SUMMARY
Upon sensing of the peptide pheromone cCF10, Enterococcus faecalis cells carrying pCF10 produce three surface adhesins (PrgA, PrgB or Aggregation Substance, PrgC) and the Prg/Pcf type IV secretion system and, in turn, conjugatively transfer the plasmid at high frequencies to recipient cells. We report that cCF10 induction is highly toxic to cells sustaining a deletion of prgU, a small orf located immediately downstream of prgB on pCF10. Upon pheromone exposure, these cells overproduce the Prg adhesins and display impaired envelope integrity, as evidenced by antibiotic susceptibility, misplaced division septa, and cell lysis. Compensatory mutations in regulatory loci controlling expression of pCF10-encoded prg/pcf genes, or constitutive PrgU overproduction, block production of the Prg adhesins and render cells insensitive to pheromone. Cells engineered to overproduce PrgB, even independently of other pCF10-encoded proteins, have severely compromised cell envelopes and strong growth defects. PrgU has an RNA-binding fold, and prgB-prgU gene pairs are widely distributed among E. faecalis isolates and other enterococcal and staphylococcal species. Together, our findings support a model in which PrgU proteins represent a novel class of RNA-binding regulators that act to mitigate toxicity accompanying overproduction of PrgB-like adhesins in E. faecalis and other clinically-important Gram-positive species.
Keywords: Gram-positive cell surface adhesins, cell death, PUA domain, sex pheromone, Enterococcus, gene regulation
Graphical Abstract
Upon sensing of sex pheromone, Enterococcus faecalis cells carrying pCF10 produce PrgB (Aggregation Substance, AS) and a type IV secretion system responsible for high-frequency plasmid transfer. We show PrgB overproduction is highly toxic to E. faecalis cells, and that PrgU mitigates toxicity by downregulating PrgB synthesis. PrgU is a predicted RNA binding protein, and prgB-prgU gene pairs are widely dispersed among enterococci suggestive of a conserved mechanism of feedback regulation of a major surface adhesin.

INTRODUCTION
Enterococcus faecalis infections are increasingly recognized as serious clinical threats due to the acquisition of multiple antibiotic resistance and the capacity of these organisms to rapidly disseminate resistance and virulence traits by lateral gene transfer (Lebreton et al., 2013, Van Tyne & Gilmore, 2014). Many E. faecalis clinical isolates harbor members of a large family of conjugative plasmids whose transmission is induced by sensing of peptide pheromones (Dunny, 2013, Dunny & Berntsson, 2016). These plasmids typically code for antibiotic resistance as well as surface proteins such as bacteriocins (cytolysin) or adhesins (PrgB, Esp) of established importance for tissue attachment and biofilm formation (Clewell et al., 2014). Many E. faecalis clinical isolates also carry fragments of the pheromone responsive plasmids encoding virulence traits in their chromosomes, underscoring both the selective advantages of the plasmid-borne elements and the plasticity of enterococcal genomes (McBride et al., 2007, Palmer et al., 2010).
One of the best-characterized representatives of the family of pheromone responsive plasmids is the tetracycline-resistance plasmid pCF10 (Dunny, 2013, Dunny & Berntsson, 2016). Detailed studies have unveiled a complex regulatory circuitry that operates to regulate pheromone-dependent expression of the plasmid-borne prgQ operon. The prgQ operon has three cassettes of genes encoding proteins of importance for plasmid transfer in natural settings such as biofilms (Bhatty et al., 2015). One cassette codes for three surface adhesins (PrgA, PrgB, PrgC) and an uncharacterized protein (PrgU), a second for the Prg/Pcf type IV secretion system (T4SS), and a third for DNA transfer and replication (Dtr) factors required for processing of pCF10 prior to its delivery through the T4SS (Fig. 1A) (Dunny, 2013). The prgQ operon is expressed from the PQ promoter, which is repressed by binding of the transcriptional regulator PrgX (Nakayama et al., 1994, Kozlowicz et al., 2006b). When cells import the cCF10 sex pheromone (LVTLVFV), secreted by plasmid-free enterococci in the vicinity, PrgX interacts with the pheromone and undergoes a structural transition resulting in PQ activation (Kozlowicz et al., 2006a, Kozlowicz et al., 2006b). Within 15 – 30 min of exposure to pheromone, donor cells undergo a burst of transcriptional activity and Prg/Pcf protein synthesis. They form intercellular aggregates primarily as a result of synthesis of PrgB (also known as Aggregation Substance or AS) and conjugatively transfer pCF10 at high frequencies approaching 1 transconjugant per donor (Hirt et al., 2005, Bhatty et al., 2015). Then, within the next 1 – 2 h, donor cells enter a shut-down phase characterized by a return of prgQ transcription to preinduction levels (Hirt et al., 2005, Chatterjee et al., 2013).
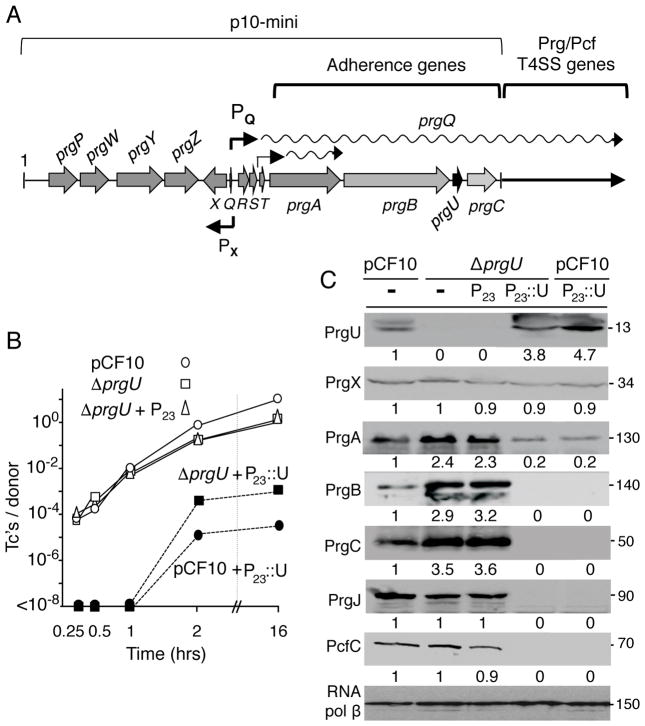
FIG. 1.
Effects of PrgU deletion and overproduction on Prg/Pcf protein synthesis and plasmid transfer. A) Schematic of the PQ regulatory region and prgQ operon carried by pCF10. The pheromone-responsive PQ promoter is regulated by flanking regulatory functions and directs expression of the downstream-encoded adherence and plasmid transfer functions. The PX promoter controls expression of prgX, which encodes the PrgX transcriptional regulator. Another promoter in the prgR/S region constitutively expresses a short prgA transcript at low levels. Plasmid p10-mini carries the ~17-kb fragment shown. B) Transfer frequencies of pCF10 plasmids in filter matings carried out for the durations indicated. Strains: OG1RF with pCF10 or pCF10ΔprgU (denoted ΔprgU) plasmids alone or additionally with the vector plasmid (P23; pDL278p23) or the P23:prgU expression plasmid (denoted P23::U; pMB11). Transfer frequencies are presented as the number of transconjugants per donor cell (Tc’s/Donor). Experiments were repeated at least three times in duplicate, and results from a representative experiment are shown. C) Steady-state levels of Prg/Pcf proteins in strains induced for 1 h with cCF10 pheromone (10 ng ml−1). OG1RF with plasmids indicated. Upper line: pCF10, ΔprgU (pCF10ΔprgU). Lower line: P23 (pDL278p23), P23::U (pMB11), - (no P23 plasmid). Immunoblots were developed with antibodies to the Prg/Pcf proteins shown or to RNA polymerase β subunit as a loading control. Protein sizes (in kilodaltons, kDa) are listed at the right. Protein extracts were loaded on a per-cell equivalent basis. Numbers below each panel correspond to relative protein abundance compared to levels detected in OG1RF(pCF10) cell extracts (set to 1), as determined by densitometry tracing using Image J software.
A number of transcriptional and posttranscriptional mechanisms control expression of the prgQ operon, presumptively to ensure tight regulation of the energetically expensive processes associated with Prg/Pcf T4SS assembly and plasmid transfer (Fig. 1A) (Johnson et al., 2010, Chatterjee et al., 2011, Dunny & Johnson, 2011). Despite this complex regulatory circuitry, we recently gained evidence that a subpopulation of pheromone induced pCF10-carrying cells undergo lysis during early stages of biofilm development as a result of overproduction of the Prg surface proteins (Bhatty et al., 2015). When exposed to pheromone, OG1RF cells carrying pCF10 form considerably thicker biofilms than plasmid-free OG1RF, and these biofilms possess abundant amounts eDNA, polysaccharides, and other matrix materials. Confirming the importance of the Prg surface adhesins, pheromone-treated OG1RF cells engineered to carry a plasmid expressing only prgA-C genes among the prgQ-regulated prg/pcf genes similarly develop thick biofilms with a pronounced accumulation of lysed cells and matrix components. In view of these findings, we hypothesized that the stochastic overproduction of Prg surface proteins in a subpopulation of pheromone-induced cells has lethal consequences (Bhatty et al., 2015).
