Abstract
The emergence of liquid biopsy using circulating tumor cells (CTCs) as a resource to identify genomic alterations in cancer presents new opportunities for diagnosis, therapy and surveillance. The presented study identified EML4-ALK gene rearrangement in expanded CTCs from one ALK positive lung adenocarcinoma patient. At the time of radiographic progression, CTCs obtained from the patient revealed a drug resistance mutation, L1196M on the ALK gene. CTCs were expanded ex-vivo and drug sensitivity testing was performed using 2 ALK inhibitors, crizotinib and ceritinib. The half maximal inhibitory concentration (IC50) of ceritinib was 1664 nM compared with crizotinib, 2268 nM showing that ceritinib was a more potent ALK inhibitor. We demonstrate that it is feasible to detect serial genetic alterations in expanded CTCs and perform in vitro drug screening. These findings support the clinical utility of CTCs not only for diagnosis, but also a potential tool for drug sensitivity testing in distinct subsets of lung cancer and for personalized precision medicine.
Introduction
Advances in sequencing have shifted the landscape of non-small cell lung cancer (NSCLC) therapy to a personalized approach that is driven by molecular alterations in each patient’s tumor, leading to improved survival for a small minority of patients. 3–7% of NSCLC patients have tumors that contain an inversion in chromosome 2 that juxtaposes the 5′ end of the echinoderm microtubule-associated protein-like 4 (EML4) gene with the 3′ end of the anaplastic lymphoma kinase (ALK) gene, resulting in the novel fusion oncogene EML4-ALK1. Therapy in this group of patients with a specific tyrosine kinase inhibitor, crizotinib, resulted in rapid (within 6 weeks), greater response rates (65%) and longer progression free survival (PFS 7.7 months), compared with chemotherapy2. However, most patients with ALK-positive lung cancer who respond to crizotinib undergo a relapse within months (3.8–21 months) due to emergence of resistance mechanisms3. Among the various resistance mechanisms, gatekeeper mutation in the ALK gene (L1196M) is identified in 9% of patients4. The second-line inhibitor, ceritinib, overcomes this crizotinib-resistant mutation. More recently, third generation ALK inhibitors with improved IC50, targeting wild-type EML4-ALK as well as resistance mutations are in clinical trials, including PF-064639225.
To enhance treatment response, it is imperative to identify the most effective tyrosine kinase inhibitor at the outset as well as identify, by longitudinal testing the emergence of resistance which typifies the molecular evolution of these driver oncogene mediated cancers. At the current time, the practice is to biopsy and re-biopsy at the time of clinical/radiographic progression, which is invasive and not without risk. ALK rearrangement is identified through fluorescence in situ hybridization (FISH) using a break-apart probe6. A separation of a red and a green signal indicates gene rearrangement. In recent years, EML4-ALK rearrangement has been detected in circulating tumor cells (CTCs) from peripheral blood of NSCLC cancer patients7, 8. CTCs can be expanded ex vivo for genotyping and drug testing, as demonstrated in breast and colon cancer9, 10.
Here we present a case study on one advanced stage NSCLC cancer patient harboring EML4-ALK rearrangement in the tumor. CTCs were isolated from the peripheral blood of this patient during three clinical visits. The cells were expanded through a microfluidic co-culture model reported in our previous study11. We found concordant ALK rearrangement and secondary resistance mutation L1196M in the CTCs similar to that in the tumor. Drug testing with crizotinib and ceritinib was also performed on the expanded CTCs.
Results
Primary tumor mutation status
A 42-year-old man with a history of Stage IV Hodgkin’s lymphoma was found to have metastatic lung adenocarcinoma on re-staging scans (Figure 1A). Biopsy of the lung mass and FISH revealed ALK gene rearrangement (Figure 1B). The patient was initially placed on crizotinib. A computerized tomography (CT) scan 4 months after crizotinib revealed near complete resolution of disease. However, a scan at 7 months showed progression in the right hilum (Figure 1A). Patient underwent a bronchoscopy and re-biopsy of the hilar mass. Whole-exome sequencing (70–100× coverage) of the biopsy revealed a secondary mutation L1196M and resultant resistance to crizotinib. He was then treated with ceritinib and his repeat scans revealed a marked decrease in the hilar mass. The patient had serial imaging of the brain with magnetic resonance imaging (MRI). At the time of first disease progression, that was 2 months prior to start of ceritinib treatment, the brain MRI was negative. The next brain imaging occurred when he had neurological symptoms. The patient then had regular brain surveillance with MRI. After 11 months, the patient had a progression of disease with brain metastasis in addition to progression in right lower lobe and right hilum. Whole-exome sequencing of the biopsy of brain metastasis revealed wild-type EML4-ALK and an absence of L1196M mutation. ALK amplification and alternative oncogenic drivers were not detected in the brain biopsy. Table 1 summarizes the mutation status of tumor biopsy at three clinical visits.
