Microorganisms and plants synthesize an immense variety of metabolites, which are generally classified into two major groups based on their function. Primary metabolites are essential for growth and universally used, whereas secondary metabolism is highly diverse and variable and plays a role for the survival of the producing organism within its natural habitat (Demain, 1989; Harborne, 1993; Hartmann, 1996; Croteau et al., 2000; Kutchan, 2001). The study of the biochemistry of the formation of all these compounds is the study of the molecular basis of life (Stryer, 1995), which explains why there is so much ongoing interest in the field. Today, the chemical mechanisms of many central processes are well understood (Michal, 1999), and it is clear that common molecular patterns and principles underlie the diverse forms of life (e.g. the use of the same building blocks to construct macromolecules and the flow of genetic information from DNA to RNA; Stryer, 1995). Nevertheless, distinct differences in the basic biosynthetic pathways exist. For example, plants and microorganisms can synthesize their amino acids from simple building blocks, whereas most animals lost this ability in the course of their adaptation to heterotrophic life. Thus, many biosynthetic pathways are regarded as specific for certain groups of organisms.
The ability to form thousands of structurally diverse natural products due to secondary metabolism is considered a typical feature of plants and microbes. These compounds are usually classified into major groups depending on the basic building blocks of the final structure, e.g. the terpenes that are formed from isoprenoid moieties, or the polyketides, which are assembled from short chain carboxylic acids. These products not only play an important role for their producers in the natural habitat, but also for human health (as opposed to their name indicating “of secondary interest”), because almost 50% of the most important medications are derived from these so-called natural products (Demain, 1999). These encompass not only antibacterials, antivirals, and antitumor compounds, but also products with immunosuppressant, antihypertensive, antidiabetic, antimalarial, and antihypercholesterolemic properties (Strohl, 1997; Grabley and Thiericke, 1999).
Although approximately 170,000 secondary metabolites are known according to the Chapman & Hall dictionary of natural products (see http://www.chemnetbase.com/scripts/dnpweb.exe), there is a clear trend as to which group of organisms produces what type of compounds. For example, alkaloids (e.g. morphine) and phenylpropanoids are considered typical plant metabolites, whereas nonribosomally biosynthesized peptides (e.g. cyclosporin) are regarded as microorganism specific. Thus, a variety of structures, biosynthetic pathways, proteins, and genes are believed to be only found in plants, raising a question of the origin of biosynthetic pathways. Did plants “invent” all of these pathways without using and adapting genetic information from prokaryotes?
In contrast to this generally accepted hypothesis, this update will show that a variety of metabolic pathways that were regarded to be present only in plants have in the recent years also been found in prokaryotes. Microorganisms with a complex life cycle and large genomes (e.g. myxobacteria [Shimkets, 1993; Pradella et al., 2002] and actinomycetes [Bentley et al., 2002]), which can frequently be isolated as soil inhabitants, seem to be particularly rich in such pathways, indicating that some “typical plant pathways” may actually originate from microorganisms. Alternatively, biosynthetic pathway genes might have been taken up by the microbes from plant material, which might be a consequence of their saprophytic lifestyle. One example is the myxobacterium Sorangium cellulosum, which actively degrades cellulose and, therefore, can be isolated from rotting plant material (Reichenbach and Dworkin, 1992). Such a close organismic association could explain the uptake of plant DNA eventually resulting in horizontal gene transfer. Here, we provide information about recent findings, mainly from actinomycetes and myxobacteria, both of which are very potent sources of secondary metabolites among the microorganisms. Because of the immense emerging body of knowledge from genome sequencing, this update is restricted to selected examples of microbial taxa and does not aim at reviewing an all-inclusive list.
PHE AMMONIUM LYASE (PAL), THE FIRST DEDICATED SECONDARY METABOLIC ENZYME IN MANY PLANT AND SOME BACTERIAL PATHWAYS
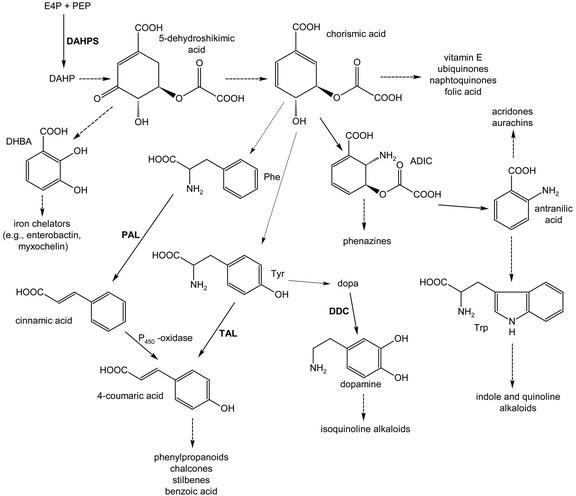
PAL is a ubiquitous enzyme in higher plants; it catalyzes the deamination of l-Phe to trans-cinnamic acid (Fig. 1). Without PAL, our world would be less colorful due to the lack of flavonoids and anthocyanins. Moreover, plants would most likely not exist in their current form due to the lack of lignin. PAL is the first enzyme in the conversion of l-Phe to benzoic acid (Fig. 1), an important structural element in a variety of natural products from plants (e.g. cocaine [Leete, 1990], paclitaxel [Walker and Croteau, 2000], and fungi, e.g. zaragozic acid [Bergstrom et al., 1995]).