Here, we tested this model by evaluating the contributions of proteins encoded within the prgA-C cassette to pheromone-mediated toxicity. Strikingly, we discovered that deletion of a small orf termed prgU located immediately downstream of prgB (Fig. 1A) confers strong growth defects when cells are exposed to cCF10 pheromone. We present several lines of evidence that PrgU functions as a suppressor of sex pheromone-mediated cell toxicity by blocking overproduction of the Prg surface adhesins. Indeed, overproduction specifically of PrgB in the presence or absence of other pCF10-encoded proteins severely compromises cell envelope integrity, as evidenced by sensitivity to antibiotics, misplaced division septa, and cell lysis. prgB-prgU gene pairs are widely distributed on plasmids or chromosomes of E. faecalis clinical isolates and other enterococci. Our cumulative findings thus suggest that feedback control by PrgU serves to maintain cell envelope homeostasis by blocking overproduction of PrgB-like adhesins in E. faecalis and related species.
RESULTS
PrgU regulates production of Prg surface adhesins and the Prg/Pcf T4SS
The PQ promoter and flanking regulatory region controls expression of the ~28-kb prgQ operon (Hirt et al., 2005). Our recent studies of the Prg adhesins encoded by the prgA-C gene cassette established their importance for intercellular aggregation and biofilm development, but surprisingly not for high-frequency transfer of pCF10 (Bhatty et al., 2015). Immediately downstream of prgB in the prgA-C cassette is an uncharacterized orf termed prgU (Fig. 1A), which is predicted to encode a 106-residue, cytosolic protein. We deleted prgU from pCF10, and determined that OG1RF cells transferred the pCF10ΔprgU mutant plasmid at frequencies comparable to wild-type (WT) pCF10 in short-term (≤ 1 h) matings and at slightly lower frequencies in longer (≥2 h) matings on solid surfaces (Fig. 1B). Interestingly, however, within 1 h of exposure to exogenous cCF10 pheromone, OG1RF(pCF10ΔprgU) cells accumulated the PrgA, PrgB, and PrgC surface adhesins at levels ≥2-fold higher than comparably-treated OG1RF(pCF10) cells (Fig. 1C). No corresponding increases in protein levels were detected for the downstream-encoded T4SS subunits, PrgJ or PcfC.
Within 30 min of exposure to cCF10 pheromone, OG1RF(pCF10) cells accumulate abundant levels of PQ transcripts spanning the length of the prgQ operon, and then over the next 1 – 2 hours transcript levels return to pre-induction levels (Hirt et al., 2005). OG1RF harboring pCF10ΔprgU displayed a similar transcription profile, with the exception that at 1 h following pheromone induction, transcript levels remained high, suggestive of a block or delay in PQ promoter shutdown (Fig. S1). It is interesting to note that we did not detect elevated levels of the T4SS subunits (Fig. 1C) even though levels of the corresponding transcripts remained high (Fig. S1). We suspect this is because these Prg/Pcf subunits assemble in specific stoichiometries as stable type IV machines, as shown for other T4SSs (Low et al., 2012), and that excess amounts of machine subunits are shunted to a degradative pathway.
OG1RF(pCF10ΔprgU) cells carrying pMB11, which expresses prgU from the constitutive P23 promoter, accumulated PrgU at levels ~4–5 fold relative to levels detected in pheromone-induced OG1RF(pCF10) cells (Fig. 1C). Strikingly, the ΔprgU mutant strain harboring pMB11 accumulated only a very low level of PrgA and undetectable amounts of PrgB and downstream-encoded Prg/Pcf proteins (Fig. 1C). Similarly, OG1RF(pCF10) cells carrying pMB11 overproduced PrgU and accumulated low or undetectable levels of the Prg/Pcf proteins. As expected, PrgU overproduction strongly suppressed transfer of both pCF10 and pCF10ΔprgU to undetectable levels in short-term (≤1 h) matings, and by 3- to 4-orders of magnitude in 2 h or overnight matings (Fig. 1B). The phenotypes associated with the ΔprgU mutation and PrgU overproduction suggested that PrgU negatively regulates Prg protein synthesis in response to pheromone induction.
The ΔprgU mutation confers pheromone toxicity
During these initial studies, we discovered that the ΔprgU mutation also confers striking growth defects upon exposure of cells to cCF10. Pheromone toxicity was shown with complementary serial dilution (Fig. 2A), pheromone spot (Fig. 2B), and growth curve (Fig. 2C) assays. As shown with each assay, pheromone treatment strongly inhibited growth of OG1RF(pCF10ΔprgU) without affecting growth of isogenic OG1RF(pCF10) cells or the ΔprgU mutant strain harboring the PrgU-overproducing plasmid pMB11 (Figs. 2A, B & C). The ΔprgU mutation was unique among our collection of prg or pcf mutations (prgA, prgB, prgU, prgC, pcfC, prgK, prgJ) in conferring pheromone growth suppression (Fig. S2) (Chen et al., 2008, Li et al., 2012, Laverde Gomez et al., 2014, Bhatty et al., 2015). Furthermore, pheromone toxicity of the ΔprgU mutant required active cell growth, as shown by the inhibitory effects of pheromone on cells in early-exponential but not stationary phases of growth (Fig. S2).
FIG. 2.
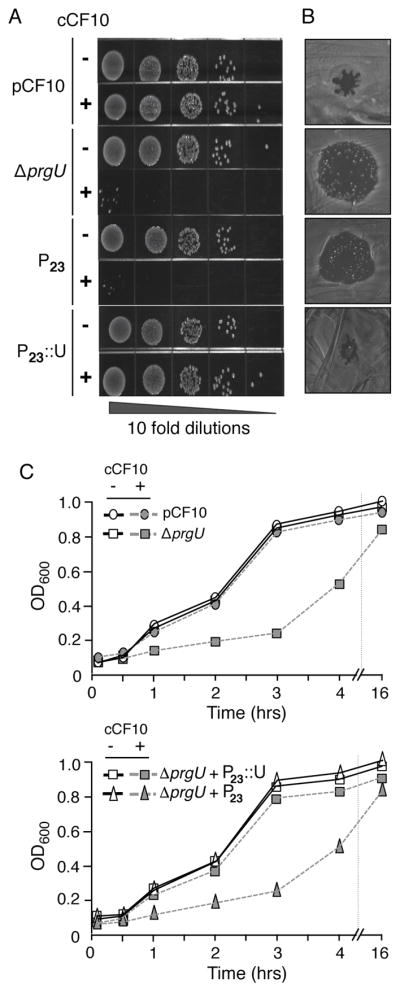
cCF10 pheromone suppresses growth of E. faecalis cells carrying the pCF10ΔprgU plasmid. A) Serial dilution plate assay. E. faecalis overnight cultures were diluted 1:100 in fresh BHI and incubated for 1 h at 37°C in the absence (−) or presence (+) of cCF10 (10 ng ml−1). Tenfold serial dilutions were inoculated onto BHI medium and assessed for growth. Strains: OG1RF with pCF10 or ΔprgU (pCF10ΔprgU) alone, or additionally with the P23 vector plasmid (pDL278P23) or the P23::prgU expression plasmid (denoted P23::U; pMB11). B) Pheromone spot assay. Overnight cultures listed at the left in Panel A were diluted 1:100 in fresh BHI and incubated for 1 h at 37°C in the absence of pheromone. Cultures were spread on BHI media, allowed to dry, and cCF10 pheromone (10 ng ml−1) was added to the center of the plate. Plates were incubated overnight at 37 °C and assessed for growth. cCF10 pheromone is solubilized in DMSO, which inhibits E. faecalis growth and causes small zones of clearance independently of cCF10-induced toxicity; for example, see spot assay for OG1RF(pCF10). C) Growth curve assay. Overnight cultures were diluted 1:50 in BHI and incubated for 1 h at 37 °C. The cells were normalized to an OD600 of 0.1 and cultures were incubated in the absence or presence of cCF10 (10 ng ml−1) at 37°C. Aliquots were removed at specified time points, 10 μl of 0.5 M EDTA was added to disaggregate the cells, and the OD600 was measured.
