Abstract
Objective
HIV-positive individuals are at higher risk than healthy persons for aging-related diseases, including myocardial infarction and non-AIDS defining cancers. Recent evidence suggests that HIV infection may modulate changes in the host cell epigenome, and these changes represent a potential mechanism through which HIV infection accelerates aging. We assessed the difference in DNAm age, an aging marker involving multiple age-related CpG sites, among antiretroviral treatment (ART) naïve HIV-positive and HIV-negative individuals in a cohort of veterans from the Veterans Aging Cohort Study (VACS).
Design
Peripheral blood samples were collected from 19 ART-naïve, HIV-positive and 19 HIV-negative male participants, matched by age and race. Blood samples were collected from HIV-positive participants 7–11 years after ART initiation.
Methods
We compared DNAm age between HIV-positive and HIV-negative groups at baseline and between HIV-positive patients at baseline and follow-up. We also performed an epigenome-wide analysis to identify CpG methylation sites associated with HIV infection.
Results
DNAm age in HIV-positive individuals is, on average, 11.2 years higher than HIV- subjects at baseline, and 2 of 10 HIV-positive individuals showed an increase in DNAm age after ART initiation. Epigenome-wide association studies showed an association of HIV infection with one site, in gene VPS37B, which approached statistical significance in our cohort (p=3.30×10−6, Bonferroni-corrected threshold=1.22×10−7) and was replicated in a second, larger cohort.
Conclusion
ART treatment-naïve HIV-positive individuals have significantly older DNAm age compared to HIV-negative individuals in the VACS cohort. Longitudinal changes in DNAm age are highly variable across individuals after initiation of antiretroviral therapy.
Keywords: HIV, epigenomics, aging, DNA methylation, antiretroviral therapy, highly active
Background
Worldwide, it is estimated that 36.9 million people are living with human immunodeficiency virus (HIV), with 2 million people newly infected in 2014 (World Health Organization, 2014). HIV has notoriously high evolutionary potential, attributable to its high mutation rate and rapid turnover upon infection of a human host [1]. This plasticity in the viral genome gives HIV sophisticated capacity to adapt to its host environment, improving its fitness. Recent evidence also suggests that HIV may induce alterations of the host genome that facilitate its survival and transmission [2].
Epigenetic modifications are changes to the genome of an organism that can alter transcriptional activity without affecting the nucleotide sequence, and recent evidence suggests that virally-mediated host epigenetic modifications play a pivotal role in the pathogenesis of HIV [2, 3]. CpG methylation, a type of epigenetic modification wherein a methyl group is attached to guanine in a G-C dinucleotide in the DNA sequence, is a dynamic process that can precipitate both transient and stable changes in gene expression. Research suggests that HIV infection represents an important driver of CpG methylation changes in the human genome through upregulation of methyltransferase proteins [4] and transcriptional effects on genes such as interleukin 2 (IL-2) [5], IGFBP6, and SATB2 [6]. At a population level, methylation in cis-regulatory regions of CCR5, the major coreceptor that mediates T-cell entry of HIV, is linked to HIV susceptibility and AIDS progression [7].
Age-associated methylation, or patterns of CpG methylation that correlate highly with chronological age in healthy individuals, are well characterized and may represent the cumulative effects of an epigenetic ‘maintenance system’ [8]. It has been suggested that activity, or lack of activity, of this maintenance system is a potential mechanism through which older individuals are at higher risk for age-related diseases. The methylation status of several hundred CpG sites associated with age has been used to create an algorithm for calculating biological age, or DNA methylation (DNAm) age[8].
HIV infection is also associated with increased risk of age-related conditions, including myocardial infarction and non-AIDS defining cancers [9]. The cause for this increased risk is not well understood, but it is thought that HIV-positive patients experience some degree of accentuated aging. Persistent, HIV-related inflammation may result in accelerated immunosenescence mediated through changes in methylation patterns of leukocytes [10]. Two recent studies that compared the DNAm ages of blood in HIV-positive and negative individuals reported that blood from HIV+ individuals is approximately five years older than that of controls [11, 12]. Similar CpG sites are affected by both natural aging and HIV infection [13], with HIV infection accelerating age-related methylation changes in blood tissue by approximately 14 years [13]. Commonalities between processes of natural aging and HIV infection may elucidate a mechanism through which HIV-positive individuals are at higher risk for aging-related diseases.