Figure 1.
Biosynthesis of several primary and secondary metabolites from plants and/or bacteria. Broken arrows indicate multistep reactions. ADIC, 2-amino-2-desoxy-isochorismate; DAHP, 3-desoxyarabinoheptulosonate-7-phosphate; DAHPS, DAHP synthase; DHBA, 2,3-dihydroxybenzoic acid; E4P, erythrose-4-phosphate; PAL, phenylalanium ammonia; PEP, phosphoenolpyruvate; TAL, tyrosine ammonia.
In prokaryotes, only very few cinnamic and benzoic acid derived metabolites have been described, presumably due to the rarity of PAL in these organisms. However, there is evidence for two PAL independent pathways to benzoate: the anaerobic degradation of l-Phe (Schneider et al., 1997; Breese et al., 1998) and, similar to plants, a benzoate biosynthesis directly from shikimate (Grond et al., 2000).
PAL is highly homologous to His ammonia lyase (HAL), the enzyme catalyzing an analogous deamination of His to trans-urocanate in pro- and eukaryotes and, therefore, initiates His degradation in almost all organisms (Michal, 1999). It is also similar to Tyr ammonia lyase (TAL), a common enzyme of various monocotylic plants that is rarely found in prokaryotes (Kyndt et al., 2002). All three enzymes have the unique prosthetic group 4-methylidene imidazol-5-one (Schwede et al., 1999; Rother et al., 2001), indicating a very similar catalytic mechanism. However, because PAL is the first enzyme in plant phenylpropanoid secondary metabolism, it is the best studied member of this enzyme family (Schuster and Retey, 1995).
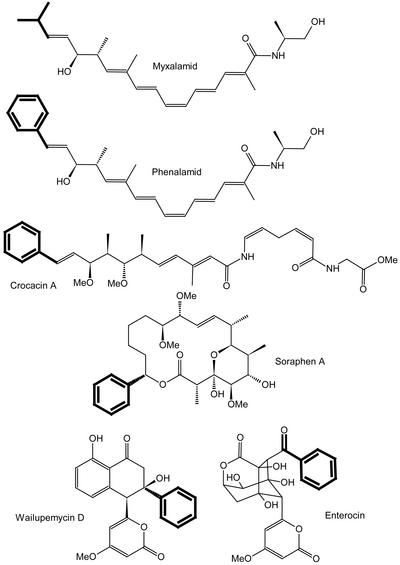
The presence of a PAL activity in prokaryotes was first described in 1970 for Streptomyces verticillatus, which produces cinnamide (Emes and Vining, 1970). Indirectly, PAL activity has been shown by the incorporation of l-Phe and benzoate into the myxobacterial metabolite soraphen (Reichenbach and Höfle, 1994; Hill et al., 1998) and into the wailupemycins and enterocin in “Streptomyces maritimus” (Hertweck and Moore, 2000; Fig. 2). Additional prokaryotic secondary metabolites have been described with a benzoic acid moiety that is used as the polyketide synthase (PKS) starting unit. Two examples from myxobacteria are the crocacins (Jansen et al., 1999) from S. cellulosum and the phenalamids from Myxococcus stipitatus (Trowitzsch-Kienast et al., 1992), which closely resemble the myxalamids (Gerth et al., 1983), but use benzoate instead of short branched-chain carboxylic acids as starter molecules (Fig. 2). Sequencing of the myxalamid biosynthetic gene cluster revealed the presence of a HAL-like gene adjacent to the biosynthetic gene cluster (Silakowski et al., 2001) that might be inactive in the myxalamid producer Stigmatella aurantiaca Sg a15 because no phenalamide is produced. However, assuming a highly similar gene cluster for phenalamide production in M. stipitatus due to the similar chemical structures of both compounds, one would postulate the HAL-like gene to be active in this strain.
Figure 2.
Structures of selected secondary metabolites from myxobacteria and streptomycetes.
The only case in which prokaryotic PAL activity has been clearly correlated to a biosynthetic protein is EncP from S. maritimus, which is needed for enterocin biosynthesis (Xiang and Moore, 2002). Although this enzyme shows higher similarities in size and sequence to prokaryotic HALs than to plant PALs (which is also the case for the above mentioned gene in S. aurantiaca), Phe was unambiguously proven as the substrate of EncP by heterologous expression of encP and production of cinammic acid in Streptomyces coelicolor. Nevertheless, the enzyme awaits a detailed characterization for a comparison of PALs from eukaryotic and prokaryotic sources.
