Abstract
Staphylococcal colonization of human skin is ubiquitous, with particular species more frequent at different body sites. Whereas Staphylococcus epidermidis can be isolated from the skin of every individual tested, Staphylococcus aureus is isolated from <5% of healthy individuals. The factors that drive staphylococcal speciation and niche selection on skin are incompletely defined. Here we show that S. aureus is inhibited to a greater extent than S. epidermidis by the sebaceous lipid sapienic acid, supporting a role for this skin antimicrobial in selection of skin staphylococci. We used RNA-Seq and comparative transcriptomics to identify the sapienic acid survival responses of S. aureus and S. epidermidis. Consistent with the membrane depolarization mode of action of sapienic acid, both species shared a common transcriptional response to counteract disruption of metabolism and transport. The species differed in their regulation of SaeRS and VraRS regulons. While S. aureus upregulated urease operon transcription, S. epidermidis upregulated arginine deiminase, the oxygen-responsive NreABC nitrogen regulation system and the nitrate and nitrite reduction pathways. The role of S. aureus ACME and chromosomal arginine deiminase pathways in sapienic acid resistance was determined through mutational studies. We speculate that ammonia production could contribute to sapienic acid resistance in staphylococci.
Keywords: RNA-Seq, Staphylococcus aureus, Staphylococcus epidermidis, sapienic acid, fatty acid, skin, colonization
Introduction
Staphylococci are major commensal colonizers of healthy human skin and leading causes of hospital-acquired infections, responsible predominantly for wound and device-associated infections. Understanding skin survival mechanisms of staphylococci is vital, as individuals colonized by Staphylococcus aureus are at greater risk of infections during hospitalization (Kluytmans et al., 1997; Davis et al., 2004).
Unlike those coagulase-negative staphylococci that are skin-dwelling, the primary human niche of coagulase-positive S. aureus is the nares, with skin colonization being transient and seeded from this location (Moss and Squire, 1948; Kluytmans and Wertheim, 2005; Cho et al., 2010). A comparison of responses and resistance mechanisms between S. aureus and these closely related, long-term skin colonizers, such as S. epidermidis, therefore provides a useful tool to investigate functionalities required for skin colonization and persistence (Coates et al., 2014). Such investigations have increasing relevance with the emergence of community-acquired MRSA lineages, such as USA300 which cause increased skin pathology (Moran et al., 2006; Li et al., 2009).
Atopic dermatitis is a disease presenting as dry, flaky skin lesions, abscesses, and unusually high levels of S. aureus skin colonization (Higaki et al., 1999; Bieber, 2008; Kong et al., 2012). Many host factors of skin are altered in atopic dermatitis, including levels of antimicrobial peptides, antimicrobial fatty acids, and sphingosines, all of which have been associated with S. aureus exclusion (Schafer and Kragballe, 1991; Arikawa et al., 2002; Cho et al., 2010). Levels of sapienic acid in particular were determined to be inversely proportional to levels of S. aureus (Takigawa et al., 2005), identifying sapienic acid as a strong candidate host factor that contributes to prevention of long-term skin colonization by S. aureus.
Recent studies revealed the effects that sapienic acid and other skin fatty acids have on S. aureus survival (Kenny et al., 2009; Cartron et al., 2014; Neumann et al., 2015). Together these studies showed that unsaturated long-chain fatty acids, including sapienic and linoleic acids, cause membrane depolarization in S. aureus leading to large transcriptional changes, especially those pathways associated with cellular energetics (Kenny et al., 2009; Neumann et al., 2015). From the transcriptomic response, it is inferred that the membrane depolarisation leads to disruption of the electron transport chain (Kenny et al., 2009; Neumann et al., 2015).
Here we show that the mean sapienic acid MIC of S. epidermidis strains is greater than S. aureus. Consequently, RNA-Seq was used to compare sapienic acid transcriptional responses with the aim of highlighting skin survival determinants. These investigations form the basis to determine whether sapienic acid responses discriminate staphylococcal species based on their skin-dwelling propensity.
Materials and Methods
Bacterial Strains and Culture
Strains used in this study are listed in Table 1. Overnight cultures were grown for 18 h at 37°C with shaking. Todd Hewitt broth (THB) or agar (THA) was used as the culture media for all experiments. Sapienic acid (Matreya) stock solution was prepared at 8 mg ml-1 in ethanol. Antibiotics were incorporated at concentrations of 12.5 μg ml-1 tetracycline, 100 μg ml-1 ampicillin, 10 μg ml-1 chloramphenicol, and 5 μg ml-1 erythromycin, when appropriate.
Table 1.
