, typically around 10 per growth zone, in contrast to the data at every pixel obtained with RootflowRT (thousands of pixels per growth zone). Most previous publications show velocity profiles averaged for a group of roots and, therefore, would smooth out abrupt behavior. To obtain measurable displacements, marking experiments typically use imaging intervals between 15 and 60 min, compared with the 80 s per stack used here. Marks moving through an abrupt transition will yield an average velocity for before and after the transition. Furthermore, if either the position of a transition or the magnitude of the relative elongation rate fluctuated during the imaging interval, then the measured displacements would also reflect average behavior. Fluctuations in overall elongation rate occur in the Arabidopsis root (Fig. 8) and appear to be widespread among plant organs (Kristie and Jolliffe, 1986; Jiang and Staude, 1989; Liptay et al., 1995; Degli Agosti et al., 1997; Walter et al., 2002), although their cause on a cellular level is not known. The fluctuations typically have periods around 30 to 60 min and, thus, are too slow to greatly affect the results from the imaging interval of 80 s used here but would affect results from the longer intervals used in marking experiments.
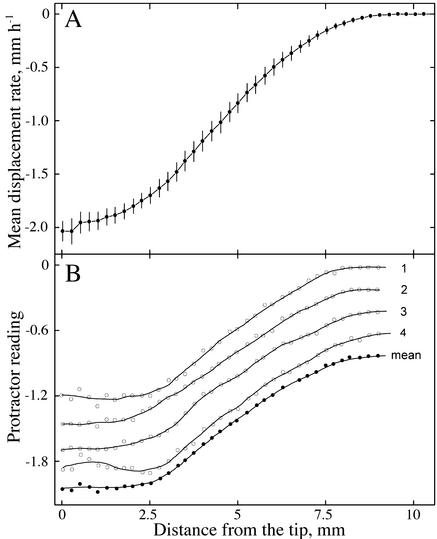
Our interpretation that the instantaneous velocity field has quite linear regions is supported directly in the classic paper of Erickson and Sax (1956). Their velocity profile of the maize root has been widely reproduced and is a sigmoid curve (Fig. 9A); this curve reflects the average profile of 10 roots, with data for each root averaged over four to six measured time points. However, they also published raw data for one root, showing velocity profiles that result from single measurements at successive times (Fig. 9B). In contrast to the sigmoid curve for the average, the profiles for the raw data have a phase of gradually increasing velocity at the tip and one of steeply increasing velocity further back, with a distinct transition region between the two phases.
Figure 9.
Figures redrawn from Erickson and Sax (1956) showing the velocity profile for maize roots measured using streak photography. A, (Fig. 4), Average data for 10 roots, ±95% confidence interval. B, (Fig. 3), Raw data measured at four different times from a single root and the mean of the four measurements. Curves are displaced for clarity. The y axis is calibrated in protractor units (actually tangents of the angles made by the streaks), which are proportional to velocity. Solid lines were obtained by numerical smoothing.
Amazingly, the method used by Erickson and Sax is in essence an analog version of the digital image processing used here. They marked a root densely by dipping it in lampblack and then photographed the longitudinal axis of the root through a narrow rectangular aperture onto film while the film moved continuously. This caused the mark images to leave streaks on the film, with the angle of the streak to the horizontal being proportional to the velocity of movement. The streaks are lines of (roughly) constant intensity and, therefore, are an analog of the structure tensor. In fact, streak photography was designed by Erickson explicitly to reach as “elemental” and “instantaneous” a level as possible, and he appears to have succeeded.
That the root growth zone has a velocity profile with linear phases separated by an abrupt transition is supported by other data. A pioneering paper by Brumfield (1942) on timothy reported a roughly constant rate of relative elongation in the zone of elongation, and early work by Hejnowicz (1959) for wheat (Triticum aestivum) reported roughly constant relative elongation rate across the meristem. Ivanov and Maximov (1999) reanalyzed the profile of metaxylem cell length in maize roots and concluded that relative elongation rate accelerates abruptly at the base of the meristem; in addition, they discuss indirect evidence supporting the idea that relative elongation rate is essentially constant across the meristem. Most recently, the spatial profiles of growth in the maize root obtained by tensor-based image processing, although not spanning the meristem, show a strikingly steep increase in relative elongation rate at the start of the elongation zone (Walter et al., 2002), similar to the results shown here.
