Abstract
CD4 T cells can differentiate into multiple effector subsets, including ThCTL that mediate MHC-II restricted cytotoxicity. Although CD4 T cell mediated cytotoxicity has been reported in multiple viral infections, their characteristics and the factors regulating their generation are unclear, in part due to a lack of a signature marker. We show here that in mice, NKG2C/E identifies the ThCTL that develop in the lung during influenza A virus (IAV) infection. ThCTL express the NKG2X/CD94 complex, in particular the NKG2C/E isoforms. NKG2C/E+ ThCTL are part of the lung CD4 effector population and they mediate IAV-specific cytotoxic activity. The phenotype of NKG2C/E+ ThCTL indicates they are highly activated effectors expressing high levels of binding to P-selectin, T-bet and Blimp-1, and that more of them secrete IFNγ and readily degranulate than non-ThCTL. ThCTL also express more cytotoxicity-associated genes including perforin and granzymes and fewer genes associated with recirculation and memory. They are found only at the site of infection and not in other peripheral sites. These data suggest ThCTL are marked by the expression of NKG2C/E and represent a unique CD4 effector population specialized for cytotoxicity.
INTRODUCTION
Activated CD4 T cells have the potential to differentiate into unique effector subsets tailored to respond to various pathogens. During viral infection, CD4 effectors can become specialized to help antibody responses, to secrete effector cytokines, and to mediate inflammation. These distinct activities are mediated by functionally and phenotypically distinct subsets that develop simultaneously, but in distinct sites and whose generation requires unique instructive signals from distinct antigen-presenting cells (APC) (1) and the microenvironment (2–4). We have studied the cytotoxic subset of CD4 T cells, which we call ThCTL (5) that are found in the lung following influenza A virus (IAV) infection. Although much is understood about how naïve CD4 T cells differentiate into Th1 (6) and TFH (7, 8), and Th17 (9), less is known about how ThCTL are generated and how they become restricted to what seems to be sites of infection.
ThCTL are found in response to multiple viral infections including lymphocytic choriomeningitis virus (LCMV) (10), poxvirus (11), γ-herpesvirus-68 (12), cytomegalovirus (13), as well as IAV (14, 15). After intranasal IAV infection in mice, they are found in the lungs but are absent from secondary lymphoid organs (SLO) (14), while in infections during which virus replicates in other sites, cytotoxic CD4 are reported in those sites (11). Our work and that of others have shown ThCTL generated against IAV infection in mice can lyse infected cells through a perforin-dependent mechanism likely using granular exocytosis (14, 16). They can protect against IAV infection on their own and in synergy with anti-IAV antibody to combat a lethal challenge of IAV (14, 15, 17). Monoclonal ThCTL effectively kill enough infected targets to drive the generation of IAV variants in vivo (17). In other studies, it has been shown that ThCTL correlate with better protection against IAV infection in humans (18) and predict better disease outcome in HIV (19). These potent functions highlight the importance of understanding how ThCTL are generated in order to design effective vaccines to best harness their potential.
ThCTL have unique requirements for generation. In vitro, generation of cytotoxic CD4 effectors requires IL-2, but not known polarizing cytokines such as IL-4, IL-12, IFNγ, TGFβ or IL-6 (20) indicating that a novel pathway regulates their generation. Recent studies show that activating CD137 (4-1BB) (21), CD134 (OX-40) (22), or both (23) can enhance the generation of cytotoxic CD4 T cells that mediate tumor rejection. Further work in an influenza model has demonstrated that ThCTL development and/or function depends on expression of the Blimp-1 transcriptional repressor and type I IFN (16), also consistent with an activated effector phenotype.
Viral infections are very effective at activating cytotoxic CD8 T cells (CTL) that play a key role in clearing virus by killing infected cells. However, viruses often employ evasion mechanisms that prevent CD8 CTL clearance, by downregulating MHC-I expression and presentation of antigen (24). MHC-II restricted cytotoxicity offers an alternate mechanism for host recognition and clearance of virally-infected cells. Indeed several groups have reported key roles for MHC-II restricted cytotoxicity during infection with IAV (25), LCMV (26), Sendai virus (27), and γ-herpesvirus-68 (28). This supports the efficacy of ThCTL killing, especially given the fact that multiple arms of the immune system often work both independently and in synergy to promote clearance of IAV (17, 25, 29, 30) and other viruses (31).
Most studies have defined ThCTL by their ability to mediate MHC-II restricted cytotoxicity or by their expression of high levels of granzyme B (GrB). Although cytotoxicity is the defining feature of ThCTL, the use of functional assays as a marker precludes the phenotyping and tracking of these cells. The serine protease GrB is commonly used to mark cytotoxic cells since among cytotoxic effectors, levels of expression of GrB correlate with cytotoxicity (20). However non-cytotoxic cells can also express GrB that mediates granular exocytosis for cytokine secretion (32), thus it is not a reliable indicator of ThCTL. Another commonly used marker is CD107a (LAMP-1) that identifies cells that have recently degranulated. Identifying ThCTL by staining with CD107a requires stimulating cells in vitro before labeling the cells and then determining if expression is related to cytotoxic function (33). Therefore, CD107a is not useful as a signature phenotypic marker. This lack of a cell surface marker to identify ThCTL has prevented the study of their phenotype and functions and of the pathways that regulate their development. Thus a phenotypic surface marker that identifies unmanipulated ThCTL would greatly facilitate these further analyses.
Here we show that expression of NKG2A/C/E, collectively termed NKG2X (34), identified by antibody clone 20d5 (35), marks cytotoxic ThCTL in IAV infected mice. We show that NKG2X+ effectors express high levels of Blimp-1 and that expression of this transcription factor is required for optimum CD4 effector differentiation to cytotoxic cells in the lung. However, ThCTL do not require NKG2X expression for their MHC-II-restricted cytotoxicity of conventional targets. ThCTL have a phenotype consistent with highly activated effector CD4 T cells and we confirm their localization to the site of infection and show they are not found in other tissue sites. The ThCTL effectors are poised to secrete IFNγ and to degranulate, and they express higher levels of multiple genes associated with increased cytotoxicity than other lung CD4 effectors, and lower levels of genes associated with memory, other CD4 subsets, and re-circulation. Thus we conclude that ThCTL represent a phenotypically and functionally unique cytotoxic subset of CD4 effectors generated at the site of acute viral infection.
MATERIALS AND METHODS
Mice
BALB/cByJ, C57BL/6 (B6), and B6.PL-Thy1a/CyJ (B6.Thy1.1) mice were obtained from The Jackson Laboratory. Prdm1fl/fl mice were originally received from Dr. Alexander Tarakhovsky (The Rockefeller University, New York) and were crossed with Cd4-cre+ (Blimp-1 CKO) (36). Prdm1gfp/+ (Blimp-1 GFP) knock-in mice were originally obtained from Dr. Stephen Nutt (The Walter and Eliza Hall Institute of Medical Research, Australia). Blimp-1 CKO OT-II cells were obtained by crossing Prdm1fl/fl Cd4-cre+ mice with OT-II TcR transgenic (Tg) mice. B6.OT-II.Thy1.1.Hcst−/−/Tyrobp−/− (OT-II.Thy1.1 DAP10/12 KO) cells were obtained by crossing Hcst−/−/Tyrobp−/− mice (kindly provided by Dr. Toshiyuki Takai, Tohoku University and Dr. Lewis Lanier, UCSF) with OT-II.Thy1.1 TcR Tg mice. H2-t23−/− (Qa-1 KO) cells were kindly provided by Dr. Harvey Cantor (Dana Farber Cancer Institute). HNT mice express a TcR recognizing aa 126–138 of A/PuertoRico/8/34 (PR8, H1N1) hemagglutinin (HA) and OT-II mice express a TcR recognizing aa 323–339 of chicken ovalbumin (OVA). OT-II.Thy1.1 TcR Tg and HNT.Thy1.1 TcR Tg mice were obtained from the animal breeding facility at Trudeau Institute or University of Massachusetts Medical School. SMARTA TcR Tg mice (kindly provided by Dr. Raymond Welsh, UMMS) express a TcR recognizing LCMV epitope GP61–80. All mice were at least 8 weeks old at time of infection. Naïve CD4+ cells were obtained from 5–8 week old mice. Experimental animal procedures were conducted in accordance with the UMMS Animal Care and Use Committee guidelines.
Naïve cell isolation
Naïve T cells from TcR Tg mice were obtained from cell suspensions prepared from pooled spleen and lymph nodes as previously described (25). Resulting TcR Tg cells were routinely >97% TcR+ and expressed a characteristic naïve phenotype (small size, CD62Lhi, CD44lo and CD25lo). Purified CD4 cells (0.5−3 × 106) were adoptively transferred in 200 μl PBS by i.v. injection and tracked by expression of congenic markers. Host mice were subsequently infected with virus.
Virus stocks and infections
Mouse-adapted influenza viruses A/Puerto Rico/8/34 (A/PR8), (H1N1) originating from stocks at St. Jude Children’s Hospital and A/PR8-OVAII (H1N1) (kindly provided by Dr. Peter Doherty) were grown in the allantoic cavity of embryonated hen eggs at the Trudeau Institute. Mice were infected intranasally (i.n.) under light isoflurane anesthesia (Piramal Healthcare) with virus in 50 μl PBS. Mice received a 0.3 LD50 dose of IAV. Mice that received adoptively transferred T cells were infected on the same day as cell transfer. For LCMV experiments, mice were infected with 5 × 104 p.f.u. of LCMV Armstrong strain intraperitoneally.
Cell preparation from various organs or tissues
At time points indicated after virus infection, mice were euthanized and lungs, spleen, and draining mediastinal lymph node (dLN) were harvested and single cell suspensions prepared by mechanical disruption of organs and passage through nylon mesh. For some experiments, mice were euthanized and bronchioalveolar lavage (BAL) was also isolated. Then peripheral blood was collected with cardiac puncture and mice were perfused with 10mL of PBS and the lungs, dLN, spleen, kidney, and liver was taken. The lungs, kidney, and liver were digested with collagenase P (Roche) and passed through a nylon membrane. Kidney and liver cell suspensions were layered over percoll (GE Healthcare) and the interface between the 40% and 70% layers was collected.
Flow cytometry and sorting
Cell suspensions were washed, resuspended in FACS buffer (PBS + 0.5% bovine serum albumin and 0.01% sodium azide; Sigma-Aldrich) and incubated on ice with 1 μg anti-FcR (2.4G2) followed by incubation with combinations of fluorochrome-labeled antibodies (Ab) for surface staining: anti - CD4 (GK1.5, RM4-4, RM4-5), CD27 (LG.7F9), CD44 (IM7), CD90.1 (Thy1.1, OX-7 and HIS51), CD103 (2E7), CD150 (SLAM, TC15-12F12.2), CD152 (CTLA-4, UC10-4B9), CD160 (CNX46-3), CD183 (CXCR3, CXCR3-173), CD184 (CXCR4, 2B11), CD186 (CXCR6, SA051D1), CD191 (CCR1, 643854), CD192 (CCR2, SA203G11), CD194 (CCR4, 2G12), CD195 (CCR5, HM-CCR5), CD223 (Lag-3, C9B7W), CD244 (2B4, ebio244F4), CD272 (BTLA, 6F7), CD279 (PD-1, 29F.1A12), CD314 (NKG2D, CX5), CD335 (NKp46, 29A1.4), CD366 (Tim-3, 8B.2C12), CX3CR1 (SA011F11), KLRG1 (2F1), Ly-49A (A1), MHC-II (I-A/I-E, M5/114), NKG2A/C/E (20d5), NKG2A (16a11), NK1.1 (PK136). Ab purchased from eBioscience, BioLegend, BD Biosciences, and R&D systems. Dead cells were excluded using Live/Dead Fixable Amine Dye (Invitrogen). For P-selectin binding, cells were incubated with mouse P-selectin-IgG fusion protein (BD Biosciences), washed and detected with fluorescent goat anti-human IgG (Jackson Immunoresearch) secondary Ab. For NKG2A and NKG2A/C/E co-staining, cells were incubated with anti-NKG2A first, washed and then incubated with anti-NKG2A/C/E to avoid steric blocking. For tetramer staining, cells were stained for 1 h at 37°C with flurochrome conjugated I-Ab-NP311–325 tetramer obtained from the NIH tetramer facility prior to surface marker staining.