Strains used in this study.
| Species | Strain | Description | Reference |
|---|---|---|---|
| S. epidermidis | Rp62a | Intravascular catheter isolate | Christensen et al., 1982, 1985 |
| Tü3298 | Epidermin producer | Allgaier et al., 1986 | |
| NCTC 1457 | PIA producer | Mack et al., 1992 | |
| A19 | Recent skin (forearm) isolate | Kelly, 2013 | |
| B19 | Recent skin (forearm) isolate | Kelly, 2013 | |
| O16 | Recent skin (forearm) isolate | Kelly, 2013 | |
| BL115 | Recent nasal isolate | Libberton, 2011 | |
| S. aureus | Newman | Osteomyelitis isolate | Duthie and Lorenz, 1952 |
| SH1000 | Lab strain (rsbU repaired 8325-4 derivative) | Horsburgh et al., 2002 | |
| MSSA476 | Osteomyelitis isolate | Holden et al., 2004 | |
| MRSA252 | Fatal bacteraemia isolate, MRSA | Holden et al., 2004 | |
| BL014 | Recent nasal isolate | Libberton, 2011 | |
| BL032 | Recent nasal isolate | Libberton, 2011 | |
| SF8300 | CA-MRSA | Diep et al., 2008 | |
| SF8300ax | SF8300 with ACME deletion | Diep et al., 2008 | |
| Liv1245 | Newman arcA::tet from Liv692 | This study | |
| Liv1247 | SF8300 arcA::tet from Liv692 | This study | |
| Liv1249 | SF8300ax arcA::tet from Liv692 | This study | |
| Liv692 | S. aureus SH1000 arcA::tet | Kenny et al., 2009 | |
| Newman tagO | tagO::ery from SA113 tagO | This study | |
| SA113 tagO | tagO::Ery | Bera et al., 2005 | |
| Newman mcrA | NWMN_0050:: Ery | This study | |
| Newman mcrA pSK5632 | Newman mcrA containing pSK5632 + mcrA | This study | |
| Liv1023 | mtlD::tet (SH1000) | Kenny et al., 2013 | |
| Liv1024 | mtlABCD::tet (SH1000) | Kenny et al., 2013 | |
| RN4220 | Restriction deficient strain | Kreiswirth et al., 1983 | |
| SH1000 mnhF | mnhF in frame unmarked deletion | Sannasiddappa et al., 2015 | |
Minimum Inhibitory Concentration Assay
Minimum inhibitory concentration (MIC) assays were performed using a broth microdilution method in 96 well plates, with final well volumes of 200 μl and a sapienic acid concentration range of 200–0.8 μg ml-1. An inoculum of ∼104 CFU ml-1 was used.
Growth and Sapienic Acid Challenge
Overnight broth cultures were adjusted to an OD600 of 0.5 then diluted 25-fold in fresh medium prior to incubation in a water bath with shaking (250 rpm) at 37°C. Sapienic acid/ethanol was added to cell cultures in mid-exponential phase (OD600 ∼0.5) with equivalent volumes of ethanol added to control cultures. For RNA-Seq experiments, cells were harvested by centrifugation 20 min after challenge and suspended in RNAlater (Qiagen).
RNA Extraction and Library Preparation
For cell lysis, bacteria were pelleted at 6,000 RCF for 5 min at 4°C and suspended in 100 μl TE containing 6 mg ml-1 lysostaphin and 400 U ml-1 mutanolysin. Lysis was performed for 15 min at 37°C for S. aureus and 30 min for S. epidermidis. Subsequently, samples were treated with 25 μl of proteinase K (Qiagen) for 30 min at 37°C. RNA was extracted using the RNeasy kit (Qiagen). Samples were DNase-treated using turbo DNase (Ambion), and the DNase removed using the RNeasy MinElute clean up kit (Qiagen).
Depletion of rRNA was achieved with a Ribo-Zero magnetic kit for Gram-positive bacteria (Epicentre). The concentration of RNA was normalized before library construction using strand specific ScriptSeq kits (Epicentre); libraries were prepared by the Centre for Genomic Research (CGR), Liverpool. RNA-Seq samples were sequenced by paired-end sequencing using the HiSeq platform (Illumina).
RNA-Seq Differential Expression Analysis
Bowtie (Langmead et al., 2009) and Edge R (Robinson and Oshlack, 2010; Robinson et al., 2010) were used to map reads and determine the differentially expressed (DE) genes, respectively. Genes with mapped transcripts that had a false discovery rate <0.05, as determined by Benjamin and Hochberg analysis, were considered differentially expressed between control and test conditions.
Gene expression changes in biosynthetic pathways were associated using KEGG mapper-search and color (Kanehisa and Goto, 2000; Kanehisa et al., 2012). The S. aureus transcriptome meta-database (SATMD) (Nagarajan and Elasri, 2007) was used to compare sapienic acid DE gene sets with existing S. aureus DE gene sets.
cDNA Generation and qPCR
The tetro cDNA synthesis kit (Bioline) was used for cDNA synthesis using random hexamer primers and 2 μg RNA per reaction. Table 2 lists the qPCR primers. Novel primers were designed using primer-BLAST (Ye et al., 2012). Primer efficiency for all primers was confirmed to be within 90–100% as described previously (Nolan et al., 2006). All qPCR reactions were performed using SensiFAST SYBR Hi-ROX kit (Bioline) with the ABI StepOnePlus (Life Technologies); data analysis used the ABI StepOnePlus software. At least two technical replicates and three biological replicates were used to determine fold change in gene expression between samples.