To the extent that the sigmoid curve reflects averaging out real phenomena such as growth fluctuations, these curves may be appropriate for understanding processes taking place in the growth zone over hours to days, such as cell division (Sacks et al., 1997; Beemster and Baskin, 1998), nutrient partitioning (Muller et al., 1998), or osmotic adjustment (Sharp et al., 1990); but for understanding the mechanism of expansion and its regulation, the sigmoid curve may be misleading.
The importance of the shape of the velocity curve lies in the implications for relative expansion rate, which is the derivative of the velocity profile. Relative elongation rate, technically a strain rate, reflects the deformation of the underlying cell wall and, thus, is the appropriate parameter for characterizing the mechanism of growth on the cellular or subcellular scale. A smooth, sigmoid-like velocity curve gives rise to a profile of relative elongation rate that is bell shaped, implying that relative elongation rate changes continuously as a cell traverses the growth zone (Erickson, 1976), whereas a velocity profile with linear phases give rise to a “step stool” derivative, implying that relative elongation rate is essentially constant within the meristem and also constant, albeit greater, in the elongation zone (Fig. 4). It is difficult to envisage the regulatory machinery needed to generate a bell-shaped variation in relative elongation rate; in contrast, the root can generate a step stool profile by regulating two rates of relative elongation (height of the steps) and two positions where growth rate changes (width of the steps). Deviation from perfect steps would occur to the extent that the transitions take a finite time to execute, and relative elongation cannot be maintained constant exactly (van der Weele, 2001; Walter et al., 2002; Ma et al., 2003).
The Idea of a Constant Relative Elongation Rate Is Particularly Appealing for the Meristem
Whatever its character, the profile of relative elongation rate must equal the profile of cell division rate because congruence of these profiles is necessary to maintain a constant cell length (per cell file) as observed (Green, 1976; Ivanov et al., 2002). The bell-shaped profiles have relative elongation rate increasing continuously across the meristem, implying that cell division rate increases in parallel. Although there is some uncertainty about the profile of cell division rate across the meristem, to our knowledge no one has reported that it increases steadily, and much evidence indicates that it is constant, apart from the quiescent center (Baskin, 2000). Constancy of cell division rate implies consistency in the underlying cell cycle engine, and a balancing relative elongation rate provides further consistency for the activities of the meristem. Our finding that relative elongation rate tends to be constant across the meristem suggests that division parameters and elongation parameters are regulated uniformly.
With our algorithm, we are zeroing in on the truly instantaneous growth behavior of the root: the growth zone comprises two zones, each with more or less uniform relative elongation, separated by a relatively abrupt transition. Future studies can now determine how these zones are maintained and modified in response to the environment and how they correspond to other processes in the root that are delimited spatially, such as cell division and differentiation.
MATERIALS AND METHODS
Plant Growth
Seeds of Arabidopsis Columbia, timothy (Phleum pretense), tomato (Lycopersicon lycopersicum Mill. var. Roma VFN), lettuce (Lactuca sativa L. var. Black Seeded Simpson), and alyssum (Aurinia saxatilis L. Desv. var. Gold Dust), the latter three obtained from a local supermarket, were surface sterilized and germinated on agar-solidified nutrient solution in 9-cm petri dishes as described previously for Arabidopsis (Baskin and Wilson, 1997). Sucrose concentration in the media was 2% (w/v) for tomato and lettuce, 0.5% (w/v) for Arabidopsis, and 0% (w/v) for timothy. Plates were put vertical in a growth chamber with constant conditions (19°C, 200 μmol m–2 s–1) permitting roots to grow along the surface of the agar.
Imaging
A petri dish was placed on the stage of a horizontal compound microscope so the plants remained vertical. Images were taken through the lid of the petri dish to prevent evaporation of the water film around the root and consequent movement of the root. To accommodate for the focal length of the objective, a dimple lid was constructed. Light from the built-in microscope lamp (12-V halogen bulb) was passed through either yellow acrylic (Plexiglas J2208, Cope Plastic, St. Louis) for broad-band yellow light or glass (Schott RG-9, Bes Optics, West Warwick, RI) for infrared light. Roots were imaged with a 10× objective and a CCD camera (C2400, Hamamatsu Co., Hamamatsu, Japan) with the infrared cutoff filter removed and coupled to the microscope with either a 5× or 2.5× intermediate tube lens. A time stamp was placed on the image with a time date generator. Images were captured on an Apple Macintosh G3 (Apple Computer, Cupertino, CA) equipped with a frame grabber board (Scion LG-3) and the image analysis program Scion Image (www.Scioncorp.com, Scion Image, Frederick, MD). Each frame is 640 × 480 pixels in size, with 0.8 to 1.5 μm pixel–1, depending on the tube lens.