For intracellular cytokine staining, donor lung CD4 T cells were isolated with CD90.1 MACS enrichment and were cultured for 4 h with peptide pulsed activated B cells (LPS and dextran sulfate), brefeldin A (Sigma), monensin (BD Golgi Stop), and anti-CD107a (LAMP-1, 1D4B, Biolegend). Cells were then surface stained and fixed for 20 min in 4% paraformaldehyde followed by permeablization for 15 min by 0.1% saponin buffer (PBS plus 1% FBS, 0.1% NaN3 and 0.1% saponin; Sigma-Aldrich) and stained with anti-IFNγ (XMG1.2, eBioscience) for 20 min. Granzyme B (GB11, Thermo Fisher) expression was determined by intracellular staining directly ex vivo. For transcription factor staining, cells were fixed and permeabilized according to manufacturer’s protocol (eBioscience) and then stained with labeled anti-Tbet (4B10), anti-Eomes (Dan11mag), anti-FoxP3 (FJK-16s), anti-Bcl6 (K112-91), anti-ROR(γ)t (B2D), anti-Gata-3 (TWAJ), and anti-ThPOK (2POK). (eBioscience and BD Biosciences).
All flow cytometry results were acquired using LSRII flow cytometers (BD Biosciences) and analyzed with FlowJo (Tree Star) analysis software.
For flow sorting, lungs were pooled and target cells were enriched by magnetic enrichment (MACS, Miltenyi Biotec) for CD90.1 according to manufacturer’s protocol. Enriched cells were then stained to isolate NKG2A/C/E+ or NKG2A/C/Eneg of CD4+ CD90.1+ CD8aneg NK1.1neg I-Ab neg cells using the FACS Aria cell sorter (BD Biosciences). We routinely get 75–90% purity of NKG2A/C/E+ effectors and >95% purity for the NKG2A/C/Eneg effectors. For SMARTA effectors, we achieved >45% purity of NKG2A/C/E+ effectors and >90% purity of NKG2A/C/Eneg effectors.
Cytotoxic Assays
For ex vivo cytotoxic assays, effector cells were isolated either through magnetic enrichment (MACS, Miltenyi Biotec) or by flow sorting from pooled lungs of IAV infected mice. Effectors were pre-incubated with anti-CD178 (Fasl, MFL3, eBioscience). Target cells were generated 2 days prior by stimulating CD19 MACS enriched spleen cells with 25 μg/ml LPS and 25 μg/ml dextran sulfate. Targets were separated into two fractions (targets and bystanders) and labeled with either 1μM or 0.4μM of dye (CFSE or CellTrace Violet, Thermo Fisher). Targets were pulsed with cognate peptide at 5 μM for 1h at 37°C, including OVA323–339 (ISQAVHAAHAEINEAGR), HA126–138 (HNTNGVTAACSHE), NP311–325 (QVYSLIRPNENPAHK) (37), NP216–230 (RIAYERMCNILKGKF) (14), or GP67–80 (IYKGVYQFKSVEFD) (10) all from New England Peptide. Targets and bystanders were mixed at a 1:1 ratio and co-cultured with effectors for 4 h at 37°C and 5% CO2. In some cases, anti-MHC-II Ab (M5/114, BioXcell) was added at 20 μg/ml or anti-NKG2A/C/E (20d5, eBioscience) was added at 10 μg/ml. Plates were then harvested, washed, and stained with Annexin V, 7-AAD (Sytox), or Live/Dead Amine (all Thermo Fisher). Specific killing was calculated as 100 × (1 − (live targets/live bystanders normalized to no effector control wells)). Peptide pulsing and cytotoxic assays were done in complete RPMI media (cRPMI) (RPMI 1640 containing 7.5% fetal bovine serum, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 100 IU penicillin, 100 μg/ml streptomycin and 10 mM HEPES).
For in vivo cytotoxic assays, CD90.2 depleted spleens (MACS) were split into targets and bystanders and labeled and pulsed with peptide like above. Targets and bystanders were mixed at a 1:1 ratio and injected i.v. into host mice. 18 h later, mice were sacrificed and cells from the lungs were isolated for staining. Specific killing was calculated as 100 × (1 − (live MHC-II+ targets/live MHC-II+ bystanders normalized to the ratio found in uninfected mice)).
Intravenous labeling
2.5 μg of anti-CD4 clone (clone RM4-5) was injected i.v. into infected host mice. Mice were euthanized 3–5 min after injection and harvested quickly. Peripheral blood was taken via cardiac puncture and the mouse was perfused with 10 mL of PBS. Organs were taken and single cell suspensions were stained with anti-CD4 (clone RM4-4) as described. Staining of peripheral blood showed CD4 positive (clone RM4-4) cells were also >95% CD4 (clone RM4-5) positive indicating successful i.v. labeling.
Real Time-PCR and microarray analysis
Isolated cell populations from flow sorting were immediately placed in RNA cell protect (Qiagen) and frozen at −80° C until extraction. RNA was extracted (Qiagen) and complementary RNAs were labeled and hybridized onto Affymetrix 2.0 ST arrays according to manufacturer’s protocols. Data was normalized with the RMA algorithm and log transformed using Affymetrix expression console. P-values were generated using unpaired ANOVA in Affymetrix Transcriptome Analysis Console 3.0. Heat maps were generated using Gene-E (The Broad Institute). Data for microarray analysis is deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE89634. Differential expression of selected genes was validated by reverse transcribing RNA and amplifying using Taqman gene expression assays (Thermo Fisher). The fold increase in expression of NKG2A/C/E+ relative to NKG2A/C/Eneg was determined with the “2−ΔΔCT” method.
Statistical analysis
Unpaired, two-tailed, Students t-tests with an ∝ = 0.05, were used to assess whether the means of two normally distributed groups differed significantly. The Welch-correction was applied when variances were found to differ. Paired analysis was done when comparing populations within the same mouse. Two-way ANOVA analyses were performed on log-transformed data with Fisher’s LSD. Analyses were performed using Prism GraphPad or JMP software (SAS Institute). Significance is indicated as * P < 0.05, ** P < 0.005, *** P < 0.001, and **** P < 0.0001. All error bars represent the standard deviation.
RESULTS
Using Blimp-1 to identify ThCTL markers
Since previous studies suggested that Blimp-1 is required during IAV infection for the generation of CD4 effectors with cytotoxic function (16), we reasoned that comparing the phenotype of WT vs. Blimp-1 deficient CD4 effectors might reveal candidate markers of ThCTL. We adoptively transferred either naïve WT (a mix of either B6 or Prdm1+/+Cd4-cre+ mice) or Prdm1fl/flCd4-cre+ (Blimp-1 CKO) (36) OT-II transgenic cells to B6.Thy1.1 mice and infected with the recombinant A/PR8-OVAII virus, which generates a robust CD4 effector response in the lung, draining lymph node, and spleen (38). We have shown previously that ThCTL are found in the lung but not SLO of IAV infected mice (14). Therefore, we isolated CD4 effectors based on their congenic marker Thy1.2 from the lungs 8 days post infection (dpi) and titrated them in an ex vivo flow cytometry-based cytotoxicity assay, which confirmed that T cell intrinsic Blimp-1 was required for optimal induction of cytotoxic CD4 effectors from donor OT-II cells (Fig. 1A). We also found that polyclonal CD4 effectors from Blimp-1 CKO mice infected with A/PR8 demonstrated impaired ex vivo cytotoxicity against targets pulsed with IAV peptides compared to WT at a ratio of 20 effectors to 1 target (Fig. 1B). The polyclonal Blimp-1 CKO effectors at 8 dpi were also defective compared to WT in in vivo IAV MHC-II-restricted killing of activated T-depleted splenocyte targets pulsed with IAV derived peptides (Fig. 1C). Because Blimp-1 was required for optimum development of ThCTL function, we reasoned that it might also be required for their differentiation, so we asked what difference in CD4 effector surface phenotype would be seen when Blimp-1 was absent. At the peak of the CD4 effector response in the lung, 8 dpi, Blimp-1 CKO CD4 effectors had reduced expression of several markers associated with effector T cells compared to WT. Lung CD4 effectors from Blimp-1 CKO mice expressed lower levels of PD-1 and CD27 (Fig. 1D, 1E). We also measured the ability of lung CD4 effectors to bind P-selectin via PSGL-1, a phenotype characteristic of effector T cells that are able to enter infected tissue sites (39). Blimp-1 CKO CD4 lung effectors bound significantly less P-selectin than WT CD4 (Fig. 1F). Taken together, these data indicate that the CD4 effectors generated in the absence of Blimp-1 express lower levels of markers associated with highly differentiated effector T cells and ThCTL, which supports the overall hypothesis that ThCTL are highly differentiated.
FIGURE 1.
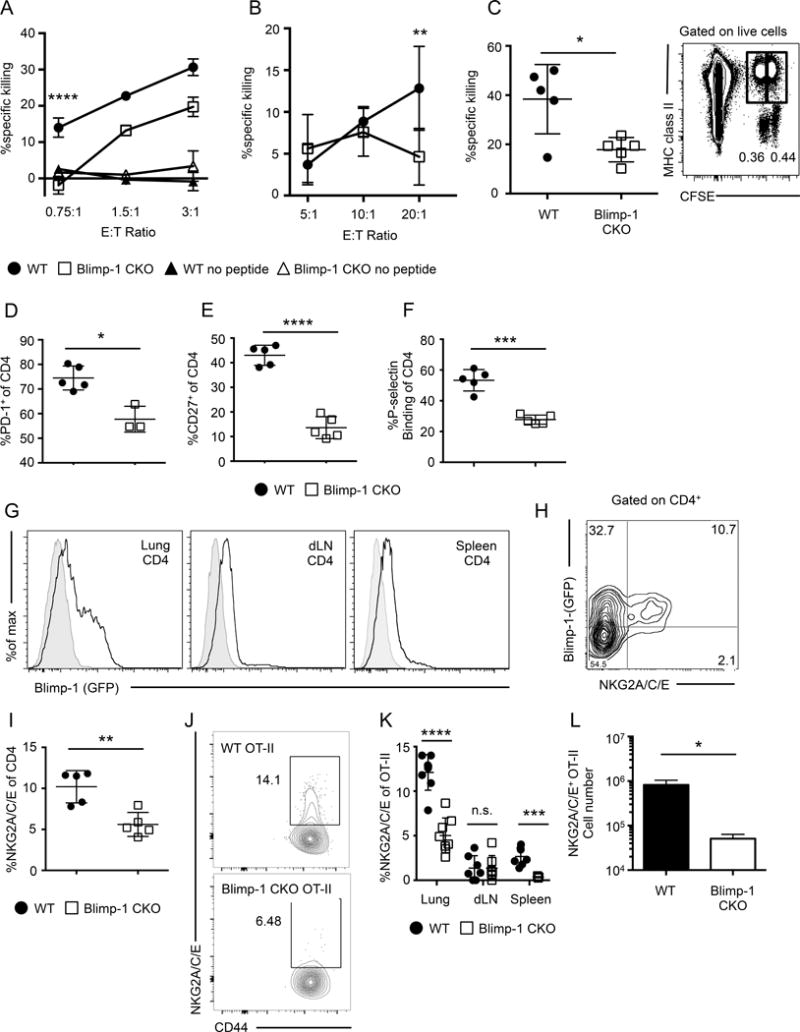
Blimp-1 is required for ThCTL differentiation. (A) WT (circle) or Blimp-1 CKO (square) naïve OT-II CD4 cells were transferred into B6.Thy1.1 mice and infected with A/PR8-OVAII. 8 dpi OT-II effectors were isolated from pooled lungs and assayed for ex vivo cytotoxicity against peptide-pulsed targets (representative of 2 independent experiments, n=5 mice per group each). Significance is shown between WT and Blimp-1 CKO OT-II. (B) CD4 cells isolated from pooled lungs (n = 10–15 mice) from WT or Blimp-1 CKO mice infected with A/PR8 at 8 dpi was assayed for ex-vivo cytotoxicity (pooled data of 2 independent experiments). (C) In vivo killing of peptide-pulsed targets in WT or Blimp-1 CKO mice infected with A/PR8 at 8 dpi (n = 5 mice). Right: Representative flow plot of targets (CFSEloMHC-II+) and bystander cells (CFSEhiMHC-II+). WT or Blimp-1 CKO mice were infected with A/PR8 and the phenotype of lung CD4 cells assayed at 8 dpi for (D) PD-1, (E) CD27, (F) binding to P-selectin (n = 3–5 mice, representative of two independent experiments). (G) Representative histograms of GFP expression of A/PR8-infected Prdm1gfp/+ (open) or Prdm1+/+ (filled) mice at 8 dpi, gated on CD4 from the lung (left), draining lymph node (middle) or spleen (right). (H) Representative flow plot of NKG2A/C/E and Blimp-1 expression of CD4 cells from Blimp-1gfp/+ mice infected with A/PR8 at 8 dpi. (I) WT or Blimp-1 CKO mice were infected with A/PR8 and the percentage of CD4 T cells expressing NKG2A/C/E on 8 dpi was quantified. (J) Same experimental setup as in (A), representative flow plot of NKG2A/C/E expression on WT or Blimp-1 CKO OT-II lung CD4 cells at 8 dpi. Percent (K) NKG2A/C/E+ of OT-II effectors recovered from the lung, dLN, and spleen. (L) Numbers of NKG2A/C/E OT-II effectors recovered from the lung. (J-L n=3–4 mice per group, representative of 2 independent experiments, K= pooled data). Error bars represent SD and significant differences were determined with two-way ANOVA and Fisher’s Least Significant Difference test (A & B) and unpaired two-tailed Student’s t-tests (C–L) where (α = 0.05, * p <0.05, ** p < 0.005, *** p < 0.001, and **** p < 0.0001).