TRUE STEROIDS FROM PROKARYOTES
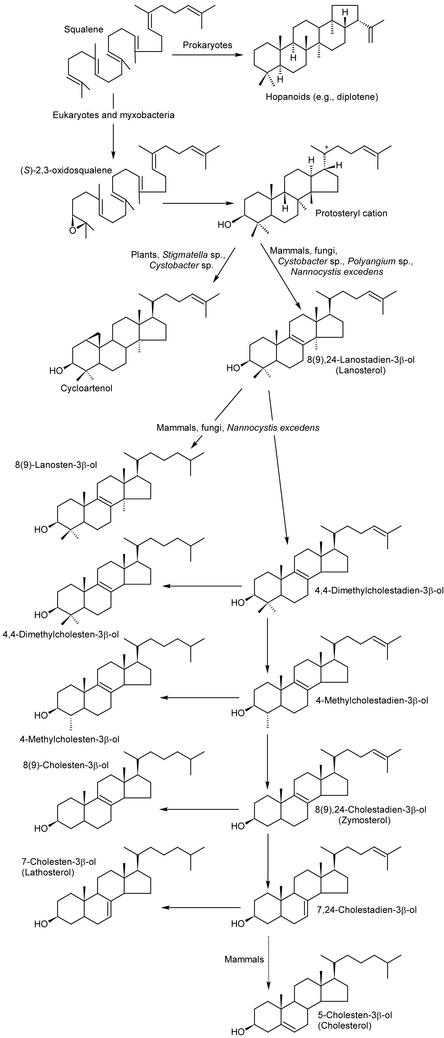
The absence of steroids in prokaryotes has been close to a dogma for many years. Until recently, only two bacterial species were known to have the triterpenes typical for eukaryotes: 4,4-dimethyl, 4-methyl, and 4-desmethylcholesterols in the methylotrophic bacterium Methylococcus capsulatus (Bird et al., 1971) and 8(9)-cholesten-3β-ol in the myxobacterium Nannocystis excedens (Kohl et al., 1983). Instead of steroids, bacteria often employ squalene-derived pentacyclic terpenes of the hopanoid type to build their membranes (Kannenberg and Poralla, 1999). Very recently, a detailed analysis of many myxobacterial strains from different genera revealed the presence of several steroids, which were previously only known from eukaryotes (Bode et al., 2003; Fig. 3).
Figure 3.
Steroid and hopanoid biosynthesis in eukaryotes and prokaryotes. Cholesterol could not be isolated from myxobacteria.
Strains of the myxobacterium N. excedens are potent prokaryotic steroid producers that contain more than 2% steroids in their cellular dry weight and 10 distinct steroids. Almost all intermediates and side products of the biosynthesis of the major steroid in mammals, 5-cholesten-3β-ol (cholesterol; see Fig. 3), except for the final product itself, could be isolated from different N. excedens strains. In addition, the well known plant or phytosterol cycloartenol could be isolated from different species of Stigmatella and Cystobacter. The latter strains produce both lanosterol and cycloartenol, which are the first cyclization products of (S)-2,3-oxidosqualene, giving rise to the huge variety of zoo-/mycosterols and phytosterol, respectively (Fig. 3; Michal, 1999). Phylogenetic analysis of the cycloartenol synthase, whose corresponding gene was described as the first prokaryotic steroid biosynthesis gene from the myxobacterium S. aurantiaca Sg a15, revealed a higher similarity of up to 59% to eukaryotic oxidosqualene cyclases (e.g. cycloartenol synthase, lanosterol synthase, or lupeol synthase) than to prokaryotic squalene/hopene cyclases (42% similarity; Bode et al., 2003). However, almost no similarity could be detected on the DNA level.
Studies with different types of steroid biosynthesis inhibitors underline the differences between the prokaryotic and the eukaryotic enzymes. No inhibition was observed in vivo in myxobacteria by terbinafin, tolnaftat, or AMO1618, well-known inhibitors of the eukaryotic squalene epoxidase. No inhibition of hydroxymethyl glutaryl-CoA reductase with fluvastatin was observed (Bode et al., 2003), indicating structural differences between eukaryotic and bacterial enzymes in mevalonate biosynthesis. Only miconazol, an inhibitor of the eukaryotic P450-dependent steroid C14-demethylase, showed the mode of action known from eukaryotes.
Although Nannocystis strains produce quantities of steroids equal to that in eukaryotes, their function in myxobacteria remains a mystery. No difference in reproduction time, ethanol tolerance, fatty acid composition, or swarming could be observed upon comparing a wild-type strain of S. aurantiaca with a cycloartenol knockout (Bode et al., 2003). This contrasts with the knowledge of hopanoids in bacteria, which are assumed to serve as steroid surrogates. Nevertheless, the physiological function of hopanoids is only poorly understood (Kannenberg and Poralla, 1999), although it is known that they are formed during transition from substrate to aerial hyphae in S. coelicolor A3(2) (Poralla et al., 2000). The strong hydrophobicity of both classes of triterpenoids suggests they are located in the membranes and might be involved in controlling membrane fluidity and rigidity (Ourisson and Nakatani, 1994). Even though myxobacteria contain a sufficiently wide variety of different fatty acids to fine tune their membrane fluidity (Ware and Dworkin, 1973; Mahmud et al., 2002), steroids may also play a role. In addition, steroids could themselves serve as signaling compounds (e.g. like hormones) or as part of signaling molecules, which has been described for the hedgehog family of secreted proteins that are characterized by an essential cholesterol residue (Porter et al., 1996). Additional strains have to be investigated to support this hypothesis.