Table 2.
Primers used in this study.
| Gene name | Primer sequences | Efficiency (%) | Reference |
|---|---|---|---|
| rpoB | F-GCGAACATGCAACGTCAAGR-GACCTCTGTGCTTAGCTGTAATAGC | 97.0 | This study |
| hu | F-TTTACGTGCAGCACGTTCACR-AAAAAGAAGCTGGTTCAGCAGTAG | 90.3 | Duquenne et al., 2010 |
| gyrB Tü3298 | F-AGAAAAGATGGGACGCCCTGR-CACCATGAAGACCGCCAGAT | 96.6 | This study |
| gyrB Newman | F-ATCGACTTCAGAGAGAGGTTGR-CCGTTATCCGTTACTTTAATCCA | 92.9 | Kenny et al., 2009 |
| capB | F-GCGATATGCGTAAGCCAACACR-GGTACAGGGCCAGCTGTTAG | 91.5 | This study |
| pyrP | F-CGATGTTTGGCGCAACAGTAR-GCTGGTATTTGCGCCTTCG | 92.5 | This study |
| clpB | F-TGGTGCACCTCCAGGTTATGR-AGAATCCGTAAGACGACCTTCA | 99.0 | This study |
| farR | F-ACGCCAGCTGTGTGGATTATR-AACGACTGCGACCTTGATGT | 93.3 | This study |
| sasF | F-TCACTCTGCGATTGAAGGCAR-TTTCCGGTGCCGAATGATCT | 95.0 | This study |
| narH | F-TGGCCTTTCCATTGCATCCTR-TTCAGTGTCGCCAGCAGTTA | 93.6 | This study |
| mcrA (gene knockout) | F1-ACATGAATTCGGAATTGGTTAAGTTCACTCR1-CCGGTACCAGAACTCATCTAATA CAGACF2-ATAACTGCGGCCGCTGTATCACTTAGGTGTATCAR2-CGACGGATCCTCCAGCTGTTACCAGTCCGA | – | This study |
| mcrA (complementation) | F-TTACGGATCCTTAAGTAACTTCTTTCAAR-TTATAAAGCTTACATCATTTCTGTCCCAG | – | This study |
Construction of Gene Mutants
An allelic replacement mutant of NWMN_0050 and complementation of this mutant were constructed using the previously described method of Horsburgh et al. (2004) using the primers listed in Table 2. Allelic replacement mutants of tagO and arcA in strain Newman were generated by phage transduction (Horsburgh et al., 2001) from previously described mutants (Table 1).
Data Accession Numbers
The complete genome sequence of S. epidermidis Tü3298 is available at http://www.ebi.ac.uk/ena/data/view/PRJEB11651 (Moran and Horsburgh, 2016). The Illumina sequence read data generated from the RNA-Seq experiments are available from ArrayExpress database1 under accession number E-MTAB-4587.
Results and Discussion
Comparative Sapienic Acid Resistance
We hypothesized that differences in sapienic acid resistance might contribute to the higher frequency and persistence of S. epidermidis on healthy human skin relative to S. aureus. Consistent with our hypothesis, we identified that the mean sapienic acid MIC of S. epidermidis was approximately three times higher than that of S. aureus strains (p = 0.023) (Figure 1).
FIGURE 1.
Sapienic acid minimum inhibitory concentration (MICs) of S. aureus and Staphylococcus epidermidis strains. MICs were determined using the broth microdilution method. MICs are the lowest concentration found to inhibit growth. The dotted line shows the mean MIC of each species.
Previous studies revealed that S. aureus colonizes the skin of atopic dermatitis sufferers, and its colonization frequency inversely correlates with sapienic acid levels (Takigawa et al., 2005). Sapienic acid is not the only factor on atopic skin to be linked with S. aureus colonization, and levels of other skin lipids and antimicrobial peptides are also linked with colonization (Schafer and Kragballe, 1991; Arikawa et al., 2002; Cho et al., 2010). Although sapienic acid is antimicrobial it remains to be demonstrated that within sebum its activity in vivo reduces S. aureus colonization. These in vitro data presented here support a hypothesis that sapienic acid resistance contributes to skin colonization and persistence. The differential survival data are consistent with the increased frequency of staphylococci, not only S. aureus, on skin of atopic dermatitis patients whose skin lipid levels are reduced (Kong et al., 2012; Soares et al., 2013).
While S. epidermidis strains exhibited a greater sapienic acid MIC compared with S. aureus, S. epidermidis growth was inhibited by lower concentrations than skin colonizing corynebacteria. For example, Corynebacterium stratum which colonizes sebaceous niches has up to 10 times greater sapienic acid MIC (Fischer et al., 2012) than the S. epidermidis strains studied here.