We next measured the expression of Blimp-1 in Prdm1gfp/+ (Blimp-1) reporter mice following A/PR8 infection. We found high Blimp-1 was expressed by CD4 effectors exclusively in the lung and not by other CD4 effectors in the SLO which belong to other subsets (Fig. 1G), indicating that high Blimp-1 correlates with the observed pattern of ThCTL and cytotoxicity that are found exclusively in the lung following IAV infection (14, 16). We searched for markers that co-stained with Blimp-1 expression. Since, cytotoxic CD4 T cells in humans can express NKG2C (40) and highly polarized in vitro Th1 effectors also can express the NKG2 complex (41), we examined the expression of NKG2X (indicating expression of any of the forms: NKG2A, C or E recognized by the 20d5 Ab). Majority of the NKG2X+ CD4 effectors expressed Blimp-1 (Fig. 1H). Thus, we hypothesized that NKG2X might be a signature marker for the ThCTL subset. To test whether Blimp-1 was required for optimum generation of CD4 effectors expressing NKG2X, we infected either WT or Blimp-1 CKO mice and found Blimp-1 CKO polyclonal CD4 lung effectors expressed significantly less NKG2X than WT effectors (Fig. 1I). Moreover, at 8 days post A/PR8-OVAII infection, donor Blimp-1 CKO OT-II lung effectors also expressed reduced NKG2X compared to WT effectors (Fig. 1J, 1K), suggesting the reduced NKG2X is due to a T cell intrinsic loss of Blimp-1. We also saw a reduction in total NKG2X+ OT-II recovered from the lungs (Fig. 1L). Importantly, the NKG2X+ Blimp-1 CKO OT-II cells were not found in the dLN or spleen (Fig. 1K), suggesting they were not stuck in the dLN as has been found for CD8 T cells lacking Blimp-1 (42). We also note the lung exclusivity of the expression of NKG2X (Fig 1G), correlates with Blimp-1 tissue expression and the tissue restriction of CD4 cytotoxicity. Thus, NKG2X marks a population of T cell effectors that express Blimp-1 at the effector stage and require Blimp-1 for their optimal generation in or migration to the lung and thus we postulate that NKG2X is a marker for ThCTL.
NKG2X marks cytotoxic ThCTL
To directly test whether NKG2X expression and ThCTL cytotoxic activity are associated, we sorted lung CD4 effector cells generated from congenically marked OT-II.Thy1.1 donor cells at 8 dpi for expression of NKG2X and compared the ability of positive and negative sorted effectors to kill targets pulsed with specific peptide. The donor NKG2X+ OT-II effectors were effective killers of MHC-II restricted OVA323–339 peptide-pulsed targets even at low effector-to-target ratios, while the NKG2X negative donor OT-II effectors had no detectable peptide-specific cytotoxic activity (Fig. 2A), confirming that the ThCTL activity was exclusively in the sub-population of lung CD4 effector expressing NKG2X. In order to rule out the possibility that NKG2X marks cytotoxic ThCTL of only one transgenic TcR, we also examined the cytotoxic activity of HNT TcR transgenic CD4 effectors that recognize HA126–138 of A/PR8 in the context of I-Ad (43) in BALB/c mice. In this case, naïve congenically marked HNT.Thy1.1 CD4 T cells were adoptively transferred into BALB/c mice infected with A/PR8. A subset of the donor HNT effectors in the lung expressed NKG2X as seen in Figure 3A. HNT CD4 effectors were then sorted after 8 dpi and as with OT-II cells the ThCTL cytotoxic activity was only in the NKG2X+ population (Fig. 2B). This population also co-expressed CD94 the known co-receptor for NKG2X. In at least 5 separate experiments, the cytotoxic activity was found only in the NKG2X-expressing CD4 effectors. Taken together, these data strongly suggest that NKG2X is a reliable signature marker of cytotoxic CD4 effectors in the lung of IAV-infected hosts. In the subsequent experiments, we will refer to NKG2X+ CD4 effectors as ThCTL.
FIGURE 2.
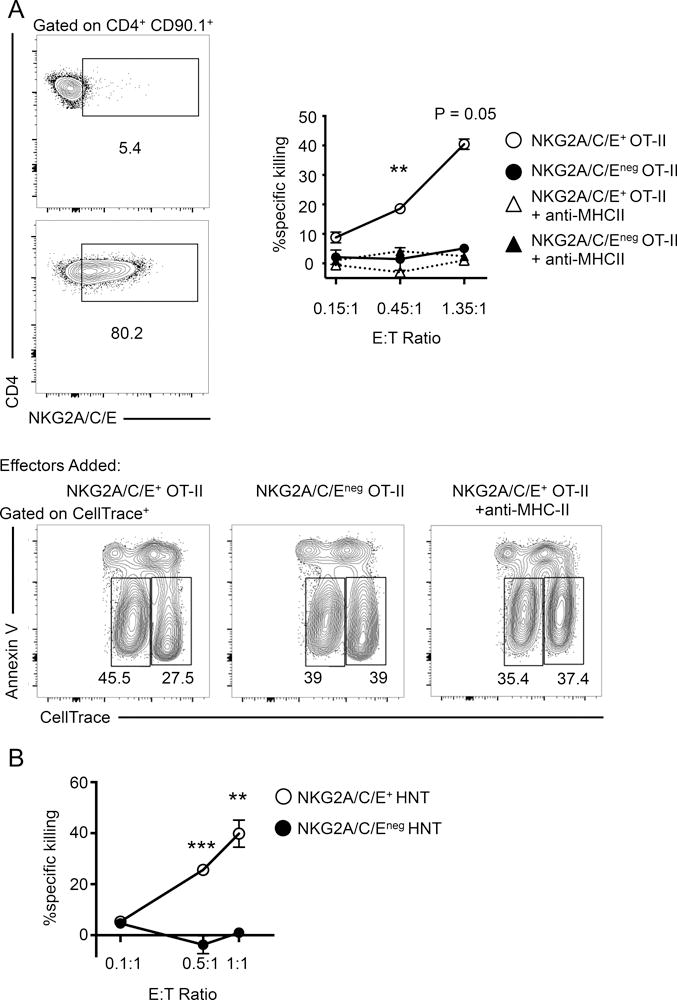
NKG2A/C/E marks cytotoxic ThCTL in the lung. (A) Naïve OT-II.Thy1.1 CD4 T cells were adoptively transferred into B6 mice and infected with A/PR8-OVAII. At 8 dpi lung donor OT-II cells were isolated and sorted based on NKG2A/C/E expression. Left: Sample flow plot of sorted purities of NKG2A/C/Eneg OT-II (top) and NKG2A/C/E+ OT-II (bottom), both are gated on donor OT-II cells (CD90.1+CD4+). Right: NKG2A/C/E+ OT-II (open circle) and NKG2A/C/Eneg OT-II (closed circle) CD4 T cells were assayed in triplicate for cytotoxicity against peptide pulsed targets. Anti-MHC-II antibody was also added to NKGA/C/E+ OT-II (open triangle) or NKG2A/C/Eneg OT-II (closed triangle), one well per E:T ratio. Significance is between NKG2A/C/E+ versus NKG2A/C/Eneg OT-II. Bottom: Representative flow plots from the cytotoxicity assay, gated on CellTrace+ cells, targets (CellTracehi) and bystanders (CellTracelo) after 4 h incubation with NKG2A/C/E+ OT-II effectors (left), NKG2A/C/Eneg OT-II effectors (middle) and NKG2A/C/E+ OT-II effectors with anti-MHC-II antibody (right). (CD4 T cells sorted lungs of n=10 mice and pooled each time, representative of 2 experiments). (B) Naïve HNT.Thy1.1 CD4 T cells were adoptively transferred into BALB/c mice and infected with A/PR8. At 8 dpi donor CD4 cells were isolated from lungs and sorted based on NKG2A/C/E expression. NKG2A/C/E+ HNT (open circle) and NKG2A/C/Eneg HNT (closed circle) CD4 T cells were assayed in triplicate for cytotoxicity against peptide pulsed targets. The E:T ratio of 0.1:1 are single wells. Data are representative of two independent experiments. Error bars represent SD and significant differences were determined with two-way ANOVA and Fisher’s Least Significant Differences test where (α = 0.05, ** p < 0.005, *** p < 0.001).
FIGURE 3.