Our study aimed to assess the difference in DNAm age and identify specific CpG sites associated with HIV infection among antiretroviral treatment (ART) naïve HIV-positive and HIV-negative individuals in a cohort of veterans from the Veterans Aging Cohort Study (VACS). We focused on ART naïve HIV-positive individuals to avoid the potentially confounding effects of ART. Additionally, we assessed DNAm age longitudinally after ART initiation.
Methods
This pilot study included 19 treatment-naïve, HIV-positive and 19 HIV-negative male participants matched by age and race. Participants completed a questionnaire at baseline that collected clinical information, including BMI, the presence of chronic health conditions such as diabetes, and information on cigarette smoking. Peripheral whole blood samples were collected from participants at baseline and for HIV-positive patients at a follow-up visit 7–11 years after baseline. The study was approved by the Veteran’s Administration (VA) Research and Development Committee and the Institutional Review Board of Atlanta VA. All participants signed an informed consent.
Methylation arrays were performed on peripheral blood mononuclear cells (PBMCs) using the Illumina 450k platform (Illumina, San Diego, California, USA) and the data were cleaned and normalized using subset-quantile within array normalization (SWAN) in the minfi package in R version 3.1.3 [14]. After CpG filtering, 409,785 autosomal sites remained for analysis. We separately analyzed 11,080 CpG sites on the X chromosome [15].
DNAm age was calculated using the algorithm developed by Horvath et al. by uploading to the web-based DNAm age calculator tool [8]. Differences of DNAm age between HIV-positive and HIV-negative groups were assessed using a paired t-test.
We performed multivariate linear regression to model DNAm age as a function of HIV infection, controlling for diabetes, BMI, smoking status, and cell type proportions, which are established epigenetic modifiers. The proportions of six different cell types, including granulocyte, monocyte, natural killer cells (NK), B cell, CD4+ and CD8+ T cells, were projected based on cell-type specific DNAm sites from a reference panel of sorted cells [16]. We modeled these calculated leukocyte proportions as covariates to control for confounding effects on epigenetic associations due to the shift of cellular proportions.
Mixed linear regression models for the effect of HIV on methylation status at individual CpG sites included a random effect for chip and were run using the nlme package in R (Pinheiro J et.al. and R Core Team 2016). The final adjusted model for the epigenome-wide analysis included BMI, diabetes status, smoking status (current smoking vs. non-current smoking), and cell type proportions (age was not included since subjects were matched on this factor). A Bonferroni correction was used to determine statistical significance, with the threshold for significance at p=1.2 ×10−7. The top associations with HIV infection were replicated in a publicly available dataset (GSE67751, [11]).
Results
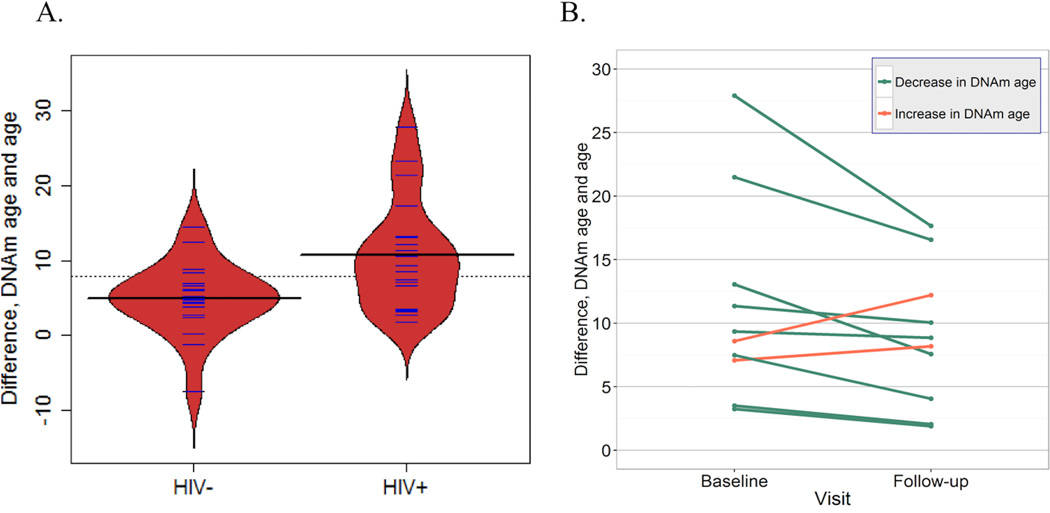
The average age of subjects at baseline was 51.1 years. HIV- subjects had, on average, higher BMI (p=0.034), and were more likely to have diabetes (p=0.047) and lipidemia (p=0.008). DNAm age was correlated with chronological age (R2=0.46) (Supp. Table 1). The difference between DNAm age and chronological age was significantly different at baseline (p=0.007) between HIV-positive and HIV-negative groups; the average difference was 11.2 years (95% CI: 7.7–14.6) and 5.0 years (95% CI: 2.7–7.3), respectively (Figure 1A). HIV infection was associated with DNAm age in a model adjusted for diabetes, BMI, and current smoking. This association diminished when adjusting for cell type proportions, including in a model only adjusted for CD4 T-cell proportion (Supp. Table 2).