Growth of Sapienic Acid-Challenged S. aureus and S. epidermidis
Similar to findings of our previous study of the S. aureus response to linoleic acid (Kenny et al., 2009), sapienic acid was reported by Neumann et al. (2015) to induce a major adaptive transcriptional response in S. aureus SH1000. We therefore hypothesized that comparison of the transcriptional responses of S. aureus with S. epidermidis might identify differential skin survival mechanisms of these species relating to antimicrobial lipids that might also account for their MIC differences. Since we were interested in the typical response of each species, strains S. aureus Newman and S. epidermidis Tü3298 were selected, being representative of each species based on their MICs.
We used a similar experimental design to our previous linoleic acid transcriptional response study (Kenny et al., 2009), substituting microarrays with RNA-Seq for transcriptomics. Both staphylococci were grown to mid-log phase and challenged with the lowest concentration of sapienic acid that caused an equivalent growth rate reduction (Figure 2), specifically, 11.25 μM for S. aureus Newman and 15 μM for S. epidermidis Tü3298. The transcriptomes of each species were determined under challenge and control conditions using RNA-Seq, with resolution of the differentially expressed genes between these conditions.
FIGURE 2.
Growth of S. aureus Newman (A) and S. epidermidis Tü3298 (B) challenged with sapienic acid. Bacteria were grown to OD600 = 0.5 before addition of sapienic acid (dotted line). Sapienic acid was added to concentrations 7.5 μM (cross), 11.25 μM (triangle) and 15 μM (circle)-Tü3298 only- or no addition, 0 μM (diamond).
Sapienic Acid Challenge Transcriptomes
In response to sapienic acid challenge, S. aureus Newman showed 1224 significantly differentially expressed (DE) genes; 630 genes were upregulated and 594 were downregulated (Supplementary Table S1A). S. epidermidis Tü3298 showed 1505 significantly DE genes in response to sapienic acid challenge; 708 genes were upregulated and 797 were downregulated (Supplementary Table S1B). A greater proportion of DE genes across the genome was observed for S. epidermidis Tü3298 (64.5% of 2332 total genes) compared with S. aureus Newman (45.6% of 2686 total genes).
Based on their homologous gene content, the sapienic acid transcriptomes of S. aureus and S. epidermidis were compared, which revealed 525 shared DE genes with a common regulation pattern (Figure 3, Supplementary Table S1C). Thus, the common transcriptomic response represents less than one half or one third of the DE genes of S. aureus and S. epidermidis, respectively. For both species, a selection of DE genes were confirmed using qPCR (Supplementary Figure S1).
FIGURE 3.
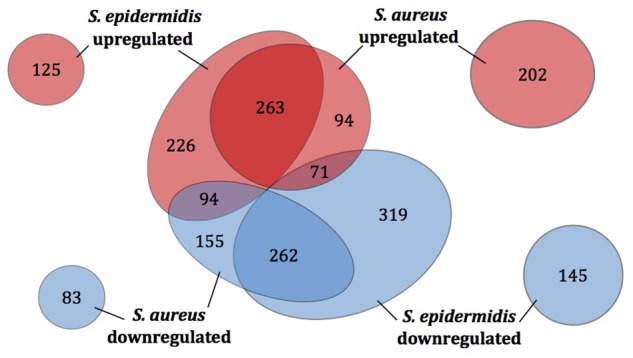
Differentially expressed genes of S. aureus and S. epidermidis post challenge with sapienic acid. The number of homologous genes of S. aureus Newman and S. epidermidis Tü3298 (central circles) that were upregulated (red) or downregulated (blue) following sapienic acid challenge were compared. Numbers of upregulated and downregulated genes with no homologue in the other species are shown by the outer circles.
Similarities in regulation within the shared responses to sapienic acid are likely to reflect a common response to the membrane depolarization mode of action that disrupts function of the electron transport chain (Cartron et al., 2014). Consistent with this mode of action, we identified upregulated transcription of genes required for sugar uptake, glycolysis, the TCA cycle, NADPH/NADP+ recycling and pyruvate metabolism (Figure 4). This shared response may enable the staphylococci to maintain ATP synthesis following disruption of energy generation at the membrane. Downregulated transcription was associated with genes for cell growth in both species (Figure 5), including peptidoglycan biosynthesis, cell membrane biosynthesis and DNA replication and repair. This response is consistent with adaptation of the bacteria to their changed environment, and with the growth lag observed following sapienic acid challenge of both S. aureus and S. epidermidis (Figure 2).
FIGURE 4.
Changes in gene expression of carbon metabolism associated genes in S. aureus and S. epidermidis challenged with sapienic acid.
FIGURE 5.
Changes in gene expression of growth associated genes in S. aureus and S. epidermidis challenged with sapienic acid.
Downregulation of cell membrane glycerophospholipid biosynthesis genes, when combined with the upregulation of fatty acid degradation genes (aldA, NWMN_1858/SETU_01602, NWMN_2090/SETU_01661, NWMN_2091/SETU_01662), supports the description of sapienic acid incorporation into membrane lipids and lipoproteins in staphylococci (Parsons et al., 2012).