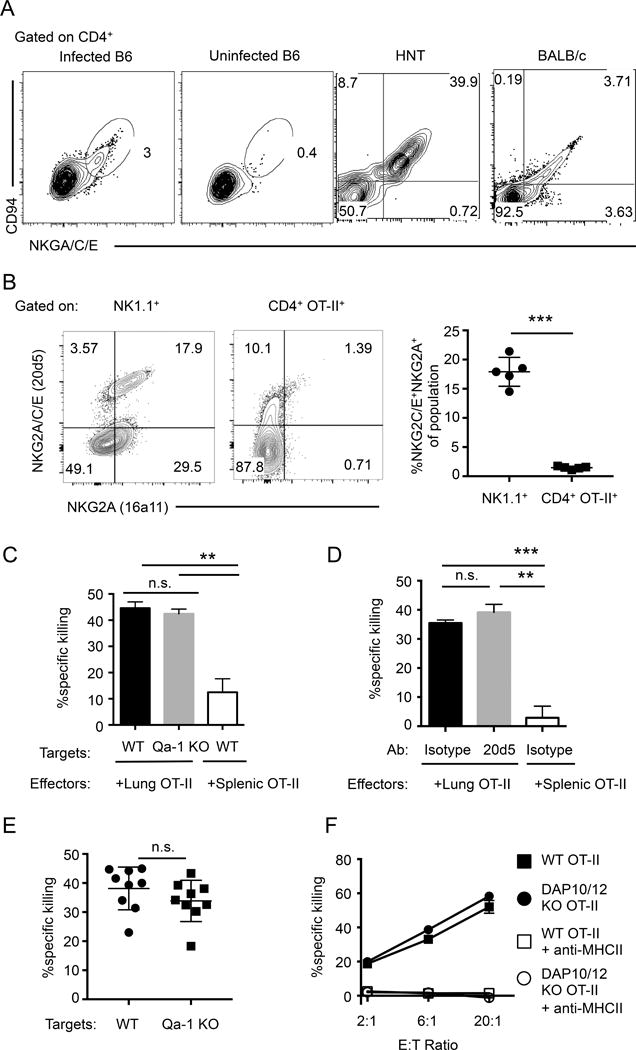
ThCTL express the NKG2C/E isoforms, which is not required for cytotoxicity. (A) Representative plots showing CD94/NKG2A/C/E co-expression on lung CD4+ cells harvested from mice 8 days post A/PR8 infection. Cells were isolated from either infected or uninfected B6 mice (left 2 panels), or infected BALB/c mice (right 2 panels). HNT indicates HNT effectors cells after adoptive transfer of naïve HNT.Thy1.1 CD4 into mice with subsequent infection, gated on CD4+CD90.1+. (B) Left 2 panels: Representative plots of NKG2A and NKG2A/C/E expression, gated on NK1.1 (left) or OT-II CD4 T cells (right) from lungs 8 days post A/PR8-OVAII infection. Right most panel: Quantification of the percent NKG2C/E+NKG2A+ of the indicated populations (representative of 2 independent experiments, n = 5 mice each). (C) Naïve OT-II.Thy1.1 CD4 cells were adoptively transferred into B6 hosts and then infected with A/PR8-OVAII. OT-II.Thy1.1 CD4 effectors from the lungs or spleen 8 dpi were isolated and assayed for ex-vivo cytotoxicity against peptide pulsed B6 targets (WT) or Qa-1 KO targets (CD4 T cells sorted from pooled lungs n=10 mice or pooled spleens n=5 mice, representative of 2 independent experiments). (D) Same OT-II effector generation as in (C), then OT-II.Thy1.1 CD4 T cells isolated from the lungs 8 dpi mice were assayed for ex-vivo cytotoxicity against peptide pulsed targets in the presence of anti-NKG2A/C/E or isotype control antibody (10 μg/ml). (CD4 T cells were isolated from pooled lungs of n=5–10 mice, or pooled spleens of n=2–5 mice, representative of 2 independent experiments). (E) WT or Qa-1 KO targets were transferred into infected host mice generated similarly like in Fig. 3C. 18 h post transfer, the ratio of live targets versus bystanders was quantified with flow cytometry. (Targets generated from pooled mice n=2 each, and data is pooled from 2 independent experiments with n=4–5 mice each). (F) WT (closed square) or DAP10/12 KO (closed circle) OT-II.Thy1.1 naïve CD4 T cells were adoptively transferred into B6 hosts and mice were infected with A/PR8-OVAII. Donor OT-II cells were isolated 8 dpi and assayed for ex-vivo cytotoxicity against peptide pulsed target cells. Anti-MHC-II antibody was added at 20 μg/ml with WT (open square) or DAP10/12 KO (open circle) OT-II CD4 T cells. (CD4 T cells isolated from pooled lungs n=5 mice per group, representative of 2 independent experiments). Error bars represent SD and significant differences were determined with unpaired two-tailed Student’s t-tests where (α = 0.05, ** p < 0.005, *** p < 0.001).
NKG2C or E, but not A are on ThCTL but they do not affect the cytotoxic activity of ThCTL against conventional targets
NKG2X receptors have been found to play a role in modulating cytotoxicity of NK cells (44, 45) and CD8 T cells (34, 46), but their role in CD4 function is less well studied. Since NKG2X forms a heterodimer with CD94, we stained CD94 on B6, BALB/c, and HNT lung effectors. Indeed the effector CD4 T cells generated to IAV co-express NKG2X and CD94 (Fig. 3A). NKG2 has three isoforms; C and E are activating receptors, while the A isoform is an inhibiting receptor for NK cytotoxicity, and all three are recognized by the 20d5 antibody (35) that we use to identify ThCTL. To determine the ThCTL, expression of NKG2A vs. C/E isoforms in the lung, we stained cells with antibody clone 16a11, a specific antibody that recognizes only the A isoform in B6 mice as well as the pan-specific 20d5 antibody. We also stained for NK cells (NK1.1+) as a positive control for NKG2A expression. The majority of 20d5+ NK cells, staining with NK1.1, also expressed NKG2A. In contrast only a few of the lung donor OT-II effectors, including those in the NKG2X+ population, expressed NKG2A and they did so at low levels. This suggests that ThCTL express mainly the NKG2C and E activating receptors (Fig. 3B) and we will from this point describe ThCTL in B6 mice as NKG2C/E+.
We reasoned that engaging the NKG2C/E receptors might costimulate ThCTL killing of MHC-II targets. The primary known ligand for NKG2X receptor is the non-classical MHC Class I molecule Qa-1 (44, 45). To probe whether the NKG2X was playing a role in killing of conventional targets, we assayed the ability of ThCTL to kill either WT targets or those lacking Qa-1 in an ex vivo cytotoxicity assay. The ThCTL effectors lysed WT and H2-t23−/− (Qa-1 KO) (45) target cells derived from activated T-depleted spleen equally well (Fig. 3C). As a second approach to determine if the NKG2C/E receptor is involved in induction or delivery of cytotoxicity, we added the clone 20d5 antibody to block NKG2A/C/E receptor interactions with cognate ligands (35) in the ex vivo cytotoxic assay. Blocking NKG2A/C/E also did not affect ex vivo MHC-II specific cytotoxicity (Fig. 3D). A third approach was to determine if in vivo killing of target cells was also similarly affected by their lack of Qa-1 expression. We isolated target cells from the spleens of either WT or Qa-1 KO mice and adoptively transferred them into host mice with OT-II CD4 effectors and assayed their in vivo cytotoxicity. We found no significant difference between WT and Qa-1 KO target cells in ability to be killed in a peptide specific manner (Fig. 3E). Because the NKG2X complexes signal through the adaptors DAP10 and 12 (47), we next asked if the ability of ThCTL to lyse conventional target cells was lost or reduced when ThCTL were generated from CD4 T cells deficient in both DAP10 and DAP12 adaptors, Hcst−/−/Tyrobp−/− (DAP10/12 KO) (48). Congenically marked, OT-II.Thy1.1 ThCTL were derived by transfer of naïve CD4 from OT-II.Thy1.1 or OT-II.Thy1.1.DAP10/12 KO donors into B6 mice. Hosts were then infected with IAV. When compared in ex vivo cytotoxicity assays, the lysis of target cells by WT and KO effectors was equivalent (Fig. 3F). In contrast, as expected, blocking MHC-II interactions abolished the cytotoxic activity of both WT and DAP10/12 KO effectors. This suggests that neither the generation of ThCTL nor their ability to kill conventional targets depends on the activating signals from NKG2C/E. Taken together, we conclude that ThCTL express NKG2C/E and that this expression is strongly correlated with their ThCTL function, but that the NKG2X receptors are not directly involved in the cytotoxic process when conventional targets are used. Thus we found no evidence that any additional MHC, other than the MHC-II restriction element seen by the OT-II TcR, was needed on conventional target cells for effective ThCTL killing.
ThCTL represent a highly activated lung effector CD4 subset
To test whether a subset of endogenous antigen specific polyclonal effectors express NKG2C/E, we isolated CD4 T cells from lungs of IAV-infected B6 mice and incubated the cells with I-Ab tetramers loaded with IAV NP311–325 peptide. As in previous polyclonal experiments, a fraction of the I-Ab-NP311–325 specific lung CD4 effectors expressed NKG2C/E (Fig. 4A). NKG2C/E+ CD4 effectors were approximately 20% of the lung CD4 effector population, similar to what we see in the donor transgenic models using OT-II and HNT naïve CD4 transfer and in polyclonal effectors in BALB/c mice (Fig. 3A, 3B).
FIGURE 4.
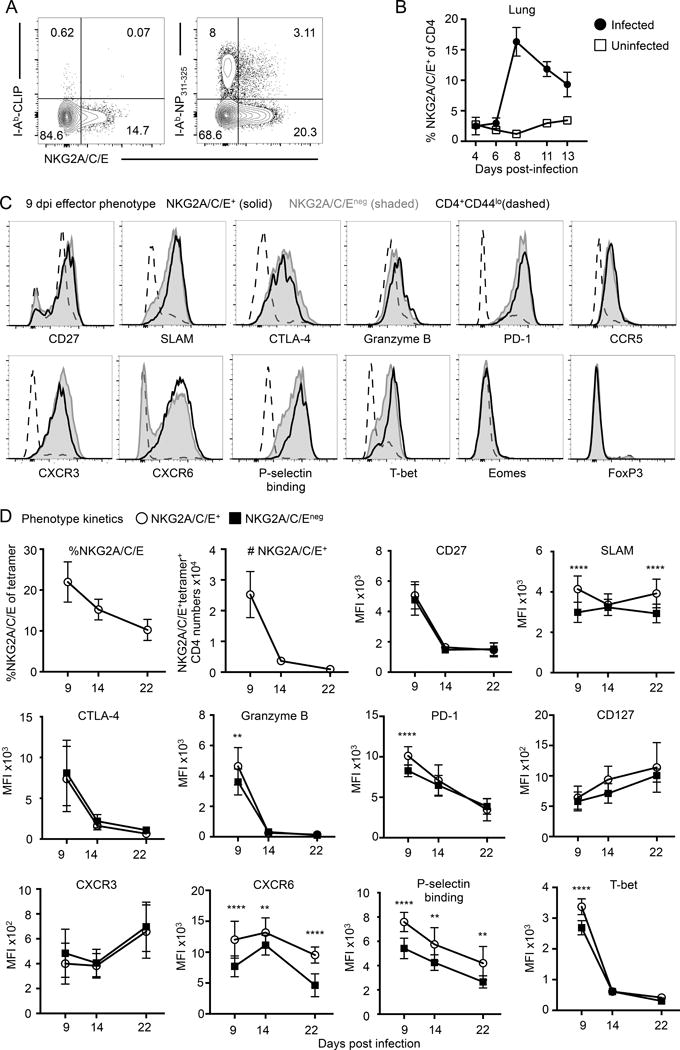
ThCTL are a highly activated lung effector subset. (A) B6 mice were infected with A/PR8-OVAII and lungs were harvested 9 dpi. NP311–325 tetramer staining of NKG2A/C/E+ ThCTL (right) compared to control CLIP loaded tetramer (left) (representative of at least 2 independent experiments, n = 3–5 each). (B) Kinetics of NKG2A/C/E expression on lung CD4 T cells control or A/PR8 infected B6 mice. (C) B6 mice were infected with A/PR8-OVAII and lungs were harvested 9 dpi. Phenotyping analysis of lung CD4 T cells gated on NKG2A/C/E+ (solid black line), NKG2A/C/Eneg (tinted gray), or CD44lo (dashed line). Histograms are normalized to the mode of each population. NKG2A/C/E+ and NKG2A/C/Eneg cells were gated on either CD44hiCD4+ cells (CXCR6 and SLAM) or NP311–325 tetramer+CD4+ (remainder of the plots). (Representative of 2–3 independent experiments, n=3–5 mice each). (D) Phenotype kinetics of CD4 T cells isolated from the lungs of B6 mice at 9, 14, and 22 dpi with A/PR8-OVAII. NKG2A/C/E+ or NKG2A/C/Eneg CD4 T cell expression of selected markers are shown. All plots are on gated NP311–325 tetramer+CD4+, except for CXCR6 and SLAM, which are gated on CD44hiCD4+ cells. (granzyme B and T-bet is representative of at least 2 experiments, remainder of the graphs are pooled from 2 experiments, n=4–5 mice each). Error bars represent SD and significant differences were determined with paired two-tailed Student’s t-tests where (α = 0.05, *** p < 0.001, and **** p < 0.0001).