ΔU-Res mutants accumulate compensatory mutations conferring pheromone insensitivity
Variants of OG1RF(pCF10ΔprgU), designated as ΔU-Res, arose as colonies within the zone of inhibition in the pheromone spot assay (Fig. 2B) and when undiluted cell cultures were plated on pheromone-containing media (Fig. 2A). The proliferation of ΔU-Res variants also accounted for the eventual increase in cell density of the ΔprgU mutant when grown in the presence of pheromone (Fig. 2C), as deduced from findings that all tested isolates from a stationary-phase (16-h) culture exhibited pheromone-insensitive growth (data not shown). We sequenced the genome of one ΔU- Res variant, designated R1, and identified a single frame-shift mutation at basepair 271 of prgR, which is a putative regulator of prgB expression (Fig. 3A) (Chung & Dunny, 1992). The ΔU- Res R1 strain accumulated detectable levels of the PrgA surface adhesin, but not of PrgB or PrgC or other Prg or Pcf proteins encoded by the distal region of the prgQ operon (Fig. 3B). Correspondingly, this variant accumulated detectable amounts of transcript for prgA but not for prgB or other downstream prg or pcf genes (Fig. S1). Previous studies established that pheromone induction of the PQ promoter generates a long prgQ transcript spanning the entire prgQ operon, and that independently of pheromone induction a distinct promoter within the prgQ regulatory region generates a transcript that spans only prgA (see Fig. 1A) (Bensing & Dunny, 1997). Our data thus suggest that the prgRΔ271 mutation blocks production of the long prgQ transcript from the pheromone-inducible PQ promoter without affecting synthesis of the short prgA transcript.
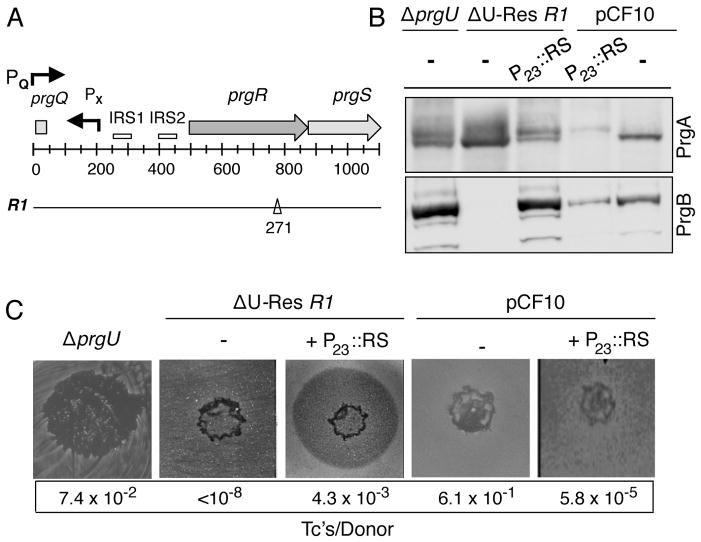
FIG. 3.
The ΔU-Res R1 compensatory mutation maps to the putative regulatory gene prgR.. A) Schematic of the prgQ regulatory region showing the locations of the PQ and PX promoters, inverted repeats IRS1 and IRS2 predicted to form stem-loop transcription terminators, and the putative regulatory genes prgR and prgS. Numbers refer to distances (in base pairs, bp’s) from the PQ promoter start-site. Below: The ΔU-Res R1 mutation is a 1 bp deletion at position 271 in the 318-bp prgR sequence. B) Modulation of PrgA and PrgB levels by the P23::prgR,prgS expression plasmid. Steady-state levels of PrgA and PrgB in strains induced for 1 h with cCF10 pheromone (10 ng ml−1). OG1RF with plasmids indicated. Upper line: pCF10, ΔprgU (pCF10ΔprgU), ΔU-Res R1 (pCF10ΔprgU,prgRΔ271). Lower line: - (no P23 plasmid), P23::RS (P23::prgR,prgS expression plasmid, pMC003). Immunoblots were developed with antibodies to PrgA or PrgB. Protein extracts were loaded on a per-cell equivalent basis with RNA polymerase β subunit as a loading control. C) Pheromone spot and mating assays. Strains analyzed are the same as in panel B. Transfer frequencies of pCF10 and its derivatives are presented as the number of transconjugants per donor cell (Tc’s/Donor).
We confirmed that the prgRΔ271 mutant allele was responsible for the observed block in production of the Prg and Pcf proteins by trans-expression of prgR alone (data not shown) or together with prgS from plasmid pMC003 (Fig. 3). Reminiscent of the ΔprgU parental strain, the complemented R1 strain exhibited pheromone sensitivity and abundant production of PrgB (Figs. 3B, C). Furthermore, whereas the R1 strain was completely defective for plasmid transfer, trans-expression of prgR,prgS in the R1 strain supported transfer at levels approaching the ΔprgU parental strain (Fig. 3C). We also analyzed the effects of prgR,prgS trans-expression in OG1RF(pCF10). OG1RF(pCF10, pMC003) did not display pheromone toxicity (Fig. 3C), but interestingly accumulated PrgA and PrgB at reduced levels relative to OG1RF(pCF10) (Fig. 3B). These findings extend earlier work implicating the prgR/S loci as regulators of prgB expression (Chung & Dunny, 1992, Bensing & Dunny, 1997). It is interesting to note, however, that the PrgR/S regulators act positively in a ΔprgU mutant and negatively in pCF10-carrying cells with respect to production of PrgB and other downstream-encoded Prg/Pcf proteins. These strains differ only in their capacity to produce PrgU, raising the possibility that PrgR and PrgS coordinate with PrgU in some way to regulate pheromone-inducible expression of the prgQ operon.
We analyzed 9 other ΔU-Res strains (designated R2 – R10) recovered from a pheromone spot assay for Prg protein production and conjugative transfer (Fig. S3A). Reminiscent of the R1 variant, these strains produced undetectable or very low levels of the PrgB or PrgC surface adhesins and of the downstream-encoded PrgJ or PcfC T4SS subunits (Fig. S3B; data not shown). In matings, only two of the ΔU-Res variants were capable of low-frequency plasmid transfer (Fig. S3C). Sequence analyses confirmed that nearly all of the R2–R10 mutant strains accumulated mutations in prgR or the adjacent putative regulator prgS, although several strains also had mutations elsewhere in the prgQ regulatory region (Fig. S3C). Together with our studies of the R1 variant, these findings suggest that compensatory mutations accumulate in prgR or prgS, or elsewhere in the prgQ regulatory region, to mitigate pheromone toxicity associated with production of one or more of the Prg/Pcf proteins..
The ΔprgU mutation confers strong growth defects
We gained four lines of evidence that pheromone induction impairs viability of the ΔprgU mutant through disruption of cell envelope integrity. First, OG1RF(pCF10ΔprgU) cells exhibited profound sensitivity to bile salts (Fig. S4). The ΔprgU mutant grew poorly upon exposure to pheromone and even more poorly with additional exposure to sodium cholate or sodium deoxycholate. By contrast, OG1RF(pCF10), OG1RF(pCF10ΔprgU) in the absence of pheromone, or OG1RF(pCF10ΔprgU) expressing P23::prgU from pMB11 were not growth suppressed in the presence of bile salts (Fig. S4).
The ΔprgU mutant also exhibited exquisite sensitivity to antibiotics (Fig 4A, B). OG1RF(pCF10) cells showed no growth impairment when exposed to antibiotics to which they encode resistance (Rif200, Fus25, Tet10) or to subinhibitory concentrations of antibiotics commonly used to treat E. faecalis infections (Dap6, Lin0.25). OG1RF(pCF10ΔprgU) also grew in these antibiotic-containing media in the absence of pheromone exposure, as did this strain when engineered to overproduce P23::prgU from pMB11 regardless of pheromone treatment. When exposed to pheromone, however, OG1RF(pCF10ΔprgU) failed to grow in the presence of all of the tested antibiotics over a 16-h incubation period (Fig. 4A). These findings suggested not only that OG1RF(pCF10ΔprgU) is highly sensitive to antibiotics, but further that the combination of pheromone and antibiotic treatments strongly suppressed even the appearance and outgrowth of the ΔU-Res variant subpopulation. The synergistic effects of pheromone and antibiotic treatments also were evident with a disc diffusion assay (Fig. 4B).
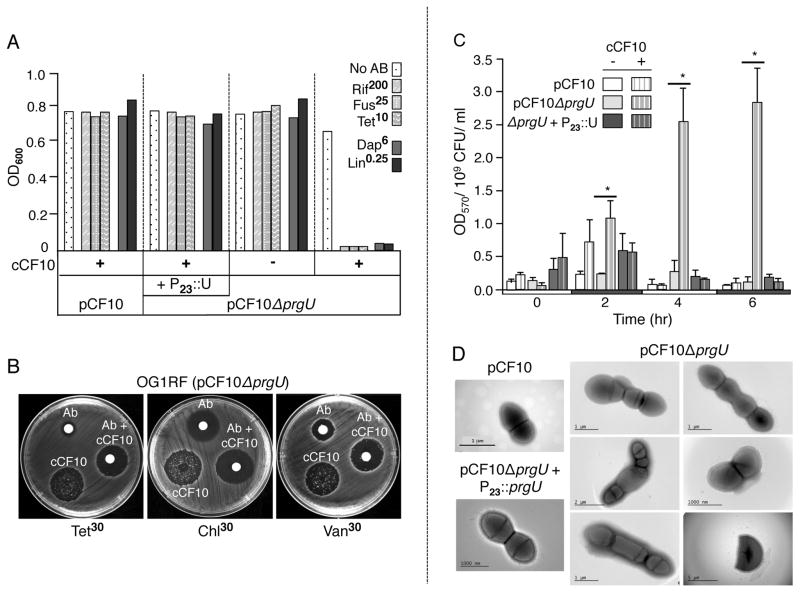
FIG. 4.