Figure 1. Tumor status of the patient.
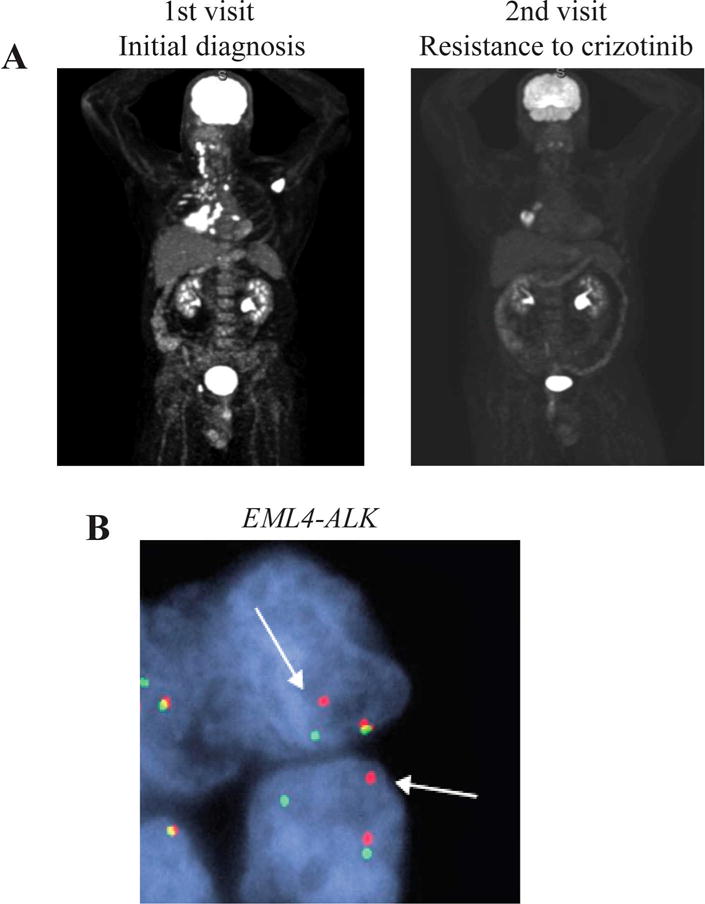
(A) Computed tomography scan at initial diagnosis and when the patient was resistant to crizotinib. (B) EML4-ALK rearrangement is detected in the tumor biopsy specimen at initial visit by FISH (1000× magnification).
Table 1.
Mutation status of primary tumor at 3 time points
| PRIMARY TUMOR | 1st visit | 2nd visit | 3rd visit |
|---|---|---|---|
| Clinical time point | Initial diagnosis | Resistance to crizotinib | Brain metastasis |
| Tumor biopsy site | Lymph node | Right hilum | Brain |
| Gene fusions | EML4-ALK | EML4-ALK | EML4-ALK |
| Somatic point mutations | N/A | ALK (L1196M) gatekeeper mutation- confers resistance to crizotinib | No ALK (p.L1196M) gatekeeper mutation detected |
Detecting ALK rearrangement in expanded CTCs
In parallel, we isolated and expanded CTCs from his blood at the time of initial diagnosis and at the time he developed resistance to crizotinib and then brain metastasis. FISH analysis identified ALK rearrangement in expanded CTCs (Figure 2). At initial diagnosis, 83 cells were counted, 15 cells had 1 copy, 6 cells had 2 copies and 4 cells had 3 or more copies of ALK (30% cells positive). At the time when patient developed resistance to crizotinib, 54 cells were counted, 5 cells had 1 copy, 3 cells had 2 copies of ALK (15% cells positive). When patient developed brain metastasis, 86 cells were counted, 10 cells had 1 copy, 2 cells had 2 copies and 2 cells had 3 or more copies of ALK (16% cells positive). The criteria for ALK positivity is at least 15% cells containing separated signals with at least 50 cells counted12. Accordingly, CTCs were positive for ALK rearrangement at all 3 time points, concordant with findings on tumor biopsies (Table 2).