Figure 1. Difference between patients’ chronological age and calculated DNAm age.
A. Difference between DNAm age and age, by HIV status, for VACS subjects. Solid horizontal black lines denote the respective means for each group; the dotted line is the overall mean. Blue lines and the red shape represent individual measurements and the density trace, respectively. B. Difference between DNAm age and age, at baseline and follow-up for HIV+ individuals only. The bottommost line reflects two lines that are nearly coincident with one another.
Follow-up visits ranged from 7.9 to 10.1 years after the baseline visit, with an average of 9.1 years between baseline and follow-up. DNAm age trajectory of patients from baseline to follow-up (controlling for age) showed high inter-patient variation (Figure 1B); two out of ten individuals had increased DNAm age difference.
Epigenome-wide association studies showed no CpG sites that reached significance in their association with HIV infection (Table 1, Supp. Figure 1, 2). In a model adjusted for current smoking, diabetes status, BMI and cell type proportions (Inflation factor: 0.946), the most significant CpG site (cg25625162, p=3.30×10−6) was in a known gene on chromosome 12, VPS37B (vacuolar protein sorting 37 homolog B), the products of which are involved in HIV life cycle and infection [17]. HIV+ individuals had, on average, 7% higher (95% CI: 4.6–9.5%) methylation at this site than HIV- individuals. The remaining four top sites could either not be mapped near or in a known gene (cg09801824, cg10580269), or were not in genes specifically associated with HIV infection (cg01424145, cg27097962). The association with cg25625162 in VPS37B persisted in the model adjusted only for BMI, diabetes status, and lipidemia (p=6.70×10−6), characteristics that were unbalanced between the HIV+ and HIV- groups at baseline, and in the unadjusted model (p=6.48×10−7) (Supp. Table 1). A regional plot of the VPS37B gene showed that only one CpG site of the 35 sites in the region of this gene interrogated by the array approached significance for the association with HIV infection (Supp. Figure 3).
Table 1.
Most significant CpG sites in EWAS of HIV infection adjusted for BMI, diabetes status, smoking, and cell proportions (VACS), adjusted for age, sex, and cell proportions and their replication in GSE67751
| CpG site | Chromosome | MAPINFO | P-value, HIV |
Regression coefficient (SE), HIV |
UCSC Gene Name |
P-value, HIV GSE67751 |
Regression coefficient (SE), HIV GSE67751 |
|---|---|---|---|---|---|---|---|
| cg25625162 | 12 | 123373173 | 3.30×10−6 | 0.0706 (0.012) | VPS37B | <0.0001 | 0.1383 (0.016) |
| cg09801824 | 10 | 74076930 | 5.43×10−6 | 0.0570 (0.010) | NA | <0.0001 | 0.1741 (0.012) |
| cg01424145 | 19 | 44285333 | 7.20×10−6 | −0.0463 (0.008) | KCNN4 | 0.8473 | 0.0027 (0.013) |
| cg27097962 | 14 | 105780695 | 1.38×10−5 | −0.0086 (0.002) | PACS2 | 0.5368 | 0.0010 (0.002) |
| cg10580269 | 2 | 69056515 | 1.51×10−5 | 0.0467 (0.009) | NA | <0.0001 | 0.0753 (0.008) |
We replicated the association of HIV infection with cg25625162 (p<0.0001) in a second cohort of 23 HIV-positive and 69 controls [11] controlling for age and sex. PBMCs were obtained from the National Neurological AIDS Bank study or Multicenter AIDS Cohort Study in Los Angeles. Cases and controls had a mean age of 45 years (range, 24–68 years) and 51 years (range, 35–64 years), respectively. A meta-analysis combining the two cohorts using a fixed effects model showed a significant combined effect (Supp. Figure 4). Five additional associations from the top ten most significant CpG sites in the VACS cohort analysis were significant at a 0.05 level when replicated in the second cohort (cg03152187, cg04067612, cg09801824, cg10580269, cg11630226) (Table 1).