Host fatty acids are metabolized in S. aureus by a fatty acid kinase consisting of FakA plus FakB1 or FakB2 subunits in the phospholipid biosynthesis pathway (Parsons et al., 2014). The fakA and fakB1 genes were downregulated in both S. aureus Newman and S. epidermidis Tü3298. Gene fakB2 was upregulated only in S. epidermidis; since FakB2 binds long chain unsaturated fatty acids this supports sapienic acid incorporation into the cell phospholipid in this species. Incorporation of AFAs into cellular lipoproteins and phospholipids was suggested as a detoxification mechanism (Desbois and Smith, 2010), while incorporation of fatty acids into lipoproteins enhances the immune response against S. aureus (Nguyen et al., 2015).
Sapienic Acid Transcriptome and Niche Colonization
The levels of topical skin lipids vary across the body surface (Lampe et al., 1983), and sebaceous richness inversely correlates with staphylococcal frequency (Costello et al., 2009; Coates et al., 2014). Cutaneous skin lipids, including sapienic acid, have potential to act as environmental cues for niche adaptation, particularly with the extensive sapienic acid-dependent transcription changes observed that indicate major transcriptome reprofiling in both staphylococci studied here.
Such an environmentally responsive pathway could correspond to the reduced virulence factor expression mediated by SaeRS in response to sapienic acid in S. aureus, which may promote its colonization over infection (Neumann et al., 2015). S. aureus Newman used in this study has an saeS mutation, resulting in constitutive expression of the SaeRS two-component system regulon (Cue et al., 2015). This mutation likely explains the lack of differential expression of hemolysins in the S. aureus Newman data set. The saeRS operon homologs in S. epidermidis (SETU_00325 and SETU_00326, respectively) were not differentially expressed in response to sapienic acid. Moreover, genes of the S. epidermidis SaeRS regulon (Handke et al., 2008) did not show any particular expression pattern that would suggest this regulon was modulated. On this basis, we propose that SaeRS is not a key regulator of the sapienic acid survival response of S. epidermidis.
Sapienic acid responsive gene expression changes included determinants that protect staphylococci from innate immune defenses of the skin. Adhesin genes implicated in colonization of the nose and skin, such as sdrC in S. aureus Newman and ebh genes in S. epidermidis Tü3298, were upregulated (7.4 and 1.3–2.6 fold, respectively) after challenge (Supplementary Table S1). In response to sapienic acid S. aureus Newman markedly upregulated (2.8–52.5 fold) capsule biosynthesis genes, approximating the response that we previously reported of S. aureus MRSA252 to linoleic acid (Kenny et al., 2009). Despite this pronounced upregulation of capsule biosynthesis genes, Neumann et al. (2015) identified that capsule deficient mutants do not have altered sapienic acid survival, at least for the laboratory strain SH1000 they studied.
Comparing each species’ response to sapienic acid there is further distinction between two-component signal transduction systems. The sapienic acid response of S. epidermidis Tü3298 includes upregulation of the vraD and vraE which encode an ABC transporter, important for bacitracin resistance. Contrastingly vraDE is not differentially expressed in S. aureus Newman. The VraDE transporter is regulated by the GraRS two-component system, that responds to cationic antimicrobial peptides (Pietiainen et al., 2009). Resistance to cationic antimicrobial peptides is an important factor in skin colonization and marks out a further distinction between the responses of each species to sapienic acid, with the potential for a coordinated antimicrobial response in S. epidermidis.
Sapienic Acid Transcriptomes and Resistance
Comparison of the sapienic acid transcriptomes of S. aureus and S. epidermidis revealed candidate genes that might be associated with sapienic acid resistance based upon their upregulation or presence in a pathway. The contribution of these genes to resistance was explored with allelic replacement mutants.
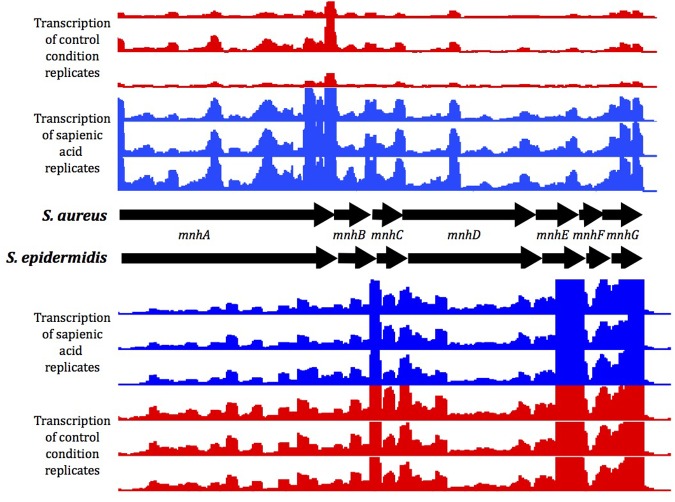
Genes of the mnhABCDEFG operon were all considerably upregulated in S. aureus (4.7–6.8 fold change), though less upregulated in S. epidermidis (1.3–1.9 fold change) after sapienic acid challenge. Analysis of transcript abundance data (Figure 6) revealed that expression of the mnh operon was high in S. epidermidis during normal growth (control) conditions (232–1341.3 FPKM), while little expression was evident in S. aureus in these conditions (22.3–124.1 FPKM). A mnhF in-frame deletion mutant was investigated here for a role in sapienic resistance and had a twofold reduction in MIC (24 μg ml-1) compared with isogenic S. aureus SH1000 (48 μg ml-1). A recent study by Sannasiddappa et al. (2015) determined that mnhF confers resistance to bile salts through efflux of cholic acid. Several mnh genes encode Mrp family secondary antiporter proteins associated with cation/proton transport which can increase the transmembrane electrical potential in staphylococci (Swartz et al., 2007).