ΔprgU mutation confers defects in cell envelope integrity and cell growth. A) Growth defect of the ΔprgU mutant in the presence of subinhibitory concentrations of antibiotics. Strains: OG1RF with pCF10, ΔprgU (pCF10ΔprgU) alone or with the P23::prgU expression plasmid (P23::U, pMB11). Strains were inoculated from glycerol stocks into BHI lacking (−) or containing (+) cCF10 and in the absence (No AB) or presence of the antibiotics at final concentrations (in μg ml−1) listed. Cultures were incubated overnight at 37°C without shaking and culture densities (OD600) were measured. B) Disc diffusion assay showing that a combination of pheromone (10 ng ml−1) and antibiotics at the indicated concentrations suppressed the appearance of ΔU-Res variants. C) E. faecalis strains containing the constitutive lacZ reporter construct pCJK205 were cultured in BHI broth supplemented with erythromycin and 40 μg ml−1 CPRG, and incubated in the absence or presence of cCF10 at 37°C. Samples were removed at the times indicated, and CPRG hydrolysis was quantitated by measuring the absorbance (OD570) of the bacterium-free culture supernatant and normalizing to CFU. The experiment was performed at least two times, and the data are represented as the mean +/− standard deviation. Statistical significance was evaluated by t test, *, P<0.05 versus cCF10-untreated culture. D) Morphological aberrations of pheromone-exposed ΔprgU mutant cells. Overnight cultures were diluted 1:100 in fresh BHI supplemented with pheromone (10 ng ml−1) and incubated without shaking for 1 h at 37°C. Cells were processed for imaging as described in the Experimental procedures.
We next used a quantitative β-galactosidase (β-Gal) release assay to test for pheromone-dependent permeabolization of OG1RF(pCF10ΔprgU) cells (Fig. 4C). OG1RF(pCF10) and OG1RF(pCF10ΔprgU) lacking or containing the P23::prgU expression plasmid were engineered to constitutively express a lacZ reporter. Cells were grown in the absence or presence of pheromone in media containing chlorophenyl red-β-D-galactopyanoside (CPRG), a membrane-impermeable substrate of β-Gal (Paradis-Bleau et al., 2014). Intact cells producing cytoplasmic β-Gal do not degrade extracellular CPRG, whereas cells with elevated permeability due to cell envelope alterations exhibit higher β-Gal activities as a result of CPRG entry or β-Gal release to the milieu. We monitored β-Gal activity in culture supernatants over 6-hr following pheromone induction, a period during which the ΔprgU mutant accumulates abundant amounts of the Prg proteins without proliferation of a ΔU-Res variant population. In the absence of pheromone induction, OG1RF(pCF10) did not appreciably hydrolyze CPRG, in line with previous findings (Djoric & Kristich, 2015, Dale et al., 2015). Within 2 h of pheromone treatment, OG1RF(pCF10) showed a slight increase in CPRG hydrolysis compared with the uninduced culture, but this increase was not statistically significant and it also was transient as shown by a reduction in CPRG hydrolysis to basal levels at 4 h and 6 h post-induction. In striking contrast, within 2 h and increasingly at 4 h and 6 h following pheromone induction, OG1RF(pCF10ΔprgU) showed statistically-significant increases in CPRG hydrolysis that were not observed in the absence of pheromone (P<0.05). OG1RF(pCF10ΔprgU) overproducing P23::prgU from pMB11 hydrolyzed CPRG at low levels regardless of pheromone exposure throughout the duration of the experiment. Pheromone induction thus strongly perturbs the ΔprgU mutant cell envelope during a period of robust prgQ expression and Prg/Pcf protein synthesis.
Finally, ultrastructural studies showed that the pheromone-exposed OG1RF(pCF10ΔprgU) cells were morphologically aberrant (Fig. 4D). OG1RF(pCF10) and OG1RF(pCF10ΔprgU) cells expressing P23::prgU from pMB11 grew as typical ovoid diplococci in short chains and clumps regardless of pheromone treatment. In striking contrast, pheromone-exposed OG1RF(pCF10ΔprgU) cells exhibited defects in placement of division septa, resulting in aberrantly large and small cells. Cells also were elongated and some possessed partial invaginations, suggestive of incomplete formation of septa. Membrane blebbing and the apparent release of cytoplasmic contents also were evident, suggestive of partial or complete disruption of the cell envelope.
PrgB (Aggregation Substance) contributes to pheromone toxicity
Having established that the ΔprgU mutation correlates with enhanced levels of the Prg surface proteins (Fig. 1C) and with a disruption of cell envelope integrity (Figs. S4 and 4), we asked whether pheromone toxicity could be recapitulated in an OG1RF strain producing only the Prg surface proteins. To this end, we constructed a miniature version of pCF10, designated p10-mini, which carries: i) prgP-prgY encoding the pheromone sensing and uptake system, ii) the prgQ regulatory region, and iii) the prgA-C gene cassette from pCF10 with or without prgU. Interestingly, OG1RF(p10-miniΔprgU) displayed phenotypes similar to those of OG1RF(pCF10ΔprgU), including pheromone-dependent overproduction of the Prg surface adhesins (Fig. S5A), growth inhibition (Fig. S5B), exquisite sensitivity to subinhibitory concentrations of antibiotics (Fig. S5C), and morphological aberrations (Fig. S5D). OG1RF(p10-mini) or OG1RF(p10-miniΔprgU) expressing P23::prgU in trans showed no discernible pheromone-mediated growth defects (Figs. S5B, C, D), strongly supporting the notion that overproduction of one or more of the Prg surface adhesins is responsible for pheromone toxicity.
Next, we constitutively expressed each of the prgA, prgB or prgC genes from the P23 promoter in otherwise plasmid-free OG1RF. At the outset, we noted that OG1RF electroporated with the P23::prgB expression plasmid formed only a few small colonies on the transformation plates, whereas transformation with the P23 vector and P23::prgA and P23::prgC expression plasmids yielded many hundreds of colonies. OG1RF cells also were rapidly cured of the P23::prgB expression plasmid but not of the P23::prgA or P23::prgC expression plasmids (Fig. 5A). In this plasmid curing assay, cultures were grown overnight in the absence of spectinomycin antibiotic selection for maintenance of the P23 expression plasmids, and then serially diluted on plates lacking or containing spectinomycin. Strikingly, OG1RF transformed with the P23::prgB expression plasmids grew well on antibiotic-free media, but formed only a few small colonies on the spectinomycin-containing media (Fig. 5A). We were unable to isolate the P23::prgB expression plasmid from colonies appearing on antibiotic-free plates, suggesting that the bulk population of cells had lost the plasmid in the absence of antibiotic selection (data not shown). In contrast to these findings, strains carrying the P23 vector or the P23::prgA or P23::prgC expression plasmids retained their plasmids following overnight cultivation in the absence of antibiotics, as shown by comparable growth on media with and without spectinomycin (Fig. 5A).
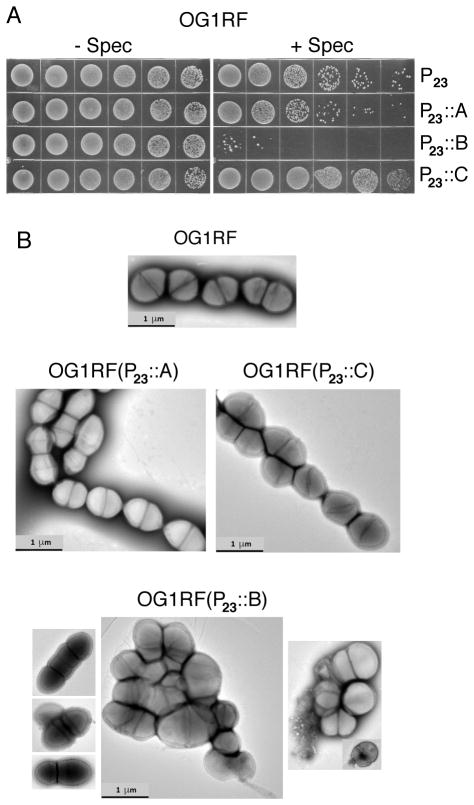
FIG. 5.