Using NKG2C/E to identify ThCTL, we analyzed the kinetics of ThCTL generation and compared the phenotype of NKG2C/E+-gated ThCTL and NKG2C/E−-gated, non-ThCTL lung effectors. When naïve B6 mice were infected with A/PR8 and the expression of NKG2X was measured 4,6, 8, 11 and 13 dpi, NKG2C/E-expressing ThCTL peaked around 8–9 dpi and declined thereafter (Fig. 4B), consistent with the peak total effector CD4 response in the lung and with studies of OT-II and HNT transfers (38, 49). Our lab has found that there is extensive inter-organ heterogeneity between CD4 effectors in different sites (49, 50), so to focus on the cytotoxic population, we compared the phenotype of ThCTL vs. non-ThCTL effectors from only the lung at 8–9 dpi (Fig. 4C, 4D). Although ThCTL (NKG2C/E+) had higher levels of granzyme B, non-ThCTL (NKG2C/E−) also expresses a high level compared to naïve cells, confirming the necessity of using markers in addition to granzyme B to identify ThCTL. The lung CD4 effectors, both positive and negative for NXG2C/E, are highly activated as indicated by forward scatter and high levels of activation markers CD27, CTLA-4, GrB and SLAM, an activation marker that is absent on TFH cells (51). Lung effectors express low levels of exhaustion markers BTLA, Lag-3, CD160 and Tim-3 (Supplemental Fig. 1), consistent with high effector function, but express high PD-1, a checkpoint regulator on T cells that is also expressed by many other activated T cells including TFH (1). This pattern connotes high levels of activation and cytotoxicity indicators without most of the signature exhaustion markers (excepting PD-1) that are often associated with terminal differentiation.
We measured various chemokine receptors and found that CD4 lung effectors, both ThCTL and non-ThCTL, express CCR5, CXCR6, and CXCR3, but not CCR1, CCR2, CCR4, CXCR4 and CX3CR1 (Fig. 4C and Supplemental Fig. 1). CCR5 and CXCR3 are associated with migration of activated T cells the lung (52, 53), while CXCR6 may be important for trafficking to inflamed sites (54). In most cases the NKG2C/E+ ThCTL and non-ThCTL had similar chemokine receptor expression patterns except that ThCTL uniformly express high levels of CXCR6 while a fraction of non-ThCTL were negative (Fig. 4C). This pattern of chemokine receptor expression suggests ThCTL and some other lung effectors may be preferentially recruited to particular sites in infected lung tissues where the ligand for CXCR6, CXCL16 is present. Consistent with this idea, ThCTL also had more uniformly high binding to P-selectin-Fc compared to non-ThCTL. P-selectin binding to its ligands is proposed to promote lung CD4 effector entry into specialized peripheral sites (39). The chemokine and adhesion receptor phenotypes of lung effectors, and the enhanced P-selectin binding and CXCR6 levels on ThCTL suggest they may be recruited to a unique tissue niche, which would be consistent with their proposed role of killing IAV-infected epithelial cells.
To further evaluate ThCTL vs. noncytotoxic lung CD4 effectors, we stained for transcription factors expressed by T cells. The ThCTL expressed more T-bet protein than non-ThCTL lung effectors, consistent with the general requirement of T-bet for effector and cytotoxicity in IAV infection (Fig. 4C, 4D) (16). This contrasted with the fact that the NKG2X+ ThCTL were negative for Eomes, Bcl6, and FoxP3 (Fig 4C) and other transcription factors (Supplemental Fig. 1). FoxP3+ Tregs can also be cytotoxic (55), however ThCTL are FoxP3 negative. Type 1 regulatory cells lack FoxP3 expression as well, however these cells often express high levels of Lag-3 (56, 57), which ThCTL are low for at the peak of the infection. The lack of transcription factors associated with the other major CD4 subsets, suggests that few if any of the ThCTL have differentiated down other polarized cytokine pathways including those expressed by Th2 (GATA-3 expressing), TFH (Bcl6-expressing), Th17 (RORγt) or FoxP3+ Treg.
We followed the contraction of CD4 effectors following influenza infection at day 14 post infection and their resulting formation of a early memory pool at day 22 post infection (Fig. 4D). CD4 T cells rapidly become resting so day 22 represents a memory time point (50, 58, 59). The percent and numbers of NKG2C/E+ ThCTL at 2–3 wk (Fig. 4D) suggest that ThCTL CD4 cells are likely retained at the same rate as other lung subsets in this memory pool. Phenotypic analysis of NKG2C/E expressing and negative cells, shows that many effector-associated markers show reduced expression from the peak at 9 to 14 and 22 days, including CD27, CTLA-4, GrB, PD-1, binding to P-selectin and T-bet. In contrast memory markers, CD127 and CXCR3 increase with time. Differences in expression of SLAM, CXCR6 and binding to P-selectin persist, with ThCTL still expressing higher levels through day 22 suggesting they retain stable programming into memory.
ThCTL localize to the site of infection
After their generation in the SLO at about 6 dpi with IAV, CD4 effectors traffic throughout the body (60) much like CD8 T cell effectors and memory cells (61, 62) and become widely dispersed (63). To analyze in more detail sites to which CD4 effectors and in particular ThCTL accumulate, we transferred naïve, congenically-marked OT-II.Thy1.1 CD4 T cells to B6 hosts, infected with A/PR8-OVAII and recovered donor OT-II effectors from various organs at 8 dpi. We found detectable numbers of donor CD4 effectors in all the organs we harvested, confirming that they migrate widely and become dispersed to most tissue sites throughout the body (Fig. 5A, 5B). After i.n. IAV infection, the total effector CD4 cells were found in largest numbers in the spleen and lung (Fig 5B). To track ThCTL, we stained donor effectors for NKG2X. ThCTL were found only in the lung and the bronchoalveolar lavage (BAL) (Fig. 5A, 5C, 5D) and were absent from the liver and blood. There were also significant numbers of ThCTL in the spleen, which had high numbers of total effectors, but they were a low fraction of that population (5C, 5D). When mice are infected i.n. with A/PR8, the virus replicates only in lung epithelial cells (64), suggesting that ThCTL are restricted to the site of infection and/or viral replication. This observation is also consistent with their proposed function of killing infected targets. To further ask whether ThCTL are associated with vascular sites or are to be found in the parenchyma less accessible to the blood, where tissue resident cells reside, we labeled CD4 T cells in vivo with fluorescently labeled monoclonal antibodies introduced i.v (65). We found ThCTL in both the i.v.-shielded and i.v.-labeled portion of CD4 effectors in the lung with a slight enrichment in the shielded fraction (Fig 5E). These data suggest that ThCTL can be found in both vascular and non-vascular associated sites in the lung, with more in the shielded sites.
FIGURE 5.
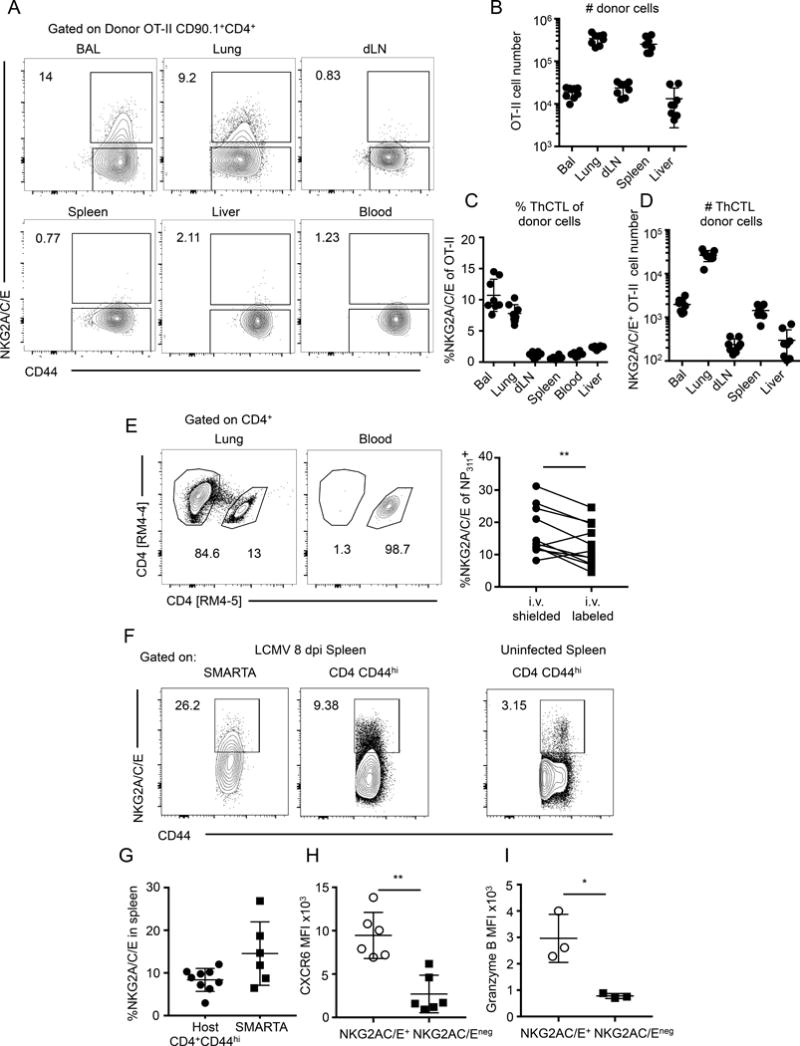
ThCTL localize to the lung, the site of IAV infection. (A) Naïve OT-II.Thy1.1 CD4 T cells were adoptively transferred into B6 mice that were then infected with A/PR8-OVAII. Representative staining of NKG2A/C/E expression on OT-II CD4 T cells recovered. Quantification of (B) donor OT-II CD4 T cells recovered from various organs, (C) percent NKG2A/C/E expression on OT-II CD4 T cells and the (D) number of NKG2A/C/E+ OT-II CD4 T cells recovered from indicated organs. (Pooled from 2 independent experiments, n=3–5 mice each). (E) At 8 dpi mice were i.v. labeled with anti-CD4 antibody and quickly harvested. Top: representative flow plot of lung CD4 T cells. Bottom: quantification of the proportion of i.v. labeled or shielded CD4 T cells expressing NKG2A/C/E. (Pooled from 3 independent experiments, n=3–4 mice each). (F) Naïve SMARTA CD4 T cells were adoptively transferred into congenically marked B6.Thy1.1 mice and then subsequently infected with LCMV Armstrong. At 8 dpi spleens were harvested and stained for ThCTL. Representative staining is shown of NKG2A/C/E expression on donor and host cells in the spleen along with uninfected mouse as control. (G) Quantification of the percent donor or host CD4 T cells expressing NKG2A/C/E. (H) CXCR6 expression on gated NKG2A/C/E positive or negative SMARTA CD4 cells in the spleen. (I) Granzyme B expression on gated NKG2A/C/E positive or negative SMARTA CD4 cells in the spleen. (F-H, data are pooled of 2 independent experiments with n=3–5 mice each. I, data is of one experiment with n=3 mice). Error bars represent SD and significant differences were determined with paired two-tailed Student’s t-tests where (α = 0.05, * p <0.05, ** p < 0.005)
Because ThCTL appear to be restricted to the site of infection, we also looked at CD4 effectors generated against LCMV to test whether we could find ThCTL in the spleen, a main site of LCMV replication(66). We transferred naïve CD4 cells from SMARTA TcR Tg mice into congenically-labeled naïve hosts and subsequently infected with LCMV Armstrong. On 8 dpi we harvested the spleen and assayed for ThCTL formation by staining for NKG2X. In contrast to what we saw after i.n. infection with IAV, we found NKG2X+ CD4 effectors in both the SMARTA and host populations in the spleen (Fig. 5F, 5G). This data suggests ThCTL can form in the spleen when the virus is also replicating in that organ. Further phenotyping shows that the splenic ThCTL generated by LCMV administered i.p., are also high in CXCR6 and GrB expression (Fig. 5H, 5I and Supplemental Figure 2A–C), similar to the ThCTL found restricted to the lung after i.n. IAV infection (Fig. 4C). We sorted NKG2A/C/E+ cells from spleen at 8 dpi with LCMV and found they are enriched for cytotoxic activity against targets expressing the GP67–80 peptide seen by SMARTA TcR Tg and this increase is blocked by Ab to MHC-II (Supplemental Figure 2D), further supporting the hypothesis that ThCTL of similar phenotype, including expression of NKG2A/C/E are generated by multiple viral infections in sites where the virus replicates.