For tip tracking, the tip was placed in the field of view and images taken every 5 or 10 s for a total of up to 200 images. For spatial analysis, a series of stacks were obtained, spanning the growth zone and including nongrowing regions of the root. Each stack has nine images, with the time interval chosen by the user (usually 10 s). The algorithm references the calculated velocity to the time of frame five, being at the center of the stack.
To concatenate the velocity output from the single stacks into a single profile, one must determine the amount of movement of the stage between the positions used to obtain each stack. Initially, this movement was determined by collecting, for each stack, a background image of the agar surface that had been marked to enable the backgrounds to be registered. As marks, good results were obtained with graphite particles or cornstarch, although care had to be taken to avoid disturbing the root. Disturbing the root could be avoided by incorporating inert, latex beads into the agar, but this required focusing into the agar and resulted in successive background images being captured at different focal planes. Because capturing a background image lengthened the time required to image the growth zone, we obtained an electro-optical position transducer (Inchworm; Burleigh Instruments, Fishers, NY), and built a cradle for the microscope stand that allowed the inchworm to engage the stage controller in the vertical direction. The inchworm moves the stage in preset increments, accurate to ±1 μm, and in this way the stage movement between stacks could be accounted for absolutely without recourse to background images. The software accepts either method for concatenation.
In addition to accounting for the movement of the stage, one must also account for the movement of the tip. For frame five of the tip stack, the user enters the coordinates of the quiescent center (in Arabidopsis, the quiescent center contains four cells, and its position is determined with reference to the columella, detectable because its abundant amyloplasts scatter light and give rise to dark bands running across the tissue—the absolute tip of the root could also be entered). The position of the quiescent center is taken as x = 0 for the velocity profile. The velocity at this region is multiplied by the time between center frames to obtain the tip displacement between a pair of stacks, and the output from each is adjusted accordingly.
Source code for RootflowRT is available for downloading from the corresponding author. Both spatial and tip-tracking versions are available. At present, the code compiles and runs under Unix and Windows operating systems with development done primarily on SGI MIPS Irix (Silicon Graphics, Mountain View, CA) and HP Intel P4 computers (Hewlett Packard, Palo Alto, CA).
Regression
The velocity profiles were fitted either with overlapping second degree polynomials as described by Beemster and Baskin (1998) or with a three-piece linear regression using the nonlinear regression module in Statistica (Statsoft Inc., Tulsa, OK). Least mean square regressions were calculated with the following model:
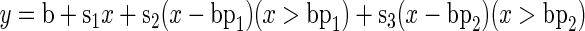 |
in which y is the predicted value, x is the distance from the quiescent center, b is the y intercept, bp1 and bp2 are the values of x at which breakpoints in the regression occur, and s1, s2, and s3 are coefficients of regression. The coefficients give the slopes of the three respective pieces, with s1 for the first slope, s1 + s2 for the second slope, and s1 + s2 + s3 for the third. The expressions x > bp1 and x > bp2 are logical multipliers: If true, it will evaluate to 1 and if false to 0. The six parameters (b, s1, s2, s3, bp1, and bp2) were estimated in the independent variable x (distance along the root) with the quasi-Newton method. Convergence was robust provided that suitable initial values were chosen.
Acknowledgments
We gratefully acknowledge Dr. Michael Keller for his help on statistics, and we thank Jan Judy-March for flawless technical assistance and Mayandi Sivaguru for the data shown in Figure 8. K.K.P. worked on this project as part of his senior year curriculum at David Hickman High School (Columbia, MO).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021345.
This paper is dedicated to Ralph O. Erickson on the occasion of his 89th birthday. This work was supported by the U.S. National Science Foundation (award no. IBN 9817132 to T.I.B.) and by the U.S. Department of Energy (award no. 94ER20146 to T.I.B.), which does not constitute endorsement by that Department of views expressed herein.