CHALCONE SYNTHASES (CHSS) AND PKSS ARE COMMON IN PLANTS AND MICROBES
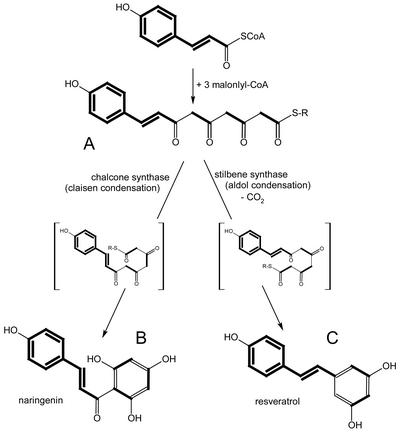
The CHS and stilbene synthase (STS) superfamily of PKSs appears to be ubiquitous in higher plants. STSs, e.g. resveratrol synthase, produce the stilbene backbone as a key reaction in the biosynthesis of stilbene type phytoalexins (Fig. 1). CHS is a key enzyme in the biosynthesis of flavonoids, which exhibit a wide range of biochemical, physiological, and ecological activities. Resveratrol and CHSs condense 4-coumaroyl-CoA with three molecules of malonyl-CoA, which results in products differing in the newly formed ring systems (resveratrol and naringenin chalcone; see Figs. 1 and 4). The same type of condensing reaction is utilized by the 2-ketoacyl-ACP synthases of fatty acid biosynthesis. However, the available data show that these enzymes share little overall homology with either resveratrol synthase or CHS (Schröder, 1999). Over the last couple of years, a series of new additions to the CHS/STS superfamily has been reported from plants, which differ from the enzymes described above by utilizing non-phenylpropanoid starter units, varying numbers of condensation reactions, and different cyclization patterns (e.g. acridone synthase [Junghanns et al., 1995] and 2-pyrone synthases [Eckermann et al., 1998; compare with Fig. 5]).
Figure 4.
Biosynthesis of chalcones and stilbenes indicating their different cyclization mechanisms. Biosynthetic building blocks (coumaryl-CoA and malonyl-CoA) are shown in bold. A to C show differently folded forms of the same tetraketide. R-S, Enzyme-bound thioester.
Figure 5.
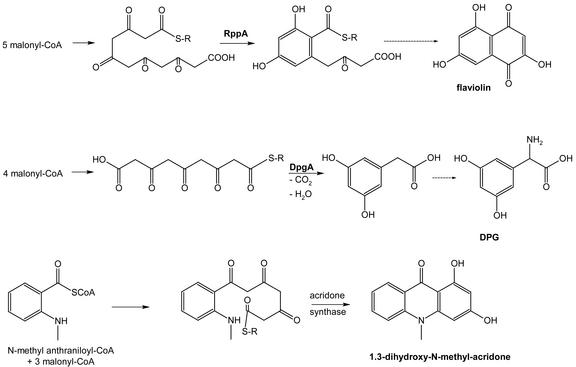
Biosynthesis of flaviolin, (S)-3,5-dihydroxyphenyl-Gly (DPG), and 1,3-dihydroxy-N-methyl-acridone. R-S, Enzyme-bound thioester. RppA and DpgA are CHSs involved in the formation of flaviolin and DPG, respectively.
Unexpectedly, in 1999, “a new type of polyketide biosynthetic pathway” was reported in bacteria. This pathway is used for the assembly of flaviolin (Fig. 5), a small aromatic metabolite in streptomycetes (Funa et al., 1999). At the same time, a similar pathway was shown to be present in pseudomonads. The PKS involved was called “type III PKS” (Bangera and Thomashow, 1999) to distinguish it from the better studied types I and II PKS from microorganisms. The latter enzymes are responsible for the biosyntheses of a large family of structurally diverse natural products with broad ranges of biological activities, reminiscent of fatty acid biosynthesis (Staunton and Weissman, 2001). Phylogenetic analyses of more bacterial type III PKS from whole-genome sequencing (e.g. S. coelicolor, Bacillus subtilis, and Mycobacterium tuberculosis) have made it clear that these enzymes belong to the CHS/STS superfamily of PKSs previously only found in plants (Schröder, 1999; Moore and Höpke, 2001). Some unusual mechanisms of PKS biochemistry have been reported for bacterial type III PKS, e.g. for the enzyme involved in the formation of the non-proteinogenic amino acid DPG (Fig. 5), which is essential for glycopeptide biosynthesis in different actinomycetes (Li et al., 2001; Pfeifer et al., 2001).
Thus far, none of the few characterized bacterial type III PKSs employs the typical plant starter molecule, 4-coumaroyl-CoA (see Figs. 4 and 5), suggesting a distinction between bacterial and plant type III PKS enzymes. This suggestion might in fact be wrong because several bacterial gene products might employ 4-coumaroyl-CoA as a starter molecule. For example, the purple photosynthetic bacterium Rhodospirillum centenum has a photoactive yellow protein (PYP) domain containing the chrompohore 4-coumarate that is part of a bacterial photoreceptor with similarity to plant phytochromes (Jiang et al., 1999; Hellingwerf, 2002). Photoreceptor regulates the expression of a type III PKS in the genome of R. centenum, which suggests that this PKS may be involved in the formation of light-protective flavonoids. In this case, 4-coumaroyl-CoA is indicated as the starter molecule because R. centenum synthesizes this compound. Recently, the characterization of a TAL (see Fig. 1 and the section about PAL-type enzymes) has been reported from Rhodobacter capsulatus, another organism encoding a PYP protein. In this case, the genes encoding TAL and PYP are located adjacent to each other, indicating that the TAL protein is used for the production of the 4-coumaroyl-CoA cofactor of PYP (Kyndt et al., 2002). Two more examples of bacterial type III PKS with close homology to CHS from plants are found in myxobacteria. In the unfinished genome sequences of Myxococcus xanthus DK1622 (see http://www.tigr.org/tdb/mdb/mdbinprogress.html) and S. cellulosum So ce56 (see http://www.genetik.uni-bielefeld.de/GenoMik/cluster6.html), one and two type III PKS genes can be found, respectively. The deduced M. xanthus protein sequence is most similar to a few bacterial proteins of unknown function (up to 36% identities), one of the closest homologs is the R. centenum protein mentioned above (32% identity). However, some plant naringenin synthases show comparable similarities to the deduced M. xanthus protein (R. Müller, unpublished data). The first type III PKS of S. cellulosum seems to encode a typical bacterial protein (up to 61% identity with RppA from different actinomycetes), whereas the second gene product is most similar to few hypothetical bacterial proteins (approximately 35% identities) and plant resveratrol and STSs (approximately 32% identities). Similar to R. centenum, S. cellulosum is able to metabolize Phe into the 4-coumarate analog cinnamate (see Fig. 1), which is used as a precursor of polyketide biosynthesis in some species (Reichenbach and Höfle, 1999). This indicates that the second type III PKS could employ 4-coumaroyl-CoA or cinnamoyl-CoA as a starter molecule. Thus, the latter examples of bacterial type III PKS genes from two myxobacterial species and from R. centenum can be expected to encode bona fide CHS enzymes.