Constitutive overproduction of PrgB confers toxicity in the absence of other pCF10-encoded factors. A) Plasmid curing assay. Colonies from transformation plates were inoculated into antibiotic-free BHI and incubated without shaking overnight at 37°C. Overnight cultures were serially diluted and spotted onto BHI agar plates containing or lacking spectinomycin (500 μg ml−1) to which the P23 plasmid confers resistance. B) PrgB overproduction confers severe growth defects. Freshly transformed cells were inoculated in fresh BHI supplemented with pheromone (10 ng ml−1) and incubated without shaking for 1 h at 37°C. Cells were processed for imaging as described in the Experimental procedures. Strains: OG1RF alone or harboring the P23::prgA, P23::prgB, or P23::prgC expression plasmids.
We attempted to assay the OG1RF cells carrying the P23::prgB expression plasmid for sensitivity to bile salts and antibiotics, but these experiments were complicated by the apparent rapid accumulation of compensatory mutations by cells grown under conditions selective for maintenance of the P23::prgB expression plasmid. However, when cells from colonies just emerging upon transformation with the P23::prgB expression plasmid were examined by transmission electron microscopy, they exhibited abundant morphological aberrations highly similar to those of pheromone-exposed cells harboring pCF10ΔprgU (compare Figs. 4D, 5B). By contrast, cells freshly transformed with the prgA or prgC expression plasmids showed very few ultrastructural abnormalities (Fig. 5B). We obtained similar results to those described above when a strain of OG1RF harboring pCF10ΔprgA-C, a plasmid deleted of the prgA-C cassette, was transformed with the P23::prgA, P23::prgB, and P23::prgC expression plasmids (Fig. S6A–C).
Interestingly, however, in a pheromone spot assay, OG1RF(pCF10ΔprgA-C) showed a slight growth suppression despite the fact that this strain does not produce PrgB or the other surface proteins (Fig. S6D). Although introduction of the P23::prgB expression plasmid disrupted growth of OG1RF(pCF10ΔprgA-C) cells even in the absence of pheromone induction, the additional exposure to pheromone imposed an additional stress that strongly suppressed further growth (Fig. S6D). The PrgB-overproducing strain formed considerably more ΔU-Res-type variants than the ΔprgU mutant in the zone of pheromone growth inhibition. We suspect that most of these variants arose through loss of the P23::prgB expression plasmid, although compensatory ΔU-Res mutations also might have arisen that enabled growth in the presence of PrgB overproduction. Overall, these data corroborate with our other findings indicating that PrgB overproduction is highly toxic to E. faecalis cells, but further suggest that one or more additional pheromone-induced factors probably encoded by pCF10 imposes an additional stress that exacerbates PrgB overproduction toxicity.
DISCUSSION
Extensive studies of the cCF10 pheromone sensing pathway of plasmid pCF10, and other pheromone plasmids, have unveiled a complex regulatory circuitry that controls expression of genes encoding surface adhesins and conjugation functions (Clewell, 2007, Dunny, 2013, Clewell et al., 2014, Hirt et al., 2005). Assembly of T4SSs is an energetically expensive process, and tight control of the sex pheromone response might have arisen to mitigate the large fitness costs associated with high-frequency plasmid transfer (Dunny & Berntsson, 2016). Here, we discovered another reason for controlling the sex pheromone response in E. faecalis. A novel pCF10-encoded regulatory factor, PrgU, appears to have evolved to specifically control production of the PrgB surface adhesin, more widely known as Aggregation Substance. Controlled synthesis of PrgB is critical because its overproduction is highly toxic, manifesting as a severe disruption of cell envelope integrity and defective cell growth. Below, we discuss the broad biological importance of PrgU regulatory control and of PrgB overproduction toxicity in enterococci and, possibly, other bacterial pathogens.
PrgU, a novel regulatory factor
Several repression mechanisms operate to control transcription of the prgQ operon in the absence of sex pheromone sensing (see Fig. 1A) (Bae et al., 2004, Kozlowicz et al., 2006b). These include: i) PrgX repression of transcription from the PQ promoter, ii) enhanced PrgX repression through binding of iCF10, a small inhibitor peptide produced from the prgQ gene located immediately downstream of the PQ promoter, and iii) production of a small antisense RNA (anti-Q) from the convergent PX promoter that binds nascent prgQ transcripts and causes them to fold into a terminator structure that blocks extension of prgQ transcription (Shokeen et al., 2010, Nakayama et al., 1994, Chatterjee et al., 2013). Upon binding and import of cCF10 pheromone produced by recipient cells in the vicinity, the pheromone binds PrgX and induces a structural change that impairs PrgX repression of PQ. This leads to increased levels of nascent short prgQ transcripts (QS) that overwhelm the anti-Q termination mechanism, and result in production of longer transcripts (QL) that can ultimately extend through the entire operon. Within ~30–60 min following pheromone induction of the PQ promoter, iCF10 levels increase to a threshold sufficient to initiate a rapid shut-down of transcription to pre-induction levels. There is also extensive genetic and biochemical evidence suggesting that polypeptides and regulatory RNAs produced from the region between prgQ and prgA, e.g., prgR and prgS, encode functions that modulate transcription elongation and translation of the downstream genes (see Figs. 3 & S3) Chung & Dunny, 1992, Bensing & Dunny, 1997, Bensing et al., 1997).
Here, we discovered that PrgU confers another layer of negative feedback regulation. One model under investigation is that PrgU coordinates with one of the PQ repression systems specifically to achieve controlled synthesis of the Prg surface adhesins in response to pheromone induction. Consistent with such a model, we found that the ΔprgU mutation correlated with enhanced accumulation of the Prg adhesins and also with a failure to undergo a shut-down phase in transcription from the PQ promoter at 60 min postinduction (Fig. S1). Conceivably, upon pheromone induction, PrgU accumulates to a threshold level necessary for activating the switch to the repression mode. This could be achieved through establishment of stabilizing or activating interactions between QS and anti-Q RNA, by inhibiting production of extended QL transcripts, or by inhibiting positive regulatory functions of the prgRS region required for expression of the downstream conjugation genes. Alternatively, PrgU might impact the formation, stability or function of PrgX/cCF10 or PrgX/iCF10 complexes, but we view this as less likely given the locations and phenotypes of the ΔU-Res compensatory mutations.
Studies of these ΔU-Res variants supplied insights into the function of PrgU and identified possible coregulators. Our detailed genome sequencing and complementation studies of one variant, ΔU-Res R1, confirmed that a single prgR frame-shift mutation was responsible for the ΔU-Res phenotype (Fig. 3). Among 9 other ΔU-Res variants analyzed, nearly all had mutations in prgR or the downstream gene prgS, although some variants acquired additional mutations elsewhere in the prgQ regulatory region. Most importantly, however, all ΔU-Res mutants were blocked for production of PrgB and the downstream-encoded Prg and Pcf proteins, establishing a correlation between Prg protein production and toxicity (Fig. S3). Interestingly, trans-expression of the putative regulators prgR and prgS exerted opposite effects on production of the Prg proteins in isogenic strains lacking or carrying prgU. PrgU thus might physically coordinate with the PrgR/PrgS regulators to fine-tune the pheromone response, although it remains formally possible that PrgU and PrgR/PrgS act at distinct points in the regulatory circuitry.
We gained further insight into the mechanism of action of PrgU through structural modeling of a crystal structure solved for a PrgU homolog (EFA0046) from the clinical isolate V583. The crystal structure presents PrgU as a tetramer where each monomer forms a six β-strand barrel with three accompanying α helices (Fig. S7A,B) (Chang C, 2006). By threading PrgU through the Phyre2 algorithm (Kelley & Sternberg, 2009), we determined that PrgU adopts a PUA (pseudouridine synthase and archaeosine transglycosylase) fold (Fig. S7). PUA domains are widely distributed among diverse types of proteins within prokaryotes, eukaryotes and archaea (Perez-Arellano et al., 2007), and typically bind RNA substrates (Sabina & Soll, 2006, Zhang et al., 2012, Tempel et al., 2013). Other characterized PUA domains invariably are physically joined to other motifs that typically catalyze posttranscriptional modifications of tRNA and rRNA, although other biochemical functions have been identified (Perez-Arellano et al., 2007). Strikingly, the PUA fold encompasses the entire PrgU sequence, making this protein unique among members of the PUA family characterized to date. It is enticing to suggest that PrgU controls Prg protein production through binding of an RNA target, potentially a regulatory trans-acting sRNA or prgQ transcripts subject to posttranscriptional control. In fact, such a regulatory function was postulated for another PUA-domain containing protein, Bacillus subtilis ProB. Although ProB is a glutamate kinase, ProB also downregulates the expression of transcription factor sigma D (σD)-dependent genes (Ogura & Tanaka, 1996). It was suggested that ProB disrupts synthesis of σD through binding of its PUA domain to σD mRNA, resulting in premature transcription termination or a block in translation (Perez-Arellano et al., 2007). By analogy, the PUA domain of PrgU might bind an mRNA target to block transcription or translation of prgB and downstream prg genes.