References
- Barron JL, Liptay A (1994) Optical flow to measure minute increments in plant growth. Bioimaging 2: 57–61 [Google Scholar]
- Barron JL, Liptay A (1997) Measuring 3-D plant growth using optical flow. Bioimaging 5: 82–86 [Google Scholar]
- Baskin TI (2000) On the constancy of cell division rate in the root meristem. Plant Mol Biol 43: 545–554 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE (1997) Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol 113: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin SS, Barron JL (1995) The computation of optical flow. ACM Comput Surv 27: 433–467 [Google Scholar]
- Beemster GTS, Baskin TI (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MJ, Anandan P (1996) The robust estimation of multiple motions: parametric and piece-wise smooth flow fields. Comput Vision Image Understanding 63: 75–104 [Google Scholar]
- Black M, Jepson AD (1996) Estimating optical flow in segmented images using variable-order parametric models with local deformations. IEEE Trans Pattern Anal Machine Intell 18: 972–986 [Google Scholar]
- Black MJ, Rangarajan A (1996) On the unification of line processes, outlier rejection, and robust statistics with applications in early vision. Int J Computer Vision 19: 57–91 [Google Scholar]
- Brumfield RT (1942) Cell growth and division in living root meristems. Am J Bot 29: 533–543 [Google Scholar]
- Degli Agosti R, Jouve L, Greppin H (1997) Computer-assisted measurements of plant growth with linear variable differential transformer (LVDT) sensors. Arch Sci Genève 50: 233–244 [Google Scholar]
- Dormann D, Siegert F, Weijer CJ (1996) Analysis of cell movement during the culmination phase of dictyostelium movement. Development 122: 761–769 [DOI] [PubMed] [Google Scholar]
- Dormann D, Weijer C, Siegert F (1997) Twisted scroll waves organize Dictyostelium mucoroides slugs. J Cell Sci 110: 1831–1837 [DOI] [PubMed] [Google Scholar]
- Erickson RO (1976) Modeling of plant growth. Ann Rev Plant Physiol 27: 407–434 [Google Scholar]
- Erickson RO, Sax K (1956) Elemental growth rate of the primary root of Zea mays. Proc Am Philos Soc 100: 487–498 [Google Scholar]
- Farnebäck G (2000) Fast and accurate motion estimation using orientation tensors and parametric motion models. In A Sanfelieu, ed, Proceedings of the 15th International Conference on Pattern Recognition, Barcelona, Spain, Vol 1. IEEE Computational Society, Los Alamitos, CA, pp 135–139 [Google Scholar]
- Fennema C, Thompson W (1979) Velocity determination in scenes containing several moving objects. Comput Graphics Image Proc 9: 301–315 [Google Scholar]
- Fox MD, Puffer (1976) Analysis of transient plant movements by holographic interferometry. Nature 261: 488–490 [Google Scholar]
- Frensch J (1997) Primary responses of root and leaf elongation to water deficits in the atmosphere and soil solution. J Exp Bot 48: 985–999 [Google Scholar]
- Germain F, Doisy A, Ronot X, Tracqui P (1999) Characterization of cell deformation and migration using a parametric estimation of image motion. IEEE Trans Biomed Eng 46: 584–599 [DOI] [PubMed] [Google Scholar]
- Gibson J (1966) The Senses Considered as Perceptual Systems. Houghton Mifflin, Boston
- Goodwin RH, Stepka W (1945) Growth and differentiation in the root tip of Phleum pratense. Am J Bot 32: 36–46 [Google Scholar]
- Green PB (1976) Growth and cell pattern formation on an axis: critique of concepts, terminology and modes of study. Bot Gaz 137: 187–202 [Google Scholar]
- Hauβecker H, Jähne B (1997) A tensor approach for precise computation of dense displacement vector fields. In A Sanfelieu, ed, Proceedings of the Mustererkennung DAGM Symposium. Springer, Germany, Braun-schweig, pp 199–208
- Hejnowicz Z (1959) Growth and cell division in the apical meristem of wheat roots. Physiol Plant 12: 124–138 [Google Scholar]
- Horn BKP, Schunck BG (1981) Determining optical flow. Art Intel 17: 185–204 [Google Scholar]
- Hsiao TC, Acevedo E, Henderson DW (1970). Maize leaf elongation: continuous measurements and the dependence on plant water status. Science 168: 590–591 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Hasenstein KH, Evans ML (1991) Computer-based video digitizer analysis of surface extension in maize roots. Planta 183: 381–390 [DOI] [PubMed] [Google Scholar]
- Ivanov VB, Dobroahaev AE, Baskin TI (2002) What the distribution of cell lengths in the root meristem does, and does not, reveal about cell division. J Plant Growth Regul 21: 60–67 [DOI] [PubMed] [Google Scholar]
- Ivanov VB, Maximov VN (1999) The change in the relative rate of cell elongation along the root meristem and the apical region of the elongation zone. Russ J Plant Physiol 46: 73–82 [Google Scholar]