FIGURE 6.
Gene expression of the mnh operon under control conditions (red) or following sapienic acid challenge (blue). Mapped reads for replicate RNA-Seq data were visualized in integrated genome browser (IGB).
In addition to the mnh operon, multiple putative cation antiporters and osmoprotectant transporters were upregulated in response to sapienic challenge in S. aureus (NWMN_2457, NWMN_2050, NWMN_2089 and NWMN_0690) and S. epidermidis (SETU_02263 and SETU_00248-00254). The transcriptional upregulation of these transporters may protect the cell from the effects of solute leakage and membrane depolarization caused by sapienic acid (Greenway and Dyke, 1979; Parsons et al., 2012).
In response to sapienic acid challenge, S. epidermidis and S. aureus upregulate expression of different metabolic pathways that generate ammonia. S. aureus upregulated the urease operon (12.2–13.4 fold) with no consistent differential expression of this operon in S. epidermidis (Figure 7). In contrast, S. epidermidis upregulated arcC (1.7-fold) of the arginine deiminase pathway while the arc operon was not DE in S. aureus. S. epidermidis also upregulated the oxygen-responsive NreABC nitrogen regulation system (1.9–2.5 fold) and likewise upregulated the nitrate and nitrite reduction pathways (1.6–3.6 and 1–2.2 fold, respectively). Nitrate and nitrite dissimilation is coupled to the generation of a proton motive force in anoxic conditions and nitrite dissimilation generates cytoplasmic ammonia (Schlag et al., 2008). The arc, nitrate and nitrite reductase operons are induced in staphylococci only in the absence of oxygen (Fedtke et al., 2002; Lindgren et al., 2014; Nilkens et al., 2014), further supporting that there is reduced uptake or altered perception of oxygen following sapienic acid challenge. S. epidermidis using nitrate as an alternative acceptor for its electron transport chain might offer considerable metabolic flexibility compared with S. aureus.
FIGURE 7.
Gene expression changes in ammonia and ammonium producing processes arginine deiminase pathway (A), urease activity (B) and the nitrate reductase pathway (C) in S. aureus and S. epidermidis. Proteins within pathways are colored for S. aureus Newman (left) and S. epidermidis Tü3298 (right) for changes in gene expression following sapienic acid challenge.
We previously determined that expression of the arginine deiminase pathway operon (arcABC) contributes to S. aureus linoleic acid resistance (Kenny et al., 2009). The arginine deiminase pathways encoded chromosomally or on the ACME element arc both result in ammonia production, so here we tested if either of these operons contribute to sapienic acid resistance. The sapienic acid MIC was determined for S. aureus SF8300 and its isogenic ACME element deletion mutant, SF8300ax; the MIC of the mutant was twofold lower than its parent strain (Figure 8). The sapienic acid MIC of the chromosomal arcA mutants of S. aureus SF8300 and Newman were also twofold lower than their parent strains. While there was no difference in the MIC of SF8300ax and SF8300ax arcA mutant, there was a consistent reduction in growth of the SF8300ax arcA mutant compared with SF8300ax (at 3 μg/ml sapienic acid in the MIC assay mean OD600= 0.41 and 0.94, respectively) (Figure 8).
FIGURE 8.

Sapienic acid MICs of S. aureus wild type and mutant strains. MICs were determined using the broth microdilution method. MICs are the lowest concentration found to inhibit growth. Mutants of genes predicted to be important in sapienic acid resistance due to RNA Seq data were contructed or obtained.
Staphylococcal myosin cross reactive antigen (McrA) homologs were previously proposed as antimicrobial lipid resistance determinants (Coates et al., 2014) due to their similarity with fatty acid hydratases of streptococci (Bevers et al., 2009; Volkov et al., 2010; Rosberg-Cody et al., 2011; Joo et al., 2012). Following sapienic acid challenge there was increased expression of the mcrA gene homologs (NWMN_0050 and SETU_00673), 7.4 and 5.4-fold, respectively in S. aureus and S. epidermidis. Fatty acid hydratase enzymes convert unsaturated fatty acids into their saturated counterparts, which is a detoxification mechanism for oleic acid in Streptococcus pyogenes (Volkov et al., 2010). The staphylococcal McrA has no obvious secretion motifs making it unlikely to act on extracellular sapienic acid, though it could act to facilitate its metabolism intracellularly. Here, allelic replacement of mcrA in S. aureus was achieved to investigate its contribution to resistance. Somewhat unexpectedly, the sapienic acid MIC of S. aureus Newman mcrA (NWMN_0050) was greater (48 μg ml-1) than its isogenic parent strain (6 μg ml-1) (Figure 8). This increased resistance phenotype was reversed in the Newman mcrA pSK5632+mcrA complementation strain, which had the same MIC as the wild type. Mutation of mcrA takes the survival of S. aureus Newman to a level of sapienic acid survival similar to a S. epidermidis strain. This indicates that an inability to saturate sapienic acid to palmitic acid through McrA acitivity, or a distinct cellular lipid conversion by McrA impacts S. aureus survival. By comparison, the mcrA gene (SETU_006730) of S. epidermidis Tü3298 contains a premature stop codon, indicating this gene activity may not be functional.