Figure 2. Mutation analysis of expanded CTCs.
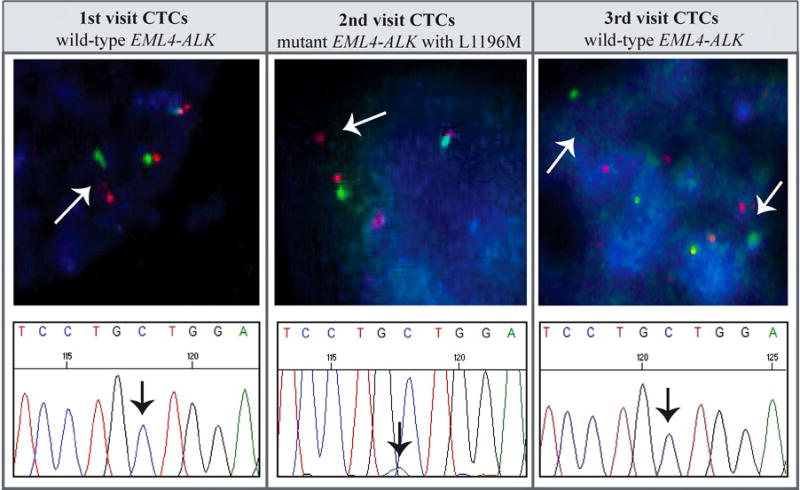
Expanded CTCs, during the three clinical visits, are positive for EML4-ALK rearrangement by FISH (1000× magnification). DNA sequencing reveals L1196M mutation in expanded CTCs when the patient developed resistance to crizotinib (2nd visit) and an absence of the mutation when brain metastasis occurred (3rd visit).
Table 2.
Mutation status of expanded CTCs at 3 time points
| CTCs | 1st visit | 2nd visit | 3rd visit |
|---|---|---|---|
| Clinical time point | Initial diagnosis | Resistance to crizotinib | Brain metastasis |
| Gene fusions | EML4-ALK | EML4-ALK | EML4-ALK |
| L1196M | No | Yes | No |
Identifying L1196M mutation in expanded CTCs
After examining ALK positivity, we performed Sanger sequencing to detect L1196M mutation on the ALK gene. It was found that the gatekeeper mutation emerged in the expanded CTC when the patient developed resistance to crizotinib and had radiographic progression. The L1196M mutation disappeared from expanded CTCs collected at the time of brain metastasis, which again mirrored the findings from the brain biopsy (Figure 2, Table 2).
Drug testing on expanded CTCs
After verifying mutational status of expanded CTCs, we performed in vitro experiments to test CTCs collected and cultured at initial visit against crizotinib and ceritinib. As the CTCs were admixed with cancer-associated fibroblasts (CAFs), we also conducted control experiments on pure CAFs, A549 (resistant negative control) and H3122 (ALK rearranged, sensitive positive control for crizotinib and ceritinib) cell lines. The IC50s to the 2 compounds, using the expanded ALK rearranged CTCs are shown in Figure 3. The IC50 for crizotinib and ceritinib was 2268 nM and 1664 nM respectively.
Figure 3. IC50s of cells treated with crizotinib and ceritinib.
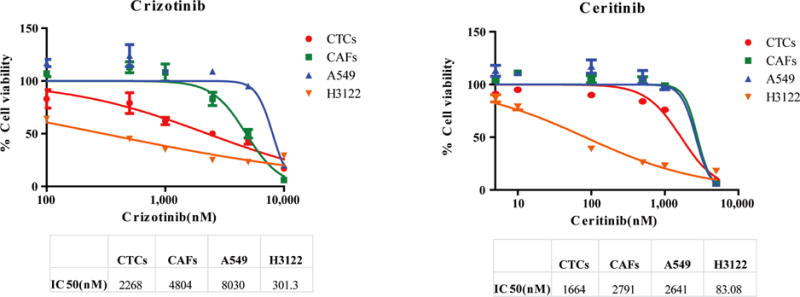
Expanded CTCs are compared with pure CAFs, a resistant cell line A549 and a sensitive cell line H3122.
Discussion
We previously reported a 3D microfluidic co-culture model for expanding small number of CTCs in lung cancer patients11. The present study demonstrates that expanded CTCs carry ALK rearrangements and dynamic detection of drug-resistant mutations by analyzing CTCs longitudinally is possible. The emergence and disappearance of L1196M mutation likely reflects the clonal evolution of tumor cells toward drug resistance against crizotinib and ceritinib. Through expansion of CTCs, drug-susceptibility testing was enabled, offering unique advantages over serum markers and circulating cell-free nucleic acids.