- Jähne B (1997) Motion. In Digital Image Processing, Ed 4. Springer, Berlin, pp 395–450
- Jiang H, Palaniappan K, Baskin TI (2003) A combined matching and tensor method to obtain high fidelity velocity fields from image sequences of the non-rigid motion of plant root growth. In MH Hamza, ed, IASTED International Conference on Biomedical Engineering, BioMED 2003, Acta Press, Calgary, Canada (in press)
- Jiang Z, Staude W (1989) An interferometric method for plant growth measurements. J Exp Bot 40: 1169–1173 [Google Scholar]
- Kristie DN, Jolliffe PA (1986) High resolution studies of growth oscillations during stem elongation. Can J Bot 64: 2399–2405 [Google Scholar]
- Liptay A, Barron JL, Jewett T, van Wesenbeeck I (1995) Oscillations in corn seedling growth as measured by optical flow. J Am Soc Hortic Sci 120: 379–385 [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxas D, Terzopoulos D (1993) Shape and nonrigid motion estimation through physics-based synthesis. IEEE Trans Pattern Anal Mach Intel 15: 580–591 [Google Scholar]
- Mullen JL, Ishikawa H, Evans ML (1998) Analysis of changes in relative elemental growth rate patterns in the elongation zone of Arabidopsis roots upon gravistimulation. Planta 206: 598–603 [DOI] [PubMed] [Google Scholar]
- Muller B, Stosser M, Tardieu F (1998) Spatial distribution of tissue expansion and cell division rates are related to irradiance and to sugar content in the growing zone of maize roots. Plant Cell Environ 21: 149–158 [Google Scholar]
- Nagel HH (2000) Image sequence evaluation: 30 years and still going strong. In 15th IEEE International Conference on Pattern Recognition, Vol 1. Computer Vision and Image Analysis. IEEE, Los Alamitos, CA, pp 149–158
- Odobez JM, Bouthemy P (1995) Robust multiresolution estimation of parametric motion models. J Visual Commun Image Represent 6: 348–365 [Google Scholar]
- Ronot X, Doisy A, Tracqui P (2000) Quantitative study of dynamic behavior of cell monolayers during in vitro wound healing by optical flow analysis. Cytometry 41: 19–30 [PubMed] [Google Scholar]
- Sacks MM, Silk WK, Burman P (1997) Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Physiol 114: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmundt D, Stitt M, Jähne B, Schurr U (1998) Quantitative analysis of the local rates of growth of dicot leaves at high temporal and spatial resolution, using image sequence analysis. Plant J 16: 505–514 [Google Scholar]
- Shapiro L, Stockman GC (2001) Computer Vision. Prentice-Hall, Engle-wood Cliffs, NJ
- Sharp RE, Hsiao TC, Silk WK (1990) Growth of the maize primary root at low water potentials: II. Role of growth and deposition of hexose and potassium in osmotic adjustment. Plant Physiol 93: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, Silk WK, Hsiao TC (1988) Growth of maize primary root at low water potentials: I. Spatial distribution of expansive growth. Plant Physiol 87: 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert F, Weijer CJ, Nomura A, Miike H (1994) A gradient method for the quantitative analysis of cell movement and tissue flow and its application to the analysis of multicellular dictyostelium development. J Cell Sci 107: 97–104 [DOI] [PubMed] [Google Scholar]
- Silk WK (1992) Steady form from changing cells. Int J Plant Sci 153: S49–S58 [Google Scholar]
- van der Weele CM (2001) Cell production, expansion, and the role of auxin in the response of the root of Arabidopsis thaliana exposed to water deficit. PhD thesis. University of Missouri, Columbia
- Vedula S, Baker S, Seitz S, Kanade T (2000) Shape and motion carving in 6D. Proc IEEE Computer Vision Pattern Recognition 2: 592–598 [Google Scholar]
- Walter A, Spies H, Terjung S, Küsters R, Kirchgeβner N, Schurr U (2002) Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. J Exp Bot 53: 689–698 [DOI] [PubMed] [Google Scholar]
- Zhou L, Kambhamettu C, Goldgof DB, Palaniappan K, Hasler AF (2001) Tracking nonrigid motion and structure from 2D satellite cloud images without correspondences. IEEE Trans Pattern Anal Machine Intel 23: 1330–1336 [Google Scholar]
- Zhuang X, Palaniappan K, Haralick RM (1999) Highly robust statistical methods based on minimum-error Bayesian classification. In CW Chen, Y-Q Zhang, eds, Visual Information Representation, Communication and Image Processing, Optical Engineering Series 64. Marcel-Dekker, New York, pp 415–430
- Zoccolan D, Giachetti A, Torre V (2001) The use of optical flow to characterize muscle contraction. J Neurosci Methods 110: 65–80 [DOI] [PubMed] [Google Scholar]