Wall teichoic acid (WTA) deficient mutants have reduced MIC for antimicrobial fatty acids (Kohler et al., 2009) and here, S. aureus Newman tagO was shown to have a very reduced sapienic acid MIC (< 0.8 μg ml-1), over eight times lower than its isogenic wild type strain (Figure 8). Despite its importance for survival, genes of the WTA biosynthesis pathway were downregulated in both species (tagA, tagG, dltX, and gtaB in S. aureus, tagB and tagF/tarF in S. epidermidis). In addition, both S. epidermidis and S. aureus downregulated dlt, mprF, and isdA genes which would be predicted to increase cell hydrophobicity, but these transcription changes may reflect peptidoglycan modification genes mirroring a reduction in cell wall biosynthesis during a period of reduced growth post sapienic acid challenge.
S. epidermidis Specific Resistance Determinants
Key sapienic acid resistance determinants that differentiate the increased S. epidermidis sapienic acid MIC from S. aureus might be identifiable from their signature of transcription upregulation and/or presence only in the S. epidermidis data set. Of those 38 genes upregulated >2-fold in S. epidermidis with no homolog in S. aureus, 12 are annotated with transport functions (Supplementary Table S2). These genes may counteract the leakage of solutes caused by sapienic acid (Greenway and Dyke, 1979; Parsons et al., 2012); transport of osmolytes is a key resistance mechanism used by staphylococci during acid stress, where there is also accompanying membrane depolarization (Bore et al., 2007). A further nine genes of the 38 were hypothetical genes, revealing a need to evaluate potential resistance determinants within this gene set.
Conclusion
S. epidermidis strains have greater sapienic resistance than S. aureus. The transcriptional responses of S. aureus and S. epidermidis to sapienic acid reveal that in addition to a shared stimulon, there are multiple distinct pathways modulated in each species. Our data identifies potential roles for the use of alternative respiration pathways, ammonia production and cation/osmolyte transport in differential survival from sapienic acid.
Author Contributions
JM designed and performed experiments and wrote the paper. JA designed and performed experiments. MH conceived and designed experiments and wrote the paper. All authors read and approved submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Centre for Genomic Research, University of Liverpool, UK carried out rRNA-Seq data generation and processing. We thank Henry F. Chambers, Friedrich Götz, and Simon R. Clarke for sharing strains.
Funding. JM was funded by BBSRC grants BB/D003563/1 and BB/L023040/1 awarded to MH, both with support from Unilever Plc. JA was funded by the Saudi Arabia Ministry of Education. The funders were not involved in the study design, collection of samples, analysis of data, interpretation of data, the writing of this report or the decision to submit this report for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00033/full#supplementary-material
References
- Allgaier H., Jung G., Werner R., Schneider U., Zahner H. (1986). Epidermin - sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur. J. Biochem. 160 9–22. 10.1111/j.1432-1033.1986.tb09933.x [DOI] [PubMed] [Google Scholar]
- Arikawa J., Ishibashi M., Kawashima M., Takagi Y., Ichikawa Y., Imokawa G. (2002). Decreased levels of sphingosine, a natural antimicrobial agent, may be associated with vulnerability of the stratum corneum from patients with atopic dermatitis to colonization by Staphylococcus aureus. J. Investig. Dermatol. 119 433–439. 10.1046/j.1523-1747.2002.01846.x [DOI] [PubMed] [Google Scholar]
- Bera A., Herbert S., Jakob A., Vollmer W., Gotz F. (2005). Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan OM acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55 778–787. 10.1111/j.1365-2958.2004.04446.x [DOI] [PubMed] [Google Scholar]
- Bevers L., Pinkse M., Verhaert P., Hagen W. (2009). Oleate hydratase catalyzes the hydration of a nonactivated carbon-carbon bond. J. Bacteriol. 191 5010–5012. 10.1128/JB.00306-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber T. (2008). Mechanisms of disease: Atopic dermatitis. N. Engl. J. Med. 358 1483–1494. 10.1056/NEJMra074081 [DOI] [PubMed] [Google Scholar]
- Bore E., Langsrud S., Langsrud O., Rode T., Holck A. (2007). Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153 2289–2303. 10.1099/mic.0.2007/005942-0 [DOI] [PubMed] [Google Scholar]
- Cartron M., England S., Chiriac A., Josten M., Turner R., Rauter Y., et al. (2014). Analysis of the bactericidal activity of the human skin fatty acid, cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob. Agents Chemotherapy 58 3599–3609. 10.1128/AAC.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J., Xuan C., Miller L. (2010). Lucky number seven: RNase 7 can prevent Staphylococcus aureus skin colonization. J. Investig. Dermatol. 130 2703–2706. 10.1038/jid.2010.294 [DOI] [PubMed] [Google Scholar]