A PROKARYOTIC DOPA-DECARBOXYLASE FROM THE MYXOBACTERIUM S. CELLULOSUM
Dopa-decarboxylase, an enzyme that was considered to be plant and animal specific, can also be found in myxobacteria. It is closely related to plant enzymes and does not group phylogenetically with bacterial amino acid decarboxylases (AADs). AADs belong to the large group of pyridoxalphosphate-dependent enzymes, which contain a conserved domain (protein families database 00282 of the Sanger Centre). They are necessary for the formation of biogenic amines with important physiological properties in animals (Voet and Voet, 1995). In plants, the AADs catalyzing Tyr, l-dopa, and Trp decarboxylation represent branching points from primary into secondary metabolism. Tyramine and dopamine serve as precursors for the formation of a variety of alkaloids such as isoquinoline alkaloids (see Fig. 1, e.g. morphine and codeine; Kutchan and Zenk, 1993; Facchini et al., 2000).
Phylogenetic analysis of nine different types of pyridoxalphosphate-dependent AADs (Sandmeier et al., 1994) grouped the enzymes into four apparently unrelated classes. Group I is represented by Gly decarboxylase; group II is represented by l-Glu decarboxylase, l-His decarboxylase, l-Dopa decarboxylase (DDC), l-Tyr/l-Dopa decarboxylase, and l-Trp decarboxylase; group III is represented by prokaryotic forms of Orn and Lys decarboxylases and the prokaryotic biodegradative type of Arg decarboxylase; and group IV is represented by eukaryotic forms of Orn and Lys decarboxylase and the prokaryotic biosynthetic type of Arg and diaminopimelate decarboxylases.
Group II contains prokaryotic and eukaryotic AADs of different function (but only l-Glu decarboxylase and l-His decarboxylase had been reported from prokaryotes before the Sandmeier study was published), which led the investigators to assume that the divergence between these enzymes occurred before the development of their catalytic specificities. Thus, one would expect a DDC of prokaryotic origin to show the highest similarities to group II AADs of prokaryotic origin, despite possible differences in substrate specificity (Müller et al., 2000).
A DDC encoded in the chromosome of the myxobacterium S. cellulosum So ce90 was found and initially characterized after functional expression in Escherichia coli. Although the function of the corresponding protein in the bacterium is still unclear, it could be shown that the DDC shares more biochemical properties with the subgroup of plant DDCs than with those of animal origin (Müller et al., 2000). Animal enzymes exhibit a preference for l-dopa but also accept the other aromatic amino acids and, to a lesser extent, derivatives thereof (Christenson et al., 1972). In contrast, plant enzymes usually show a more distinct substrate specificity (Kawalleck et al., 1993). Similarily, of six substrates tested, the DDC of S. cellulosum So ce90 only converts l-dopa to dopamine. In contrast to the theory mentioned in the last paragraph, amino acid sequence analyses revealed top scores of DDC from S. cellulosum So ce56 grouping with plant and animal but not with bacterial group-II aromatic amino acid decarboxylases. Phylogenetic analysis of the DDC amino acid sequences suggests a closer link to the plant branch of AADs as well, which indicates that DDC in S. cellulosum is the result of a horizontal gene transfer event (Müller et al., 2000). Hybridization studies with other myxobacteria and data bank searches show that AADs are not commonly found in bacteria. When compared with the list of 189 finished and unfinished bacterial genomes at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi), DDC from S. cellulosum So ce56 shows highest homologies (39% and 37% identities, respectively) only to two other putative AADs, which are encoded in the genomes of the plant symbiont Mesorhizobium loti and the animal pathogen Yersinia pestis. Due to their lifestyle, these two microorganisms could have acquired their corresponding genes in horizontal gene transfer processes as well. All other homologies within the 189 bacterial genomes were to non-AADs.