In this context, it is of interest to note that the prgB - prgU genetic linkage is not restricted to pCF10. We found that prgU genes are widely distributed on plasmids and chromosomes of E. faecalis and other enterococci, e.g., Enterococcus faecium, Enterococcus raffinosus, as well as in the genomes of Streptococcus agalactiae and Staphylococcus aureus (Fig. S8). In all cases we have examined thus far, prgU-like genes are genetically linked to prgB-like genes. Most often, this gene pair is part of a larger prgA-C-like cassette, but other genetic contexts exist in which prgA or prgC are absent or nearby genes instead encode cell wall hydrolases. We therefore suggest that the prgB-prgU genetic linkages might have evolved to ensure the controlled synthesis of PrgB-like adhesins or, possibly, cell wall metabolizing enzymes in various Gram-positive pathogens. If so, it is reasonable to predict that PrgU exerts its control by binding regulatory DNA or mRNA motifs positioned upstream or within prgB or, possibly, other target genes.
The biological importance and mechanism of PrgB overproduction toxicity
Sex pheromone-mediated toxicity in E. faecalis was first reported by M. Gilmore and his colleagues (Gilmore et al., 2015). In that study, commensal isolates of enterococci were shown to effectively kill the clinical strain V583 through release of an inducing sex pheromone, cOB1. cOB1-mediated killing of V583 requires two genetic elements, the pheromone-dependent plasmid pTEF2, which is highly similar in gene composition to pCF10, and an integrated chromosomal (IS-like) element. The mechanism of killing was not defined, but the following observations suggest cOB1-mediated killing might result from the dysregulated overproduction of one or more PrgB adhesins. V583 carries four copies of prgB-like genes, one on each of the resident pTEF1 (EFA0047) and pTEF2 (EFB0011) plasmids, one in the pathogenicity island (PAI, EF0485) and one in the IS-like element (EF0149). prgU genes are linked to three prgB genes (pTEF1, EFA0046; pTEF2, unannotated; PAI, EF0486), but, interestingly, not EF0149 in the IS-like element. These observations and results of a microarray analysis showing that cOB1 induces expression of prg- and pcf-like genes on pTEF2 (Gilmore et al., 2015) suggest the possibility that V583 accumulates one or more of the PrgB adhesins at toxic levels when exposed to cOB1 sex pheromone. Conceivably, pTEF2 encodes a regulatory factor that activates expression of chromosomal-borne EF0149, which in the absence prgU regulator, leads to excess accumulation of the PrgB adhesin. Alternatively, V583 cells might produce EF0149 endogenously at subinhibitory levels, but cOB1 induction of pTEF2-encoded EFB0011 confers PrgB overproduction toxicity due to the combined production of EF0149 and EFB0011.
PrgB is a well-characterized adhesin that mediates extensive cellular clumping and attachment to abiotic and biotic surfaces as a critical early step in establishment of robust biofilms (Olmsted et al., 1991, Chuang et al., 2009). Our present findings add to results of our recent study in which we showed that OG1RF cells carrying a miniaturized version of pCF10 that expresses only the prgA-C cassette form robust biofilms with an accumulation of dead or permeabolized cells (Bhatty et al., 2015). Although chromosomally-encoded autolytic or fratricidal mechanisms might account for the observed cell death (Thomas & Hancock, 2009, Thomas et al., 2009), we suggest instead that a subset of pCF10-carrying cells might stochastically overproduce PrgB with lethal consequences. In natural settings composed of mixed communities of plasmid-carrying and -free enterococci as well as other species, PrgB-mediated cell lysis could provide a source of extracellular matrix components, e.g., lipoteichoic acids, polysaccharides, eDNA, of importance for establishment of robust biofilms (Dunny et al., 2014). Consistent with this proposal, single cell analyses (Cook et al., 2011) and our additional unpublished findings have shown that prgB expression is highly variable among individual cells in cCF10-exposed populations.
By scanning electron microscopy, PrgA and PrgB were shown to colocalize on the E. faecalis cell surface, initially at the equatorial region of dividing cells, and then more uniformly around the polar caps of older cells (Olmsted et al., 1993). Both proteins were associated with extended filaments that form a fibrous mesh around the cell periphery. More recently, it was shown that the general secretion (Sec) system and the sortase machinery colocalize as discrete foci in the equatorial regions of E. faecalis cells, suggestive of a coordination of cell envelope biogenesis and cell division (Kline et al., 2009, Kandaswamy et al., 2013). Abundant production of PrgB, possibly with contributions by PrgA at or near the growing septum might perturb deposition of the cell wall, septal placement, and cell division. It is also noteworthy that eDNA is localized around the equatorial regions of dividing cells (Barnes et al., 2012), and recently we reported that PrgB induces extensive clumping of cells and biofilm formation by an eDNA-dependent mechanism (Bhatty et al., 2015). Interestingly, we have observed that DNase I treatment diminishes the toxic effect of pheromone on growth of PrgB-overproducing strains (data not shown). These findings suggest that the association of eDNA with abundant PrgB fibers at or near the equatorial plane of actively dividing cells might disrupt cell wall biogenesis and septal placement. Such complexes might impede cell growth through steric effects or alterations in surface hydrophobicity or charge.
Although PrgA or PrgC overproduction did not confer toxicity, arguing against the notion that overproduction of surface proteins is generally toxic to enterococci, it remains possible that excessive accumulation of PrgB disrupts export of other surface proteins through the general secretory (Sec) or their sorting at the cell surface. In E. faecalis, the Fst toxin consists of a single transmembrane (TM) domain, and its overproduction yields growth defects similar to those shown here to be associated with PrgB overproduction (Weaver et al., 2003, Patel & Weaver, 2006). It is conceivable that insertion of the C-terminal TM domains of abundantly produced PrgB, either cleaved or not by sortase, has toxic consequences similar to those accompanying Fst toxin overproduction. Recent studies have shown that excessive accumulation of certain surface proteins, e.g., Streptococcus pyogenes M protein, Actinomyces oralis AcaC surface glycoprotein, in sortase mutants results in loss of cell envelope integrity and defects in cell growth (Raz et al., 2015, Nobbs et al., 2007, Wu et al., 2014). We have confirmed that PrgB overproduction toxicity occurs in the strain OG1RF (Figs. 5 & S6) and is not affected by sortase mutations (data not shown). To our knowledge, this is the first example of surface protein overproduction toxicity in a sortase-proficient strain, although further studies are needed to define the mechanism by which PrgB confers toxicity.
In summary, we identified a novel regulator, PrgU, that controls sex pheromone toxicity, and we determined that excessive accumulation of PrgB adhesin, also termed Aggregation Substance (AS) (Dunny et al., 1985), is responsible for toxicity. In the streptococci, it is well established that small peptide quorum signals induce death of a subpopulation of cells during development of genetic competence, an activity thought to provide a source of DNA for the competent cell population (Cook and Federle, 2014, Leung et al., 2015). Our findings expand the biological functions of peptide-mediated cell death among Gram-positive species, and further establish the potential for dysregulated synthesis of a surface adhesin, as opposed to induced synthesis of an autolytic murein hydrolase (Claverys et al., 2007, Dufour & Levesque, 2013), as a basis for toxicity. Finally, despite the fact that PrgB is a well-established virulence factor (Schlievert et al., 1998, Rakita et al., 1999), our results suggest the interesting possibility that intervention strategies aimed at transient overproduction of at least certain types of cell surface adhesins early during a treatment regimen might prove efficacious in sensitizing enterococci and, possibly other Gram-positive species, to antibiotics.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
The bacterial strains, plasmids, and oligonucleotides used in the study are listed in Tables S1, S2, and S3. For plasmid construction, E. coli DH5α and EC1000, a strain that produces the pWV01 RepA protein (Leenhouts et al., 1996), were used as hosts. E. coli strains were grown at 37°C with shaking in Lysogeny Broth (LB) Broth (Difco Laboratories). Brain heart infusion broth (BHI; Difco Laboratories) was used to grow E. faecalis strains at 37°C without shaking. E. coli strains were grown with the following antibiotics as needed: chloramphenicol (20 μg ml−1), erythromycin (100 μg ml−1), spectinomycin (50 μg ml−1). The following antibiotics were used as needed for E. faecalis: erythromycin (100 μg ml−1 final concentration for plasmid markers, 10 μg ml−1 for chromosomal markers), fusidic acid (25 μg ml−1), rifampin (200 μg ml−1), spectinomycin (1,000 μg ml−1 for plasmid markers, 250 μg ml−1 for chromosomal markers), streptomycin (1,000 μg ml−1), tetracycline (10 μg ml−1), chloramphenicol (10 μg ml−1), daptomycin (6 μg ml−1) in media supplemented with 50 μg ml−1 CaCl2, linezolid (0.25 μg ml−1). All antibiotics were obtained from Sigma-Aldrich.