Discussion
Using DNAm age as a marker of biological aging, the peripheral blood cells of HIV-positive individuals are, on average, ‘older’ than HIV-negative individuals, but changes in DNAm age are highly variable across individuals after initiation of ART. Modeling HIV as a predictor of DNAm age indicated that the association between HIV and DNAm age is perhaps driven by methylation patterns in CD4 T cells, since controlling for the proportion of CD4 T cells attenuated the association between HIV status and DNAm age. Using an epigenome-wide approach, we found that the most significant CpG site in both our adjusted and unadjusted models was located in a gene related to HIV infection and the viral life cycle: VPS37B, a component of the ESCRT-1 complex that regulates vesicular trafficking processes and thus viral budding and transport. Methylation in the region of this gene may alter ESCRT-1 activity, potentially enhancing the budding of HIV virions from the host cell. The emergence of this CpG site in our genome-wide screen provides evidence for the theory that HIV infection may indeed manipulate methylation patterns in this region.
These associations are consistent with previous work showing that methylation patterns are different in HIV-positive as compared to healthy individuals [11, 13], and support the theory that infection precipitates changes in CpG methylation. It is plausible that host cell DNA methylation changes represent an adaptive technique to increase the virus’ ability to survive, replicate, and establish latency in human cells. These methylation changes, coupled with previous evidence that HIV infection and aging are associated with changes at similar CpG sites, may begin to explain the increase in aging-related outcomes observed in HIV-positive individuals.
This study selected treatment-naïve HIV+ patients for comparison to HIV-negative patients, since ART has been shown to have an effect on CpG methylation through its effect on viremia [3]. This design allows us to examine the methylation effects of HIV infection itself, rather than the combination of HIV infection and therapy. Further, this study allowed us to assess changes in DNAm age before and several years after ART initiation. The potential beneficial effects of ART on DNAm age is suggested by the observation that the majority of HIV-positive individuals (80%) showed improved DNAm age difference after long-term ART.
The sample size was restricted, which affects detection of significant epigenome-wide associations across a large number of CpG sites. However, we were able to replicate the association for cg25625162 (VPS37B) in another cohort. Our sample size also limited our ability to control for important confounding factors, including cell type in the DNAm age analysis, without an unacceptable loss of precision in our models. However, we controlled for smoking behaviors, BMI, and diabetes status, three of the strongest markers of lifestyle and environmental factors expected to influence CpG methylation. Our sample was composed of patients from the VA medical system, which provides comprehensive care to patients of any age testing positive for HIV. However, the HIV-negative persons recruited in this cohort were those seeking primary care at the VA. Notably, the DNAm age in this group was 5.0 years older than the chronological age on average, which likely reflects the high burden of age-related diseases among HIV-negative individuals who seek primary care at VA hospitals. Furthermore, a lack of CpG coverage in the 450k panel in genes known to be associated with HIV infection, such as CCR5 and FOXP3, prevented us from assessing methylation at these sites. Lastly, we assessed PMBCs as an aggregated summary of multiple cell types to identify any DNAm site associated with HIV infection. Although several subtypes of PBMCs may be affected by HIV infection, we were not able to investigate the cell type-specific epigenetic association using sorted cells in this study. Future functional studies should consider cell type-specific epigenetic associations with HIV infection, various stages of cellular differentiation, and molecular mechanisms mediating the epigenetic modification among PLHIV.
These associations are the result of data from a small and unique sample and should be regarded with caution. However, it is notable that under such circumstances, associations with CpG sites in genes relevant to HIV infection were nonetheless observed. Additional research should aim to replicate these associations in larger longitudinal cohorts that can provide more comprehensive and robust information on HIV infection-related epigenetic changes.
Supplementary Material
Negative log10 p-values for the association between HIV infection and methylation status at each CpG site, adjusted for BMI, diabetes, and smoking status, plotted against genomic location by chromosome. The horizontal red line indicates a Bonferroni-corrected significance level of 0.05.
Observed -log10 p-values plotted against expected -log10 p-values for each CpG site in the epigenome-wide analysis of HIV infection. The estimated inflation factor is 0.946.
log10 p-values from mixed model linear regression analysis adjusting for BMI, diabetes, and smoking status, for all CpG sites tested in the VPS37B gene, plotted against their genomic location. The CpG site identified in out screen, cg25625162, is labeled.