Uniquely, we demonstrate the feasibility of performing drug testing on expanded lung CTCs, which is a challenge due to small numbers of the cells. Performing drug screening by isolating CTCs from peripheral blood is meaningful because it is less invasive than solid tumor biopsy and more accessible. In this study, the IC50 for crizotinib was higher than ceritinib (2268 nM v 1664 nM), as reported by others4. These values were higher than reported IC50s against the H3122 cell line, likely due to the presence of CAFs. However, the ratio of IC50 favored ceritinib as the more potent ALK inhibitor against ALK-rearranged CTCs.
Personalized drug testing predicts therapeutic responses especially in lung cancer. It was reported that 64% of driver mutations were detected in patients with lung adenocarcinoma, of which nearly 30% were selected for targeted therapies13. It is imperative to monitor acquired mutations rendering targeted therapies ineffective early in disease progression. CTCs as “liquid biopsy” can potentially serve as drug testing platforms to guide therapeutic selection.
Materials and Methods
CTC isolation and expansion
Five mL of peripheral blood was drawn from the lung cancer patient into purple top EDTA tubes during each visit. Blood was drawn at University of Michigan Hospital under an IRB-approved protocol. CTCs were captured with a cocktail of antibodies against EpCAM and CD44 in three CTC-capture devices and cultured with CAFs in the devices for two weeks and then outside devices for one month. The detailed procedure can be found in our previous study11.
FISH analysis on CTCs
CTCs were spun onto polylysine-coated slides, and fixed with 1:3 acetic acid and methanol followed by air-drying. FISH test with two-color and break-apart probes were performed on the slides. The probes used were green 5′ ALK probe (RP11-993C21) and red 3′ ALK probe (RP11-984I21). Co-localizing red and green signal indicated normal chromosome and separate red and green signal indicated ALK rearrangement. All images were captured under 100× oil immersion objective using Zeiss Axioplan microscope equipped with CCD camera. Captured images were processed using In situ Imaging System (ISIS) software (Metasystems, Germany).
Drug testing
1000 CTCs, CAFs, A549, or H3122 were plated in each well on a 96-well plate. Each drug concentration had wells in triplicate. Six different concentrations were tested for each drug. Cells were incubated with drugs for 72hrs. After treatment, each well was incubated with Cell Proliferation Reagent WST-1 assay (Roche). Absorbance was measured with Biotek-Synergy Neo-plate Reader. IC50s were determined by nonlinear regression model by Prism Graphpad.
Supplementary Material
Acknowledgments
We acknowledge Ms. Shari Barnett, for her assistance with coordinating blood draws and sample delivery.
Funding source: University of Michigan Cancer Center Support Grant (CCSG) Development (SN and NR) National Institute of Health and United States Department of Defense (SN)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors declare no conflict of interest.
References
- 1.Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:2081–2086. doi: 10.1158/1078-0432.CCR-10-1591. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. The New England journal of medicine. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 3.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. The New England journal of medicine. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 4.Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi L, Janne PA. Crizotinib for ALK-rearranged non-small cell lung cancer: a new targeted therapy for a new target. Clin Cancer Res. 2012;18:3737–3742. doi: 10.1158/1078-0432.CCR-11-2393. [DOI] [PubMed] [Google Scholar]
- 7.Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 8.Faugeroux V, Pailler E, Auger N, et al. Clinical Utility of Circulating Tumor Cells in ALK-Positive Non-Small-Cell Lung Cancer. Front Oncol. 2014;4:281. doi: 10.3389/fonc.2014.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cayrefourcq L, Mazard T, Joosse S, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015;75:892–901. doi: 10.1158/0008-5472.CAN-14-2613. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Shiratsuchi H, Lin J, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014;5:12383–12397. doi: 10.18632/oncotarget.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Shim HS, Kim L, et al. Guideline Recommendations for Testing of ALK Gene Rearrangement in Lung Cancer: A Proposal of the Korean Cardiopulmonary Pathology Study Group. Korean J Pathol. 2014;48:1–9. doi: 10.4132/KoreanJPathol.2014.48.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kris MG, Johnson BE, Berry LD, et al. Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. Jama-J Am Med Assoc. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