ThCTL secrete IFNγ and degranulate in response to antigen stimulation
ThCTL exhibit cytotoxic function, but it is unclear whether they also secrete IFNγ, a cytokine that contributes to CD4 effector T cell protective function (17). We showed previously that ThCTL do not require IFNγ for their function or generation (15). To evaluate IFNγ production, we stimulated OT-II.Thy1.1 effector cells isolated from the lung at 8 dpi with OVA323–339 pulsed APC, and measured intracellular IFNγ expression in NKG2C/E positive and negative populations. Notably, a high proportion of ThCTL (over 70%) secreted IFNγ and they produced more IFNγ on a per cell basis than the non-ThCTL (Fig. 6A–C). To compare their relative dose response to TcR stimulation for IFNγ production, we stimulated ThCTL and non-ThCTL with a range of peptide doses, using activated splenic APC. We found that compared to the non-ThCTL, more ThCTL secreted more IFNγ over several logs of peptide dose (Fig. 6A–C), suggesting ThCTL are likely somewhat more activated and hence better poised to carry out effector function at lower density of peptide than other lung CD4 effectors.
FIGURE 6.
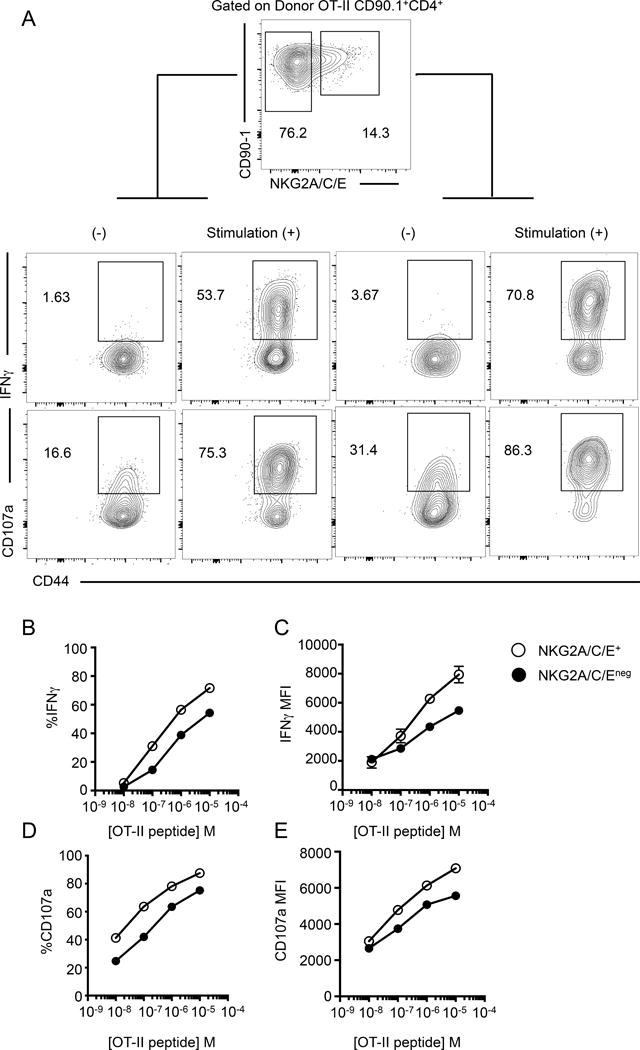
ThCTL show increased IFNγ secretion and degranulation than the non-ThCTL lung CD4 T cells. Lung OT-II.Thy1.1 CD4 T cells were isolated 8 dpi and stimulated with peptide pulsed activated B cells. (A) Representative staining plots of IFNγ and CD107a expression of donor cells after 10−5 M OT-II peptide stimulation. (B) Quantification of the percent IFNγ producing OT-II CD4 T cells gated on NKG2A/C/E+ (open circle) or NKG2A/C/Eneg (closed circle). (C) Median fluorescence values of IFNγ, gated on IFNγ+ cells of indicated populations. (D) Quantification of percent CD107a+ cells. (E) Median fluorescence values of CD107a, gated on CD107a+ cells. (CD4 T cells isolated from pooled lungs, representative of 2 independent experiments, n=5 mice each).
We also assessed the degranulation of lung CD4 effectors by staining for CD107a. Both cytotoxic cells and cells secreting cytokines are known to degranulate (33). ThCTL and non-ThCTL populations degranulated in a peptide-dose dependent manner as measured by induction of CD107a expression. A greater proportion of ThCTL expressed more CD107a compared to non-ThCTL lung effectors across a wide range of peptide doses (Fig. 6D, 6E) consistent with their cytotoxic and higher cytokine production potential. Since those CD4 lung effectors not expressing NKG2C/E have no cytotoxic activity (Fig 2A, 2B) despite having substantial CD107a expression, this confirms that like Grb, CD107a alone is insufficient to identify ThCTL. Thus in addition to possessing all the cytotoxic activity, the ThCTL, defined by NKG2C/E expression, are more readily stimulated to secrete IFNγ and degranulate than other lung effectors.
ThCTL upregulate genes associated with cytotoxic function and down regulate genes associated with recirculation
To further characterize ThCTL, we compared gene expression profiles of isolated ThCTL to non-ThCTL population in the lung. Congenically marked HNT.Thy1.1 lung effectors were flow sorted at 7 dpi from BALB/c mice based on NKG2A/C/E expression and their RNA was isolated for microarray analysis. Importantly, both populations were isolated from the same organ ruling out any tissue dependent differences, and both populations have the same TcR ruling out differences in avidity. Comparing the gene expression profiles revealed that only a limited number of genes differed between the two populations, with 58 genes significantly different (P < 0.05) using a 2-fold relative change cutoff (Fig. 7A). Additionally shown is a heat map of selected genes associated with cytotoxicity and T helper differentiation (67) that did not meet the criteria of significance or 2-fold relative change. The ThCTL population was enriched in expression of genes associated with cytotoxic function including genes encoding perforin (Prf1) and granzymes a, c and f (Gzma, Gzmc, and Gzmf). Tbx21 was upregulated, consistent with the higher levels of T-bet protein in ThCTL (Fig. 4C, 4D). Id2, a transcription factor associated with effector function in CD8 T cells (68, 69), was also up-regulated. Prdm1 was expressed equally in both groups, suggesting Blimp-1 may be required for both ThCTL and non-ThCTL lung effectors. Notably, genes associated with recirculation such as Klf2, Ccr7, and S1p1r (70–72) were downregulated, suggesting ThCTL are not part of the recirculating pool, but instead remain in the lung at least during the peak of the CD4 response when this analysis was done. This may explain the lack of ThCTL found in other peripheral sites. ThCTL also had lower expression of memory-associated genes Tcf7 and Id3 (63, 67) and Il2, which we find is required for the transition to memory (38). This pattern is consistent with a highly differentiated effector phenotype, as non-ThCTL are also full fledged effectors, that may be suppressing genes that would drive the cells to rest and progress to memory.
FIGURE 7.
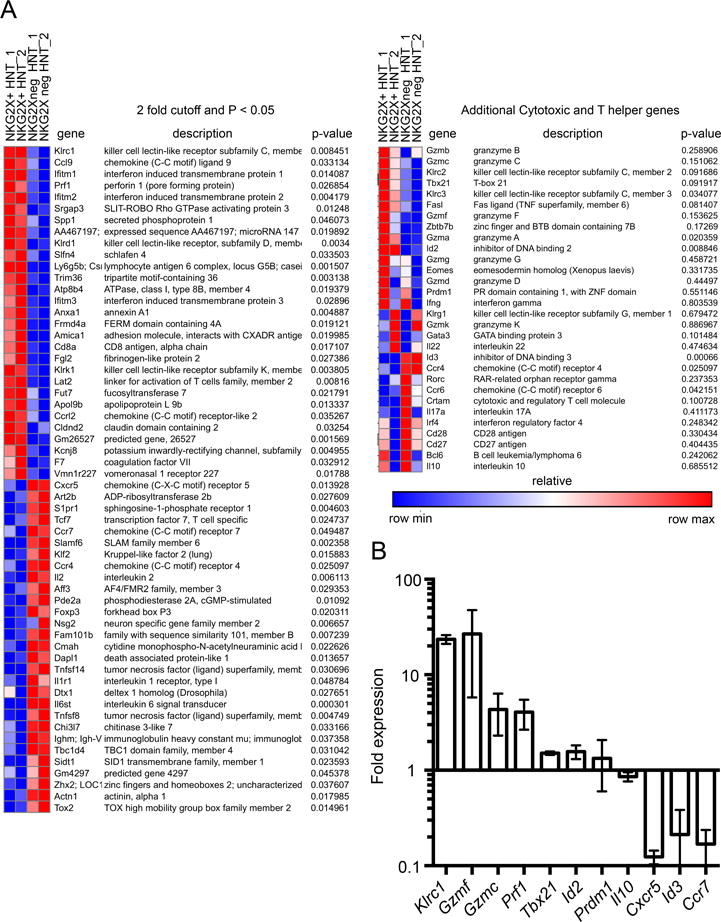
ThCTL express effector genes and suppress circulation and memory genes compared to non-ThCTL. (A) Naïve HNT.Thy1.1 CD4 T cells were adoptively transferred into BALB/cByJ mice that were then infected with A/PR8. Left: Microarray heat map showing gene expression profiles of NKG2A/C/E+ or NKG2A/C/Eneg HNT.Thy1.1 CD4 T cells sorted from pooled lungs at 7 dpi. Right: Additional genes associated with cytotoxicity and T helper differentiation. Microarray ran twice with n=10 mice each time, indicated as 1 & 2. (B) Naïve OT-II.Thy1.1 CD4 T cells were adoptively transferred into B6 mice that were then infected with A/PR8-OVAII. Fold change in expression of indicated genes from quantitative real time PCR expressed by NKG2A/C/E+ over NKG2A/C/Eneg CD4 T cells 8 dpi. Data shown is pooled from 3 independent experiments, with RNA from NKG2A/C/E+ or NKG2A/C/Eneg OT-II CD4 T cells sorted from pooled lungs, n=10 mice each experiment.
We used qRT-PCR to analyze the difference in expression of selected genes in NKG2X-sorted OT-II.Thy1.1 donor effectors to confirm that the differences in expression were also found in another TcR Tg in C57BL/6 mice (Fig. 7B). We noted that by qRT-PCR the NKG2X+ lung effectors expressed less mRNA for CCR7 and CXCR5 relative to the non-ThCTL lung effectors further distinguishing them. Taken together, these data suggest that ThCTL express a cytotoxic effector program compared to other lung effectors, that they express high levels of effector-associated and low levels of resting cell-associated genes and low levels of genes associated with recirculation. This gene expression pattern emphasizes the unique identity of ThCTL compared to other CD4 subsets in the lung.