Forest plot of the HIV-related DNAm sites from the discovery (VACS) and a replication sample (GSE67751) using β coefficients from linear regression models. The grey box represents the variance of the estimate of the beta value, and the center of the red diamond represents the summary beta point estimate from the two studies, assuming a fixed effects model. The tips of the diamond represent the associated confidence interval.
Acknowledgments
Y.V.S. and V.C.M. developed the research question, K.N.N., Q.H., and Y.V.S. contributed to the data analysis. K.N.N., D.M., K.X., M.S.F., A.C.J, V.C.M. and Y.V.S. wrote the manuscript. Y.V.S. is supported by the American Heart Association (Grant number 13GRNT17060002). This research was supported in part by a developmental grant to Y.V.S. from the NIH Center for AIDS Research at Emory University (P30 AI050409).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. 2005;79(14):9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Hameed EA, Ji H, Shata MT. HIV-Induced Epigenetic Alterations in Host Cells. Adv Exp Med Biol. 2016;879:27–38. doi: 10.1007/978-3-319-24738-0_2. [DOI] [PubMed] [Google Scholar]
- 3.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, et al. CpG methylation controls reactivation of HIV from latency. PLoS pathogens. 2009;5(8):e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pion M, Jaramillo-Ruiz D, Martinez A, Munoz-Fernandez MA, Correa-Rocha R. HIV infection of human regulatory T cells downregulates Foxp3 expression by increasing DNMT3b levels and DNA methylation in the FOXP3 gene. AIDS. 2013;27(13):2019–2029. doi: 10.1097/QAD.0b013e32836253fd. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama-Hosoya K, Ishida T, Youngblood B, Nakamura H, Hosoya N, Koga M, et al. Epigenetic Repression of Interleukin 2 Expression in Senescent CD4+ T Cells During Chronic HIV Type 1 Infection. The Journal of infectious diseases. 2015;211(1):28–39. doi: 10.1093/infdis/jiu376. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Li SK, Yi Yang K, Liu M, Lee N, Tang X, et al. Whole genome methylation array reveals the down-regulation of IGFBP6 and SATB2 by HIV-1. Sci Rep. 2015;5:10806. doi: 10.1038/srep10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gornalusse GG, Mummidi S, Gaitan AA, Jimenez F, Ramsuran V, Picton A, et al. Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(34):E4762–E4771. doi: 10.1073/pnas.1423228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Althoff KN, McGinnis KA, Wyatt CM, Freiberg MS, Gilbert C, Oursler KK, et al. Comparison of risk and age at diagnosis of myocardial infarction, end-stage renal disease, and non-AIDS-defining cancer in HIV-infected versus uninfected adults. Clinical Infectious Diseases. 2015;60(4):627–638. doi: 10.1093/cid/ciu869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath SLA. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. Journal of Infectious Disease. 2015;212(10):1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross AM, Jaeger PA, Kreisberg JF, Licon K, Jepsen KL, Khosroheidari M, et al. Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA. Mol Cell. 2016;62(2):157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rickabaugh TM, Baxter RM, Sehl M, Sinsheimer JS, Hultin PM, Hultin LE, et al. Acceleration of age-associated methylation patterns in HIV-1-infected adults. PloS one. 2015;10(3):e0119201. doi: 10.1371/journal.pone.0119201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klebaner D, Huang Y, Hui Q, Taylor JY, Goldberg J, Vaccarino V, et al. X chromosome-wide analysis identifies DNA methylation sites influenced by cigarette smoking. Clinical epigenetics. 2016;8:20. doi: 10.1186/s13148-016-0189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, et al. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. The Journal of biological chemistry. 2004;279(34):36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative log10 p-values for the association between HIV infection and methylation status at each CpG site, adjusted for BMI, diabetes, and smoking status, plotted against genomic location by chromosome. The horizontal red line indicates a Bonferroni-corrected significance level of 0.05.
Observed -log10 p-values plotted against expected -log10 p-values for each CpG site in the epigenome-wide analysis of HIV infection. The estimated inflation factor is 0.946.
log10 p-values from mixed model linear regression analysis adjusting for BMI, diabetes, and smoking status, for all CpG sites tested in the VPS37B gene, plotted against their genomic location. The CpG site identified in out screen, cg25625162, is labeled.
Forest plot of the HIV-related DNAm sites from the discovery (VACS) and a replication sample (GSE67751) using β coefficients from linear regression models. The grey box represents the variance of the estimate of the beta value, and the center of the red diamond represents the summary beta point estimate from the two studies, assuming a fixed effects model. The tips of the diamond represent the associated confidence interval.