CONCLUSIONS
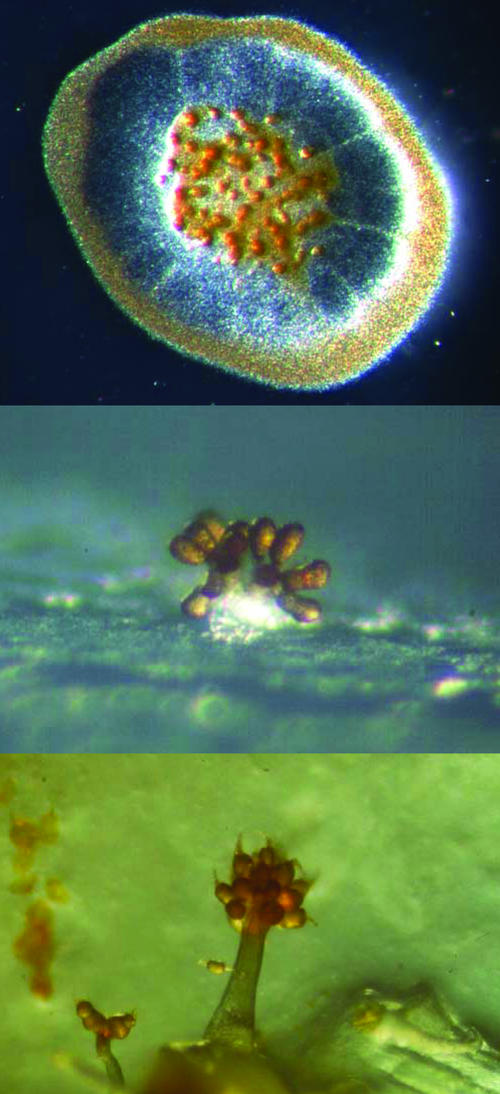
We show the presence of what was formerly regarded as plant-specific biosyntheses and pathways in prokaryotes, mostly in actinomycetes and myxobacteria. The question as to which class of organisms invented these genes, enzymes, and pathways cannot yet be answered because we are only beginning to reveal these features common to prokaryotes and eukaryotes. However, there is certainly a possibility that at least some of the genes that were regarded as typical for plants might originate from prokaryotes. What is the function of these pathways in prokaryotes, and why were they discovered only recently? The answer to the second part of the question is the wealth of information only now becoming available from various sequencing projects of prokaryotic organisms. The first part of the question is more difficult to answer. First, the accumulation of typical plant biosynthetic pathways in myxobacteria and actinomycetes might be due to their saprophytic life style and close proximity to plants in their soil environment as mentioned in the introduction. However, several other soil-inhabiting bacteria have been sequenced (e.g. B. subtilis) that do not show the observed accumulation of plant-like genes. Secondly, myxobacteria and actinomycetes have the largest genomes of all bacteria known so far with 12.2 Mb for S. cellulosum (Pradella et al., 2002) and 8.6 Mb for S. coelicolor (Bentley et al., 2002). These larger sized genomes might have resulted from “gene picking” from various sources during their evolution. However, integrating and maintaining additional genes should result in some benefit for the organism because a large genome requires more energy to maintain. Why are there not more soil-inhabiting genera of prokaryotes with large genomes? Perhaps the reason is that we only know a small fraction of the microorganisms present in the soil environment. This is due to our inability to isolate and grow most of them (Zengler et al., 2002). The lifestyle of myxobacteria and streptomycetes might be an additional reason because they can be regarded as two of the most complex prokaryotes. Myxobacteria build tree-like fruiting bodies (Reichenbach, 1993; Fig. 6) and streptomycetes form aerial mycelium (Hopwood, 1999) under starvation conditions, and their development culminates in the formation of spores for survival under stressful conditions.
Figure 6.
Fruiting bodies of the myxobacteria S. cellulosum (top, photography by Klaus Gerth), S. aurantiaca (middle), and Chondromyces crocatus (bottom, photography by Hans Reichenbach).
These are exciting times to study metabolism in plants and microbes because we are currently gaining a deeper insight into the processes underlying development and evolution. In the future, we will be able to base our research on genome and proteome data, which will hopefully reveal further details of the molecular basis of complex developmental processes and the function of “plant-like” genes in microorganisms.
Acknowledgments
We would like to thank Eric Nudleman and Lars Jelsbak for critical reading of the manuscript and Thomas Hartmann and Dale Kaiser for helpful comments.
This work was supported by the Deutsche Forschungsgemeinschaft (research grant nos. Mu 1254/3, Mu 1254/4, and Bo 1834/1).