DISCUSSION
Viral infection induces the generation of a remarkably heterogeneous population of CD4 effectors composed of multiple subsets, each possessing an array of distinct effector functions that play multiple roles in combating pathogens. The functionally distinct subsets generated can be characterized by different polarizing cytokines, by distinct patterns of transcription factors, and by their migration to different locations. CD4 T cells with cytotoxic function, ThCTL, are less well characterized than most CD4 subsets, but their role is likely to become more appreciated as their unique features and functions are uncovered. There is growing appreciation that cytotoxic CD4 T cells play important roles in clearing viral pathogens in humans, including IAV (18), HIV (19) and Dengue virus (74). They have been shown to be important for tumor rejection (21, 22) and protective immunity to viral infections in mice, indicate they warranting further study of this critical subset. Studies of ThCTL have been hampered by the lack of a signature marker to identify the subset.
Our results here establish that ThCTL generated by IAV infection in mice can be identified with a surface marker NKG2A/C/E, collectively called NKG2X. We use this marker to thoroughly characterize ThCTL by phenotype, function, gene expression, and localization. These studies reveal that ThCTL are a unique subset of CD4 T cells that share multiple aspects of a program of cytotoxic effector function with CD8 CTL. Of particular interest is their restriction to the lung and their status as highly activated and functional effectors with strong cytotoxic activity in the lung and BAL.
Staining analysis indicates that ThCTL express mainly NKG2C/E isoforms of the NKG2X family. We show that NKG2C/E is strongly correlated with the cytotoxic function of CD4 T cells and serves as a reliable marker superior to other phenotypes used previously including GrB (16) and CD107a (75), which while expressed by ThCTL, also are present on many non-ThCTL effectors in the lung (Fig. 4 and 6). Using NKG2C/E expression to identify ThCTL and compare them to non-ThCTL in the lung, we found ThCTL express a highly activated effector phenotype including high levels T-bet and Blimp-1 and of multiple indicators of cytotoxic function including high levels of GrB protein and the CD107a degranulation marker as well as granzymes and perforin genes. Functionally, they secrete high levels of IFNγ in response to low peptide dose and express low levels of inhibitory receptors CD160, Lag-3, BTLA and Tim-3.
ThCTL are found restricted to the lung, including the BAL, but are not found in other tissue sites after i.n. IAV infection. Consistent with this finding, ThCTL have increased expression of molecules associated with lung retention including the glycosylated PSGL-1 adhesion molecule that binds P-selectin, CXCR3, CCR5 and CXCR6 while having low levels of CCR7, CXCR5 and S1p1r. Other CD4 effectors in the IAV-infected lung share many of these properties, suggesting a common pathway for effector CD4 T cells that accumulate in sites of infection and viral replication.
It was disappointing that we could find no evidence that NKG2C/E itself is needed for ThCTL differentiation or killing of the targets we tested. It is possible that the putative targets involved in protection, lung epithelial cells infected with IAV, have a distinct requirement for lysis that does require this pathway and we are investigating this possibility. It is also possible that one or more other NK receptor family members are also expressed by ThCTL and are necessary or play redundant roles in cytotoxicity of IAV-infected targets. Another possibility is that NKG2X receptors are needed in some other as yet unidentified context.
Since IAV generates a heterogeneous population of effector CD4 T cells, the fact that all the ThCTL activity was found in the NKG2C/E-positive population indicates that only a fraction of the CD4 effector population in the lung are ThCTL. This suggests that unique signals or programs are in place during CD4 effector differentiation to guide and promote ThCTL generation over other alternate subset pathways, such as non-ThCTL in lung and TFH in follicular sites. Like ThCTL, the TFH represent a tissue-restricted subset, but they contrast sharply with ThCTL, developing in the follicular environment of the SLO, and expressing distinct signature surface markers such as high CXCR5 and they require Bcl-6 but not Blimp-1 (76). Exactly how these functionally-defined subsets (ThCTL and TFH) relate to the cytokine-polarized subsets such as Th1, Th2 and Th17 remains unclear at this time, but the cytokine polarized subsets appear somewhat earlier during the immune response and are generally more widely dispersed, consistent with the idea that the functionally-defined ThCTL and TFH subsets require further differentiation. This is indeed shown to be the case with TFH, that require multiple interactions with antigen-presenting specific B cells first at the border of the follicles and subsequently in the germinal centers to develop into the germinal center-TFH that then drive B cell isotype switching and somatic hypermutation (1). We favor the hypothesis that ThCTL require some form of additional differentiation from CD4 effectors that most likely occurs in the infected tissue, in this case the lung.
The lack of expression of transcription factors associated Th1, Th2, Th17 and FoxP3+ Treg subsets seem most compatible with the idea that ThCTL arise from either non-polarized Th0 or Th1 precursors. Investigators examining ThCTL in other settings have identified suppression of Thpok (67) and the increase of Eomes (21–23) transcription factors as key mediators driving ThCTL in the gut and in a tumor site, respectively. These organs may present a different inflammatory environment than that of an IAV infected lung. IAV infection induces a strong systemic Th1 polarizing environment, and we found that ThCTL, expressed a Th1 cytokine profile with more T-bet than the non-ThCTL lung effectors. This could suggest they are derived from Th1-polarized cells or that multiple CD4 subsets with distinct function share T-bet expression and IFNγ production. We have shown previously that ThCTL can be generated independently of IFNγ (14) and in vitro are optimally generated under Th0 conditions (20), suggesting the cytotoxic program and Th1 cytokine production may be independently regulated.
Our results agree with those of Sun et al. who suggested Blimp-1 is required (16) for ThCTL. We found intrinsic Blimp-1 is required broadly for several features of ThCTL in addition to cytotoxicity. Thus we speculate that ThCTL require unique combinations of transcription factors that drive cytotoxic function as well as retention at the site of infection.
ThCTL control the level of IAV and contribute to protection against lethal IAV challenge. They can synergize with Ab to protect naïve hosts from lethal doses of IAV (15). All of these activities have been shown previously to be perforin dependent. Thus we assume that the key function of ThCTL is to kill infected lung epithelial cells. While normally MHC-II negative, IAV infected lung epithelial cells express MHC-II (14). Indeed compared to other lung effectors ThCTL express high levels of perforin and granzyme. There is a broad array of effector CD4 mechanisms of viral control that develop after IAV infection, and when the response is deconstructed it is clear that at least one of these is IFNγ-mediated (15), so ThCTL may contribute to clearing infected cells through both cytotoxic and IFNγ pathways. While it might seem redundant to mount both CD8 and CD4 cytotoxic cells, we point out that viruses have evolved multiple evasion strategies to escape CD8 cytotoxicity (24) including mechanisms to down-regulate MHC-I. Having CD4 cytotoxic cells provides a way to subvert such strategies. Indeed in cytomegalovirus-based vector vaccinated animals, the immune system targets HLA-E restricted responses as a possible mechanism to counter pathogen immune evasion mechanisms like blocking Class I restricted killing (77), supporting the need for countering viral evasion strategies that target MHC-I expression.
In summary, the data presented here suggests NKG2C/E can be used as a signature marker for ThCTL and that using this to characterize ThCTL reveals their unique surface phenotype, gene expression profile, lung/site infection-restricted location and strongly supports the concept that they represent a unique lineage of CD4 T cells. The fact that many of these properties and phenotypes optimally require Blimp-1, suggests the transcriptional repressor plays a central role in specifying their development. The data showing that they are poised to degranulate and can secrete IFNγ when restimulated with lower peptide dose, combined with their signature ability to kill targets via perforin and that their killing is restricted only by MHC-II, altogether support their ability to perform unique roles in combating viruses and tumors. We predict further identification of the mechanisms involved in their generation will yield further insights into their unique protective functions and additional clues as to how we can harness their potential activities by vaccination or in immune therapies for tumors.
Supplementary Material
Acknowledgments
We would like to thank Dr. Harvey Cantor, Dr. Lewis Lanier, Dr. Toshiyuki Takai, and Dr. Jianmei Leavenworth for reagents and mice. We thank the UMMS flow cytometry core for help with cell sorting and the UMMS genomics core for help with the microarray. We thank all members of the Swain Dutton lab for helpful discussions throughout.
Abbreviations used
- A/PR8
A/Puerto Rico/8/34
- A/PR8-OVAII
A/Puerto Rico/8/34-OVA323–339
- B6
C57BL/6
- BAL
bronchioalveolar lavage
- dLN
draining mediastinal lymph node
- dpi
days post infection
- Grb
granzyme B
- HA
hemagglutinin
- IAV
influenza A virus
- LCMV
lymphocytic choriomeningitis virus
- NP
nucleoprotein
- SLO
secondary lymphoid organs
- Tg
transgenic
- UMMS
University of Massachusetts Medical School
- WT
wild type
Footnotes
Supported by grants from the NIH, including P01AI046539 and R01AI118820 to S.L.S., U19AI109858 to Dr. Raymond M. Welsh, and T32-AI 007349 to A.M.V.
DISCLOSURES
The authors declare no conflict of interest.