Construction of prgU deletion mutants
For the construction of pCF10ΔprgU, ~500–600 base-pair (bp) regions immediately upstream and downstream of prgU were amplified with forward and reverse (F/R) primers designated prgUup-XbaI or -XmaI and prgUdown-XmaI or -NcoI. These fragments were joined by overlapping PCR using the primers F-prgUup-XbaI and R-prgU down-NcoI, and the resulting fragment was digested with XbaI and NcoI for introduction into similarly-digested pCJK47, resulting in pMCM3. prgU was deleted from pCF10 using a marker-less recombination strategy (Kristich et al., 2007), resulting in plasmid pCF10ΔprgU. prgU also was deleted from plasmid p10-mini, which contains the prgQ regulatory region and the downstream prgA - prgC gene cassette on shuttle vector pCI372 (see below). p10-miniΔprgU was constructed by inverse PCR using 5′ phosphorylated primers F-prgUdown5′phos and R-prgUup5′phos. The amplified product was ligated and introduced into E. faecalis strain OG1RF by electroporation (Dunny et al., 1991). prgU deletions in pCF10 and p10-mini were confirmed by PCR and nucleotide sequence analysis.
prg expression plasmids
pMB11 constitutively expresses prgU from the P23 promoter. It was constructed by amplifying prgU from pCF10 using gene specific F/R primers containing BamHI/SphI restriction sites. The PCR product was digested using the specified restriction enzymes and introduced into similarly-digested plasmid pDL278p23 downstream of the constitutive promoter P23 from Lactococcus lactis (Chen et al., 2007). This plasmid was introduced into E. faecalis strain OG1RF by electroporation (Dunny et al., 1991). Plasmids pMC001, pMC002, and pMB4 constitutively expressing prgA, prgB, and prgC from the P23 promoter were constructed similarly by use of pCF10 as a template and F/R primers listed in Table S3 for the gene amplifications. Plasmid MC003 constitutively coexpresses prgR and prgS from the P23 promoter. It was constructed as above using pCF10 as a template and F/R primers listed in Table S3.
Conjugation assays
Overnight liquid cultures of E. faecalis donor and recipient strains were diluted 1:10 in BHI and incubated for 1 h at 37°C. Donor and recipient cells were then mixed in a ratio of 1:10 and allowed to mate for the specified time periods at 37°C in liquid or on filters placed on BHI plates. Serial dilutions of the mating mixture plated on selective BHI agar plates were used to obtain the donor and transconjugant counts. The plasmid transfer frequencies were calculated as the number of transconjugants per donor cell (Chen et al., 2008). The results are reported as an average of at least three replicates of each experiment.
Detection of Prg/Pcf proteins by immunoblotting
Overnight liquid cultures of E. faecalis strains were diluted 1:20 in fresh BHI and grown for 1 h at 37°C. Cultures (5–10 ml) were normalized to an OD600 of 0.3 and induced with 10 ng ml−1 of peptide cCF10 for 1 h at 37°C. Cells were pelleted by centrifugation at 13,200 × g for 15 min at 4°C and washed once with cold 1X physiological buffer saline (PBS). The pellet was resuspended in 250 μl of SMM buffer (0.5M sucrose, 0.02 M MgCl2, 0.02 M maleate, pH 6.5) containing 500 U ml−1 of mutanolysin (Sigma-Aldrich) and 10 mg ml−1 of lysozyme (Sigma-Aldrich), and the resulting mix was incubated for 1 h at 37°C with shaking. The mix was centrifuged at 13,200 × g for 15 min at 4°C to separate the cell wall (supernatant) and cellular (pellet) fractions. The pellet fraction was suspended in 1 X PBS, and both fractions were analyzed on a per cell equivalent basis for the presence of Prg/Pcf proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, western blot transfer and immunostaining with anti-Prg/Pcf antibodies (Christie et al., 1988, Chen et al., 2008, Li et al., 2012, Bhatty et al., 2015). As a loading control, blots were developed with antibodies to the RNA polymerase β subunit (Santa Cruz Biotechnology).
Whole genome sequencing and analysis
E. faecalis DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Inc., Valencia, CA) following the pre-treatment for Gram-positive bacteria protocol. Purified DNA was submitted to the University of Minnesota Genomics Center for whole genome sequencing using the MiSeqV2 platform (Illumina, Inc.) and 150-bp paired-end reads. Sequencing data from the OG1RF(pCF10ΔprgU) and OG1RF(pCF10ΔU-Res R1) were compared to the OG1RF(pCF10) reference genome (NCBI accession numbers NC_017316 and NC_006827) using breseq version 0.24rc6 (Deatherage & Barrick, 2014).
qRT-PCR
Three colonies of each strain were inoculated into 10 ml M9-YE (Dunny & Clewell, 1975) and incubated at 37°C for 14.5–16 h. Cultures were diluted 1:10 into 10 ml fresh M9-YE, further incubated for 1 h, and then induced with 5 ng ml−1 cCF10. Aliquots of cells collected at 0, 30 and 60 min postinduction were treated with RNAprotect Bacteria Reagent (Qiagen, Inc., Valencia, CA) according to the manufacturer’s instructions. Cell pellets were resuspended in TE (10 mM Tris-HCl pH 8, 1 mM EDTA) containing 500 U ml−1 mutanolysin and 30 mg ml−1 lysozyme and incubated at 37°C for 10 min. RNA was extracted with the RNeasy Mini Kit (Qiagen, Inc.) according to the manufacturer’s instructions. Contaminating DNA was removed with the Turbo-DNA free kit (Ambion). cDNA was synthesized with random hexamers using the Superscript III First-Strand Synthesis System (Invitrogen Corp.) according to the manufacturer’s instructions. Quantitative PCR was carried out as previously described (Frank et al., 2012). gyrB was used as a reference gene. Two biological replicates were performed with similar results.
Bile susceptibility assays
Overnight liquid cultures of E. faecalis strains were diluted 1:100 and incubated for 1 h at 37 °C. Cultures were serially diluted and spotted (5 μl) onto BHI or BHI containing cCF10 (2 ng/ml) with or without sodium cholate (4% w/v) or sodium deoxycholate (0.06% w/v) (Dale et al., 2015). Colony forming units (CFUs) per ml were determined after overnight incubation at 37°C.
Pheromone spot assay
Overnight liquid cultures of E. faecalis strains were diluted 1:10 in BHI and incubated for 1 h at 37°C. Cultures (200 μl) were spread on BHI plates and cCF10 (10 ng ml−1 final concentration) was added to the center of the plates. To monitor the effect of DNase I on pheromone toxicity, DNase I was added (500 U ml−1 final concentration) to a BHI plate prior to spreading of the cultures and addition of pheromone to the center of the plates. Pheromone toxicity was evidenced by a zone of clearance after overnight incubation at 37°C.
Growth curve determinations
Overnight liquid cultures of E. faecalis strains were diluted 1:50 in BHI and incubated for 1 h at 37°C. The cells were normalized to an OD600 of 0.1 using fresh BHI, and then incubated in the absence or presence of cCF10 (10 ng ml−1) at 37°C. Aliquots were removed at specified time points, 10 μl of 0.5 M EDTA was added to disaggregate the cells, and the OD600 was measured. All experiments were performed in duplicate and results represent the average of three experiments.
CPRG (chlorophenol red-β-D-galactopyranoside) β galactosidase assay
CPRG hydrolysis was measured as described previously (Dale et al., 2015, Djoric & Kristich, 2015). Briefly, overnight cultures of E. faecalis strains were diluted to a starting OD600 of 0.05 in BHI supplemented with erythromycin and 25 μg ml−1 CPRG in the presence or absence of cCF10 (10 ng ml−1). Cultures were incubated statically at 37°C for 0, 2, 4 or 6 h. At each time point, 1 ml culture aliquots were removed for determinations of viable bacteria (CFUs) by serial dilution. CPRG hydrolysis was quantitated by measuring the absorbance at 570 nm (OD570) following removal of bacteria by centrifugation.
Antibiotic susceptibility assays
E. faecalis strains were inoculated from frozen glycerol stocks stored at −80°C. Each strain was inoculated into BHI containing antibiotics with or without cCF10 (10ng ml−1). Cultures were incubated overnight, 10 μl of 0.5 M EDTA was added to disaggregate the cells, and OD600 values were determined. All the experiments were performed in duplicate and the results represent the average of two different experiments.
Transmission Electron microscopy
Overnight liquid cultures of E. faecalis strains grown in BHI were diluted 1:10 in BHI lacking or containing 10 ng ml−1 of cCF10 and incubated statically for 1 h at 37°C. To prepare the samples for visualization, 10 μl aliquots of cultures were placed on carbon-coated nickel or copper grids (Electron Microscopy Sciences) for 5 min, washed 3 times in sterile H20, and stained with 0.2% ammonium molybdate for 1 min. The grids were imaged using a JEOL JEM-1400 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank Dr. Barbara Murray for providing strains used in the study and Maria Camilla Montealegre for help with strain and plasmid constructions. We thank members of our respective laboratories for helpful discussions. These studies were supported by NIH grants R01GM48746 (P.J.C) and R21AI105454 (P.J.C & G.M.D).