References
- Bangera MG, Thomashow LS (1999) Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J Bacteriol 181: 3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147 [DOI] [PubMed] [Google Scholar]
- Bergstrom JD, Dufresne C, Bills GF, Nallin-Omstead M, Byrne K (1995) Discovery, biosynthesis, and mechanism of action of the zaragozic acids: potent inhibitors of squalene synthase. Annu Rev Microbiol 49: 607–639 [DOI] [PubMed] [Google Scholar]
- Bird C, Lynch J, Pirt F, Reid W, Brooks C (1971) Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 230 [DOI] [PubMed]
- Bode HB, Zeggel B, Silakowski B, Wenzel SC, Reichenbach H, Müller R (2003) Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol Microbiol 47: 471–481 [DOI] [PubMed] [Google Scholar]
- Breese K, Boll M, Alt-Morbe J, Schagger H, Fuchs G (1998) Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur J Biochem 256: 148–154 [DOI] [PubMed] [Google Scholar]
- Christenson J, Dairman W, Udenfired S (1972) On the identity of DOPA decarboxylase and 5-hydroxytryptophan decarboxylase. Proc Natl Acad Sci USA 69: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Kutchan T, Lewis N (2000) Natural products (secondary metabolites). In B Buchanan, W Gruissem, R Joneas, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 1250–1268
- Demain A (1989) Functions of secondary metabolites. In C Hirshberger, S Queener, G Hegemann, eds, Genetics and Molecular Biology of Industrial Microorganisms. American Society for Microbiology, Washington, DC, pp 1–11
- Demain A (1999) Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol 52: 455–463 [DOI] [PubMed] [Google Scholar]
- Eckermann S, Schröder G, Schmidt J, Strack D, Edrada RA, Helariutta Y, Elomaa P, Kotilainen M, Kilpeläinen I, Proksch P et al. (1998) New pathway to polyketides in plants. Nature 396: 387–390 [Google Scholar]
- Emes AV, Vining LC (1970) Partial purification and properties of l-phenylalanine ammonia-lyase from Streptomyces verticillatus. Can J Biochem 48: 613–622 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Huber-Allanach KL, Tari LW (2000) Plant aromatic-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54: 121–138 [DOI] [PubMed] [Google Scholar]
- Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S (1999) A new pathway for polyketide synthesis in microorganisms. Nature 400: 897–899 [DOI] [PubMed] [Google Scholar]
- Gerth K, Jansen R, Reifenstahl G, Höfle G, Irschik H, Kunze B, Reichenbach H, Thierbach G (1983) The myxalamids, new antibiotics from Myxococcus xanthus (Myxobacterales): I. Production, physico-chemical and biological properties, and mechanism of action. J Antibiotics 36: 1150–1156 [DOI] [PubMed] [Google Scholar]
- Grabley S, Thiericke R (1999) The impact of natural products on drug discovery. In S Grabley, R Thiericke, eds, Drug Discovery from Nature. Springer, Berlin, pp 3–37
- Grond S, Langer HJ, Henne P, Sattler I, Thiericke R, Grabley S, Zähner H, Zeeck A (2000) Secondary metabolites by chemical screening: 39. Acyl alpha-l-rhamnopyranosides, a novel family of secondary metabolites from Streptomyces sp.: isolation and biosynthesis. Eur J Org Chem 6: 929–937 [Google Scholar]
- Harborne J (1993) Introduction to Ecological Biochemistry, Ed 4. Academic Press, London
- Hartmann T (1996) Diversity and variability of plant secondary metabolism: a mechanistic view. Entomol Exp Applic 80: 177–188 [Google Scholar]
- Hellingwerf KJ (2002) The molecular basis of sensing and responding to light in microorganisms. Int J Gen Mol Microbiol 81: 51–59 [DOI] [PubMed] [Google Scholar]
- Hertweck C, Moore BS (2000) A plant-like biosynthesis of benzoyl-CoA in the marine bacterium “Streptomyces maritimus.” Tetrahedron 56: 9115–9120 [Google Scholar]
- Hill AM, Harris JP, Siskos AP (1998) Investigation of glycerol incorporation into soraphen A. Chem Commun : 2361–2362
- Hopwood DA (1999) Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145: 2183–2202 [DOI] [PubMed] [Google Scholar]
- Jansen R, Washausen P, Kunze B, Reichenbach H, Höfle G (1999) Antibiotics from gliding bacteria: LXXXIII. The crocacins, novel antifungal and cytotoxic antibiotics from Chondromyces crocatus and Chondromyces pediculatus (Myxobacteria): isolation and structure elucidation. Eur J Org Chem 5: 1085–1089 [Google Scholar]
- Jiang Z, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE (1999) Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285: 406–409 [DOI] [PubMed] [Google Scholar]
- Junghanns KT, Kneusel RE, Baumert A, Maier W, Gröger D, Matern U (1995) Molecular cloning and heterologous expression of acridone synthase from elicited Ruta graveolens L. cell suspension cultures. Plant Mol Biol 27: 681–692 [DOI] [PubMed] [Google Scholar]
- Kannenberg EL, Poralla K (1999) Hopanoid biosynthesis and function in bacteria. Naturwissenschaften 86: 168–176 [Google Scholar]
- Kawalleck P, Keller H, Hahlbrock K, Scheel D, Somssich I (1993) A pathogen-responsive gene of parsley encodes tyrosine decarboxylase. J. Biol. Chem. 268: 2189–2194 [PubMed] [Google Scholar]
- Kohl W, Gloe A, Reichenbach H (1983) Steroids from the myxobacterium Nannocystis exedens. J Gen Microbiol 129: 1629–1635 [Google Scholar]
- Kutchan T, Zenk M (1993) Enzymology and molecular biology of benzophenanthridine alkaloid biosynthesis. J Plant Res 3: 165–173 [Google Scholar]
- Kutchan TM (2001) Ecological arsenal and developmental dispatcher. The paradigm of secondary metabolism. Plant Physiology 125: 58–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ (2002) Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett 512: 240–244 [DOI] [PubMed] [Google Scholar]