References
- 1.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston RJ, Choi YS, Diamond Ja, Yang Ja, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–9. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T Follicular Helper Cells Is Mediated by Interleukin-21 but Independent of T Helper 1, 2, or 17 Cell Lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall NB, Swain SL. Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol. 2011:954602. doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing Signals from the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, Poholek AC, Ditoro D, Yusuf I, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang Y, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 10.Jellison ER, Kim S, Welsh RM. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol. 2005;174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 11.Fang M, Siciliano Na, Hersperger AR, Roscoe F, Hu A, Ma X, Shamsedeen AR, Eisenlohr LC, Sigal LJ. Perforin-dependent CD4+ T-cell cytotoxicity contributes to control a murine poxvirus infection. Proc Natl Acad Sci USA. 2012;109:9983–9988. doi: 10.1073/pnas.1202143109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuller Ka, Flaño E. CD4 T cells mediate killing during persistent gammaherpesvirus 68 infection. J Virol. 2009;83:4700–4703. doi: 10.1128/JVI.02240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma S, Weiskopf D, Gupta A, McDonald B, Peters B, Sette A, Benedict Ca. Cytomegalovirus-specific CD4 T cells are cytolytic and mediate vaccine protection. J Virol. 2016;90:650–658. doi: 10.1128/JVI.02123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DM, Lee S, de la Garcia-Hernandez ML, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol. 2012;86:6792–803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T Cell-Mediated Protection from Lethal Influenza: Perforin and Antibody-Mediated Mechanisms Give a One-Two Punch. J Immunol. 2006;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 16.Hua L, Yao S, Pham D, Jiang L, Wright J, Sawant D, Dent AL, Braciale TJ, Kaplan MH, Sun J. Cytokine-dependent induction of CD4+ T cells with cytotoxic potential during influenza virus infection. J Virol. 2013;87:11884–93. doi: 10.1128/JVI.01461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mckinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW, Swain SL. Memory CD4 + T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest. 2012;122:2847–2856. doi: 10.1172/JCI63689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson TMa, Li CK, Chui CSC, Huang AKY, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu X-N. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:276–282. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 19.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, Rychert J, Rosenberg ES, Piechocka-Trocha A, Brass AL, Brenchley JM, Walker BD, Streeck H. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med. 2012;4:123ra25. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DM, Kamperschroer C, Dilzer AM, Roberts DM, Swain SL. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell Immunol. 2009;257:69–79. doi: 10.1016/j.cellimm.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curran Ma, Geiger TL, Montalvo W, Kim M, Reiner SL, Al-Shamkhani A, Sun JC, Allison JP. Systemic 4-1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J Exp Med. 2013;210:743–755. doi: 10.1084/jem.20121190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschhorn-Cymerman D, Budhu S, Kitano S, Liu C, Zhao F, Zhong H, Lesokhin AM, Avogadri-Connors F, Yuan J, Li Y, Houghton AN, Merghoub T, Wolchok JD. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med. 2012;209:2113–2126. doi: 10.1084/jem.20120532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qui HZ, Hagymasi AT, Bandyopadhyay S, St Rose M-C, Ramanarasimhaiah R, Ménoret A, Mittler RS, Gordon SM, Reiner SL, Vella AT, Adler AJ. CD134 plus CD137 dual costimulation induces Eomesodermin in CD4 T cells to program cytotoxic Th1 differentiation. J Immunol. 2011;187:3555–3564. doi: 10.4049/jimmunol.1101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen JL, Morris CR, Solheim JC. Virus evasion of MHC class I molecule presentation. J Immunol. 2003;171:4473–4478. doi: 10.4049/jimmunol.171.9.4473. [DOI] [PubMed] [Google Scholar]
- 25.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty PC. Clearance of Influenza Virus Respiratory Infection in Mice Lacking Class I Major Histocompatibility Complex-restricted CD8+ T cells. J Exp Med. 1991;174:875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller D, Koller BH, Whitron JL, Lapan KE, Brigman KK, Frelingert JA. LCMV-Specific, Class II-Restricted Cytotoxic T Cells in beta2-Microglobulin-Deficient Mice. Science. 1992;255:1576–1578. doi: 10.1126/science.1347959. [DOI] [PubMed] [Google Scholar]
- 27.Hou S, Doherty PC, Zijlstra M, Jaenisch R, Katz JM. Delayed clearance of Sendai virus in mice lacking class I MHC-restricted CD8+ T cells. J Immunol. 1992;149:1319–1325. [PubMed] [Google Scholar]
- 28.Freeman ML, Burkum CE, Cookenham T, Roberts AD, Lanzer KG, Huston GE, Jensen MK, Sidney J, Peters B, Kohlmeier JE, Woodland DL, van Dyk LF, Sette A, Blackman MA. CD4 T Cells Specific for a Latency-Associated γ-Herpesvirus Epitope Are Polyfunctional and Cytotoxic. J Immunol. 2014;193:5827–5834. doi: 10.4049/jimmunol.1302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topham DJ, Doherty PC. Clearance of an influenza A virus by CD4+ T cells is inefficient in the absence of B cells. J Virol. 1998;72:882–885. doi: 10.1128/jvi.72.1.882-885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mozdzanowska K, Furchner M, Maiese K, Gerhard W. CD4+ T cells are ineffective in clearing a pulmonary infection with influenza type A virus in the absence of B cells. Virology. 1997;239:217–225. doi: 10.1006/viro.1997.8882. [DOI] [PubMed] [Google Scholar]
- 31.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afonina IS, Cullen SP, Martin SJ. Cytotoxic and non-cytotoxic roles of the CTL/NK protease granzyme B. Immunol Rev. 2010;235:105–116. doi: 10.1111/j.0105-2896.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- 33.Wolint P, Betts MR, Koup RA, Oxenius A. Immediate Cytotoxicity But Not Degranulation Distinguishes Effector and Memory Subsets of CD8+ T Cells. J Exp Med. 2004;199:925–936. doi: 10.1084/jem.20031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapaport AS, Schriewer J, Gilfillan S, Hembrador E, Crump R, Plougastel BF, Wang Y, Le Friec G, Gao J, Cella M, Pircher H, Yokoyama WM, Buller RML, Colonna M. The Inhibitory Receptor NKG2A Sustains Virus-Specific CD8+ T Cells in Response to a Lethal Poxvirus Infection. Immunity. 2015;43:1112–1124. doi: 10.1016/j.immuni.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vance RE, Jamieson AM, Raulet DH. Recognition of the Class Ib Molecule Qa-1 b by Putative Activating Receptors CD94/NKG2C and CD94/NKG2E on Mouse Natural Killer Cells. J Exp Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin H, Kapoor V, Guan T, Kaech S, Welsh R, Berg L. Epigenetic modifications induced by blimp-1 regulate CD8+ T cell memory progression during acute virus infection. Immunity. 2013;39:661–675. doi: 10.1016/j.immuni.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crowe SR, Miller SC, Brown DM, Adams PS, Dutton RW, Harmsen AG, Lund FE, Randall TD, Swain SL, Woodland DL. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24:457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 38.Mckinstry KK, Strutt TM, Bautista B, Zhang W, Kuang Y, Cooper AM, Swain SL. Effector CD4 T-cell transition to memory requires late cognate interactions that induce autocrine IL-2. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haddad W, Cooper CJ, Zhang Z, Brown JB, Zhu Y, Issekutz A, Fuss I, Lee H, Kansas GS, Barrett Ta. P-selectin and P-selectin glycoprotein ligand 1 are major determinants for Th1 cell recruitment to nonlymphoid effector sites in the intestinal lamina propria. J Exp Med. 2003;198:369–377. doi: 10.1084/jem.20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaguia F, Saikali P, Ludwin S, Newcombe J, Beauseigle D, McCrea E, Duquette P, Prat A, Antel JP, Arbour N. Cytotoxic NKG2C+ CD4 T cells target oligodendrocytes in multiple sclerosis. J Immunol. 2013;190:2510–2518. doi: 10.4049/jimmunol.1202725. [DOI] [PubMed] [Google Scholar]
- 41.Meyers JH, Ryu A, Monney L, Nguyen K, Greenfield Ea, Freeman GJ, Kuchroo VK. Cutting edge: CD94/NKG2 is expressed on Th1 but not Th2 cells and costimulates Th1 effector functions. J Immunol. 2002;169:5382–5386. doi: 10.4049/jimmunol.169.10.5382. [DOI] [PubMed] [Google Scholar]
- 42.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 Transcription Factor Is Required for the Differentiation of Effector CD8+ T Cells and Memory Responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Scott B, Liblau R, Degermann S, Marconi LA, Ogata L, Caton AJ, McDevitt HO, Lo D. A role for non-MHC genetic polymorphism in susceptibility to spontaneous autoimmunity. Immunity. 1994;1:73–82. doi: 10.1016/1074-7613(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 44.Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A Is a Natural Killer Cell Receptor for. J Exp Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L, Ikizawa K, Hu D, Werneck MBF, Wucherpfennig KW, Cantor H. Regulation of Activated CD4+ T Cells by NK Cells via the Qa-1-NKG2A Inhibitory Pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moser JM, Gibbs J, Jensen PE, Lukacher AE. CD94-NKG2A receptors regulate antiviral CD8(+) T cell responses. Nat Immunol. 2002;3:189–195. doi: 10.1038/ni757. [DOI] [PubMed] [Google Scholar]
- 47.Saether PC, Hoelsbrekken SE, Fossum S, Dissen E. Rat and mouse CD94 associate directly with the activating transmembrane adaptor proteins DAP12 and DAP10 and activate NK cell cytotoxicity. J Immunol. 2011;187:6365–6373. doi: 10.4049/jimmunol.1102345. [DOI] [PubMed] [Google Scholar]
- 48.Inui M, Kikuchi Y, Aoki N, Endo S, Maeda T, Sugahara-Tobinai A, Fujimura S, Nakamura A, Kumanogoh A, Colonna M, Takai T. Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc Natl Acad Sci USA. 2009;106:4816–4821. doi: 10.1073/pnas.0900463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strutt TM, McKinstry KK, Kuang Y, Bradley LM, Swain SL. Memory CD4+ T-cell-mediated protection depends on secondary effectors that are distinct from and superior to primary effectors. Proc Natl Acad Sci USA. 2012;109:E2551–E2560. doi: 10.1073/pnas.1205894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Román E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G, Swain SL. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med. 2002;196:957–968. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohlmeier JE, Miller SC, Smith J, Lu B, Gerard C, Cookenham T, Roberts AD, Woodland DL. The Chemokine Receptor CCR5 Plays a Key Role in the Early Memory CD8+ T Cell Response to Respiratory Virus Infections. Immunity. 2008;29:101–113. doi: 10.1016/j.immuni.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohlmeier JE, Cookenham T, Miller SC, Roberts AD, Christensen JP, Thomsen AR, Woodland DL. CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J Immunol. 2009;183:4378–4384. doi: 10.4049/jimmunol.0902022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, Greenberg HB, Butcher EC. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, Ley TJ. Granzyme B and Perforin Are Important for Regulatory T Cell-Mediated Suppression of Tumor Clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Yao Y, Vent-Schmidt J, McGeough MD, Wong M, Hoffman HM, Steiner TS, Levings MK. Tr1 Cells, but Not Foxp3+ Regulatory T Cells, Suppress NLRP3 Inflammasome Activation via an IL-10-Dependent Mechanism. J Immunol. 2015;195:488–497. doi: 10.4049/jimmunol.1403225. [DOI] [PubMed] [Google Scholar]
- 57.Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, Guo B, Herbert DR, Bulfone A, Trentini F, Di Serio C, Bacchetta R, Andreani M, Brockmann L, Gregori S, Flavell Ra, Roncarolo M-G. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. 2013;19:739–746. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 58.McKinstry KK, Golech S, Lee W, Huston G, Weng N, Swain SL. Rapid default transition of CD4 T cell effectors to functional memory cells. J Exp Med. 2007;204:2199–2211. doi: 10.1084/jem.20070041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bautista BL, Devarajan P, McKinstry KK, Strutt TM, Vong AM, Jones MC, Kuang Y, Mott D, Swain SL. Short-Lived Antigen Recognition but Not Viral Infection at a Defined Checkpoint Programs Effector CD4 T Cells To Become Protective Memory. J Immunol. 2016;197:3936–3949. doi: 10.4049/jimmunol.1600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrewala JN, Brown DM, Lepak NM, Duso D, Huston G, Swain SL. Unique ability of activated CD4+ T cells but not rested effectors to migrate to non-lymphoid sites in the absence of inflammation. J Biol Chem. 2007;282:6106–6115. doi: 10.1074/jbc.M608266200. [DOI] [PubMed] [Google Scholar]
- 61.Masopust D, Vezys V, Usherwood EJ, Cauley LS, Olson S, Marzo AL, Ward RL, Woodland DL, Lefrancois L. Activated primary and memory CD8 T cells migrate to nonlymphoid tissues regardless of site of activation or tissue of origin. J Immunol. 2004;172:4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
- 62.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential Localization of Effector Memory Cells in Nonlymphoid Tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 63.Marshall DR, Turner SJ, Belz GT, Wingo S, Andreansky S, Sangster MY, Riberdy JM, Liu T, Tan M, Doherty PC. Measuring the diaspora for virus-specific CD8+ T cells. Proc Natl Acad Sci USA. 2001;98:6313–6318. doi: 10.1073/pnas.101132698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, García-Sastre A, Merad M. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, Farber DL. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7:501–10. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olson MR, McDermott DS, Varga SM. The Initial Draining Lymph Node Primes the Bulk of the CD8 T Cell Response and Influences Memory T Cell Trafficking after a Systemic Viral Infection. PLoS Pathog. 2012;8:e1003054. doi: 10.1371/journal.ppat.1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, Attinger A, Shui J-W, Kim G, Lena CJ, Sakaguchi S, Miyamoto C, Wang P, Atarashi K, Park Y, Nakayama T, Honda K, Ellmeier W, Kronenberg M, Taniuchi I, Cheroutre H. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cannarile Ma, Lind Na, Rivera R, Sheridan AD, Camfield Ka, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 69.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich Ma, Walsh ER, Wani Ma, Lingrel JB, Hogquist Ka, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 71.Skon CN, Lee J-Y, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JY, Skon CN, Lee YJ, Oh S, Taylor JJ, Malhotra D, Jenkins MK, Rosenfeld MG, Hogquist KA, Jameson SC. The Transcription Factor KLF2 Restrains CD4+ T Follicular Helper Cell Differentiation. Immunity. 2015;42:252–264. doi: 10.1016/j.immuni.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou X, Yu S, Zhao D-M, Harty JT, Badovinac VP, Xue H-H. Differentiation and Persistence of Memory CD8(+) T Cells Depend on T Cell Factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Workman AM, Jacobs AK, Vogel AJ, Condon S, Brown DM. Inflammation enhances IL-2 driven differentiation of cytolytic CD4 T cells. PLoS One. 2014;9:1–12. doi: 10.1371/journal.pone.0089010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crotty S. T Follicular Helper Cell Differentiation, Function, and Roles in Disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, López CA, Früh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science. 2016;351:714–20. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