References
- Bae T, Kozlowicz BK, Dunny GM. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol Microbiol. 2004;51:271–281. doi: 10.1046/j.1365-2958.2003.03832.x. [DOI] [PubMed] [Google Scholar]
- Barnes AM, Ballering KS, Leibman RS, Wells CL, Dunny GM. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio. 2012;3:e00193–00112. doi: 10.1128/mBio.00193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Dunny GM. Pheromone-inducible expression of an aggregation protein in Enterococcus faecalis requires interaction of a plasmid-encoded RNA with components of the ribosome. Mol Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3311709.x. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Manias DA, Dunny GM. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- Bhatty M, Cruz MR, Frank KL, Gomez JA, Andrade F, Garsin DA, et al. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 2015;95:660–677. doi: 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189:6490–6493. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Volkart L, Moy S, Joachimiak A. RCSV Protein Data Bank: Midwest Center for Structural Genomics. 2006. Crystal structure of protein EF0006 from Enterococcus faecalis. [Google Scholar]
- Chatterjee A, Cook LC, Shu CC, Chen Y, Manias DA, Ramkrishna D, et al. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci USA. 2013;110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci. 2011;108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Staddon JH, Dunny GM. Specificity determinants of conjugative DNA processing in the Enterococcus faecalis plasmid pCF10 and the Lactococcus lactis plasmid pRS01. Mol Microbiol. 2007;63:1549–1564. doi: 10.1111/j.1365-2958.2007.05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang X, Manias D, Yeo HJ, Dunny GM, Christie PJ. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J Bacteriol. 2008;190:3632–3645. doi: 10.1128/JB.01999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Kao SM, Adsit JC, Dunny GM. Cloning and expression of genes encoding pheromone-inducible antigens of Enterococcus (Streptococcus) faecalis. J Bacteriol. 1988;170:5161–5168. doi: 10.1128/jb.170.11.5161-5168.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun. 2009;77:539–548. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JW, Dunny GM. cis-acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc Natl Acad Sci USA. 1992;89:9020–9024. doi: 10.1073/pnas.89.19.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- Clewell DB. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid. 2007;58:205–227. doi: 10.1016/j.plasmid.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. Extrachromosomal and mobile elements in enterococci: Transmission, maintenance, and epidemiology. In: Gilmore MS, Clewell B, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. Internet. http://www.ncbi.nlm.nih.gov/books/NBK190433/ [PubMed] [Google Scholar]
- Cook LC, Chatterjee A, Barnes A, Yarwood J, Hu WS, Dunny GM. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol. 2011;81:1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev. 2014;38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JL, Cagnazzo J, Phan CQ, Barnes AM, Dunny GM. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother. 2015;59:4094–4105. doi: 10.1128/AAC.00344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoric D, Kristich CJ. Oxidative stress enhances cephalosporin resistance of Enterococcus faecalis through activation of a two-component signaling system. Antimicrob Agents Chemother. 2015;59:159–169. doi: 10.1128/AAC.03984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Levesque CM. Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol. 2013;195:105–114. doi: 10.1128/JB.00926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G, Funk C, Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981;6:270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- Dunny GM. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet. 2013;47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- Dunny GM, Berntsson RP. Enterococcal sex pheromones: Evolutionary pathways to complex, two-signal systems. J Bacteriol. 2016;198:1556–1562. doi: 10.1128/JB.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Clewell DB. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J Bacteriol. 1975;124:784–790. doi: 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Hancock LE, Shankar N. Enterococcal biofilm structure and role in colonization and disease. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. http://www.ncbi.nlm.nih.gov/books/NBK190433/ [PubMed] [Google Scholar]
- Dunny GM, Johnson CM. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Op Microbiol. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for Gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Zimmerman DL, Tortorello ML. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc Natl Acad Sci USA. 1985;82:8582–8586. doi: 10.1073/pnas.82.24.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, Dunny GM. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immun. 2012;80:539–549. doi: 10.1128/IAI.05964-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore MS, Rauch M, Ramsey MM, Himes PR, Varahan S, Manson JM, et al. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc Natl Acad Sci USA. 2015;112:7273–7278. doi: 10.1073/pnas.1500553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F, Daly C, Fitzgerald GF. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl Environ Microbiol. 1990;56:202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, et al. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol. 2005;187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Manias DA, Haemig HA, Shokeen S, Weaver KE, Henkin TM, Dunny GM. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J Bacteriol. 2010;192:1634–1642. doi: 10.1128/JB.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, et al. Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci USA. 2013;110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocals. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, et al. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz BK, Dworkin M, Dunny GM. Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Inter J Med Microbiol: IJMM. 2006a;296:141–147. doi: 10.1016/j.ijmm.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol. 2006b;62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Chandler JR, Dunny GM. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid. 2007;57:131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverde Gomez JA, Bhatty M, Christie PJ. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J Bacteriol. 2014;196:527–539. doi: 10.1128/JB.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio. 2013:4. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, et al. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet. 1996;253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- Leung V, Dufour D, Levesque CM. Death and survival in Streptococcus mutans: differing outcomes of a quorum-sensing signaling peptide. Front Microbiol. 2015;6:1176. doi: 10.3389/fmicb.2015.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Alvarez-Martinez C, Chen Y, Choi KJ, Yeo HJ, Christie PJ. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J Bacteriol. 2012;194:4041–4051. doi: 10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr, Gilmore MS. Genetic diversity among Enterococcus faecalis. PLoS ONE. 2007;2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vajna RM, Johnson JR, Zhang Y, Erlandsen SL, Oli MW, et al. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology. 2007;153:4088–4097. doi: 10.1099/mic.0.2007/007252-0. [DOI] [PubMed] [Google Scholar]
- Ogura M, Tanaka T. Transcription of Bacillus subtilis degR is sigma D dependent and suppressed by multicopy proB through sigma D. J Bacteriol. 1996;178:216–222. doi: 10.1128/jb.178.1.216-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted SB, Erlandsen SL, Dunny GM, Wells CL. High-resolution visualization by field emission scanning electron microscopy of Enterococcus faecalis surface proteins encoded by the pheromone-inducible conjugative plasmid pCF10. J Bacteriol. 1993;175:6229–6237. doi: 10.1128/jb.175.19.6229-6237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted SB, Kao SM, van Putte LJ, Gallo JC, Dunny GM. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J Bacteriol. 1991;173:7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13:632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Kritikos G, Orlova K, Typas A, Bernhardt TG. A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet. 2014;10:e1004056. doi: 10.1371/journal.pgen.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Weaver KE. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J Bacteriol. 2006;188:5374–5384. doi: 10.1128/JB.00513-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Arellano I, Gallego J, Cervera J. The PUA domain - a structural and functional overview. FEBS J. 2007;274:4972–4984. doi: 10.1111/j.1742-4658.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, et al. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun. 1999;67:6067–6075. doi: 10.1128/iai.67.11.6067-6075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Tanasescu AM, Zhao AM, Serrano A, Alston T, Sol A, et al. Streptococcus pyogenes sortase mutants are highly susceptible to killing by host factors due to aberrant envelope physiology. PLoS One. 2015;10:e0140784. doi: 10.1371/journal.pone.0140784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina J, Soll D. The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation. J Biol Chem. 2006;281:6993–7001. doi: 10.1074/jbc.M512841200. [DOI] [PubMed] [Google Scholar]
- Schlievert PM, Gahr PJ, Assimacopoulos AP, Dinges MM, Stoehr JA, Harmala JW, et al. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokeen S, Johnson CM, Greenfield TJ, Manias DA, Dunny GM, Weaver KE. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid. 2010;64:26–35. doi: 10.1016/j.plasmid.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon JH, Bryan EM, Manias DA, Chen Y, Dunny GM. Genetic characterization of the conjugative DNA processing system of enterococcal plasmid pCF10. Plasmid. 2006;56:102–111. doi: 10.1016/j.plasmid.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W, Dimov S, Tong Y, Park HW, Hong BS. Crystal structure of human multiple copies in T-cell lymphoma-1 oncoprotein. Proteins. 2013;81:519–525. doi: 10.1002/prot.24198. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Hancock LE. Suicide and fratricide in bacterial biofilms. Int J Artif Organs. 2009;32:537–544. doi: 10.1177/039139880903200902. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D, Gilmore MS. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. 2014;68:337–356. doi: 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KE, Weaver DM, Wells CL, Waters CM, Gardner ME, Ehli EA. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J Bacteriol. 2003;185:2169–2177. doi: 10.1128/JB.185.7.2169-2177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol. 2014;94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wan H, Gao ZQ, Wei Y, Wang WJ, Liu GF, et al. Insights into the catalytic mechanism of 16S rRNA methyltransferase RsmE (m3U1498) from crystal and solution structures. J Mol Microbiol. 2012;423:576–589. doi: 10.1016/j.jmb.2012.08.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.