- Leete E (1990) Recent developments in the biosynthesis of the tropane alkaloids. Planta Med 56: 339–352 [DOI] [PubMed] [Google Scholar]
- Li T, Choroba O, Hong H, Williams D, Spencer J (2001) Biosynthesis of the vancomycin group of antibiotics: characterisation of a type III polyketide synthase in the pathway to (S)-3,5-dihydroxyphenylglycine. Chem Commun 20: 2156–2157 [DOI] [PubMed] [Google Scholar]
- Mahmud T, Bode HB, Silakowski B, Kroppenstedt RM, Xu M, Nordhoff S, Höfle G, Müller R (2002) A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria. J Biol Chem 277: 23768–32774 [DOI] [PubMed] [Google Scholar]
- Michal G (1999) Biochemical Pathways, Vol 1: Spektrum Akad. Verlag, Heidelberg
- Moore BS, Höpke JN (2001) Discovery of a new bacterial polyketide biosynthetic pathway. ChemBioChem 2: 35–38 [DOI] [PubMed] [Google Scholar]
- Müller R, Gerth K, Brandt P, Blöcker H, Beyer S (2000) Identification of an l-dopa decarboxylase gene from Sorangium cellulosum So ce90. Arch Microbiol 173: 303–306 [DOI] [PubMed] [Google Scholar]
- Ourisson G, Nakatani Y (1994) The terpenoid theory of the origin of cellular life: the evolution of terpenoids to cholesterol. Chem Biol 1: 11–23 [DOI] [PubMed] [Google Scholar]
- Pfeifer V, Nicholson GJ, Ries J, Recktenwald J, Schefer AB, Shawky RM, Schroder J, Wohlleben W, Pelzer S (2001) A polyketide synthase in glycopeptide biosynthesis: the biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J Biol Chem 276: 38370–38377 [DOI] [PubMed] [Google Scholar]
- Poralla K, Muth G, Hartner T (2000) Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol Lett 189: 93–95 [DOI] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA (1996) Cholesterol modification of hedgehog signaling proteins in animal development. Science 274: 255–259 [DOI] [PubMed] [Google Scholar]
- Pradella S, Hans A, Sproer C, Reichenbach H, Gerth K, Beyer S (2002) Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch Microbiol 178: 484–492 [DOI] [PubMed] [Google Scholar]
- Reichenbach H (1993) Biology of the myxobacteria: ecology and taxonomy. In M Dworkin, D Kaiser, eds, Myxobacteria II. American Society for Microbiology, Washington, DC, pp 13–62
- Reichenbach H, Dworkin M (1992) The Myxobacteria. In A Balows, HG Trüper, M Dworkin, W Harder, K-H Schleifer, eds, The Procaryotes, Ed 2. Springer-Verlag, New York, pp 3416–3487
- Reichenbach H, Höfle G (1994) Discovery of a new antifungal mechanism of action, soraphen: an almost-success story. In JH Walsdorff, ed, Scientific Annual Report. Gesellschaft für Biotechnologische Forschung mbH, Braunschweig, Germany, pp 5–22
- Reichenbach H, Höfle G (1999) Myxobacteria as producers of secondary metabolites. In S Grabley, R Thieriecke, eds, Drug Discovery from Nature. Springer Verlag, Berlin, pp 149–179
- Rother R, Poppe L, Viergutz S, Langer B, Retey J (2001) Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. Eur J Biochem 268: 6011–6019 [DOI] [PubMed] [Google Scholar]
- Sandmeier E, Hale T, Christen P (1994) Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur J Biochem 221: 997–1002 [DOI] [PubMed] [Google Scholar]
- Schneider S, Mohamed ME, Fuchs G (1997) Anaerobic metabolism of l-phenylalanine via benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Arch Microbiol 168: 310–320 [DOI] [PubMed] [Google Scholar]
- Schröder J (1999) The chalcone/stilbene synthase-type family of condensing enzymes. In D Barton, K Nakanishi, O Meth-Cohn, eds, Comprehensive Natural Products Chemistry, Vol 1. Elsevier, Amsterdam, pp 749–771 [Google Scholar]
- Schuster B, Retey J (1995) The mechanism of action of phenylalanine ammonia-lyase: the role of prosthetic dehydroalanine. Proc Natl Acad Sci U S A 92: 8433–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede TF, Retey J, Schulz GE (1999) Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38: 5355–5361 [DOI] [PubMed] [Google Scholar]
- Shimkets L (1993) The myxobacterial Genome. In M Dworkin, D Kaiser, eds, Myxobacteria II. American Society for Microbiology, Washington, DC, pp 85–108
- Silakowski B, Nordsiek G, Kunze B, Blöcker H, Müller R (2001) Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sg a15. Chem Biol 8: 59–69 [DOI] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ (2001) Polyketide biosynthesis: a millennium review. Nat Prod Rep 18: 380–416 [DOI] [PubMed] [Google Scholar]
- Strohl W (1997) Biotechnology of Antibiotics, Ed 2, Vol 82. Marcel Dekker, New York
- Stryer L (1995) Biochemistry, Ed 4. W. H. Freeman and Company, New York
- Trowitzsch-Kienast W, Forche E, Wray V, Reichenbach H, Jurkiewicz E, Hunsmann G, Höfle G (1992) Phenalamide, neue HIV-Inhibitoren aus Myxococcus stipitatus Mx s40. Liebigs Ann Chem 16: 659–664 [Google Scholar]
- Voet D, Voet J (1995) Biochemistry, Ed 2. Wiley, New York
- Walker K, Croteau R (2000) Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2alpha-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli. Proc Natl Acad Sci USA 97: 13591–13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware J, Dworkin M (1973) Fatty acids of Myxococcus xanthus. J Bacteriol 115: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang LK, Moore BS (2002) Inactivation, complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J Biol Chem 277: 32505–32509 [DOI] [PubMed] [Google Scholar]
- Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M (2002) Cultivating the uncultured. Proc Natl Acad Sci USA 99: 15681–15686 [DOI] [PMC free article] [PubMed] [Google Scholar]