- Christensen G., Simpson W., Bisno A., Beachey E. (1982). Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen G., Simpson W., Younger J., Baddour L., Barrett F., Melton D., et al. (1985). Adherence of coagulase negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22 966–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates R., Moran J., Horsburgh M. (2014). Staphylococci: colonizers and pathogens of human skin. Future Microbiol. 9 75–91. 10.2217/fmb.13.145 [DOI] [PubMed] [Google Scholar]
- Costello E., Lauber C., Hamady M., Fierer N., Gordon J., Knight R. (2009). Bacterial community variation in human body habitats across space and time. Science 326 1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Junecko J., Lei M., Blevins J., Smeltzer M., Lee C. (2015). SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS ONE 10:e0123027 10.1371/journal.pone.0123027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K., Stewart J., Crouch H., Florez C., Hospenthal D. (2004). Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin. Infect. Dis. 39 776–782. 10.1086/422997 [DOI] [PubMed] [Google Scholar]
- Desbois A., Smith V. (2010). Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 85 1629–1642. 10.1007/s00253-009-2355-3 [DOI] [PubMed] [Google Scholar]
- Diep B., Stone G., Basuino L., Graber C., Miller A., Des Etages S., et al. (2008). The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197 1523–U35. 10.1086/587907 [DOI] [PubMed] [Google Scholar]
- Duquenne M., Fleurot I., Aigle M., Darrigo C., Borezée-Durant E., Derzelle S., et al. (2010). Tool for quantification of staphylococcal enterotoxin gene expression in cheese. Appl. Environ. Microbiol. 76 1367–1374. 10.1128/AEM.01736-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthie E., Lorenz L. (1952). Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6 95–107. 10.1099/00221287-6-1-2-95 [DOI] [PubMed] [Google Scholar]
- Fedtke I., Kamps A., Krismer B., Gotz F. (2002). The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 184 6624–6634. 10.1128/JB.184.23.6624-6634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C., Drake D., Dawson D., Blanchette D., Brogden K., Wertz P. (2012). Antibacterial activity of sphingoid bases and fatty acids against Gram positive and Gram negative bacteria. Antimicrob. Agents Chemother. 56 1157–1161. 10.1128/AAC.05151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway D., Dyke K. (1979). Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J. Gen. Microbiol. 115 233–245. 10.1099/00221287-115-1-233 [DOI] [PubMed] [Google Scholar]
- Handke L. D., Rogers K. L., Olson M. E., Somerville G. A., Jerrells T. J., Rupp M. E., et al. (2008). Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infect. Immun. 76 141–152. 10.1128/IAI.00556-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki S., Morohashi M., Yamagishi T., Hasegawa Y. (1999). Comparative study of staphylococci from the skin of atopic dermatitis patients and from healthy subjects. Int. J. Dermatol. 38 265–269. 10.1046/j.1365-4362.1999.00686.x [DOI] [PubMed] [Google Scholar]
- Holden M., Feil E., Lindsay J., Peacock S., Day N., Enright M., et al. (2004). Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. PNAS 101 9786–9791. 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M., Aish J., White I., Shaw L., Lithgow J., Foster S. (2002). Sigma B modulates virulence determinant expression and stress resistance: characterization of a Functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184 5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M., Wiltshire M., Crossley H., Ingham E., Foster S. (2004). PheP, a putative amino acid permease of Staphylococcus aureus, contributes to survival in vivo and during starvation. Infect. Immun. 72 3073–3076. 10.1128/IAI.72.5.3073-3076.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. (2001). PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69 3744–3754. 10.1128/IAI.69.6.3744-3754.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo Y., Jeong K., Yeom S., Kim Y., Kim Y., Oh D. (2012). Biochemical characterization and FAD-binding analysis of oleate hydratase from Macrococcus caseolyticus. Biochimie 94 907–915. 10.1016/j.biochi.2011.12.011 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40 D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. (2013). Genomic and Metagenomic Analysis of the Skin Microbiota. Ph.D. thesis, University of Liverpool, Liverpool. [Google Scholar]
- Kenny J., Moran J., Kolar S., Ulanov A., Li Z., Shaw L., et al. (2013). Mannitol utilisation is required for protection of Staphylococcus aureus from human skin antimicrobial fatty acids. PLoS ONE 8:e67698 10.1371/journal.pone.0067698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny J., Ward D., Josefsson E., Jonsson I., Hinds J., Rees H., et al. (2009). The Staphylococcus aureus response to unsaturated long-chain free fatty acids: survival mechanisms and virulence implications. PLoS ONE 4:e4344 10.1371/journal.pone.0004344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans J., Wertheim H. (2005). Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 33 3–8. 10.1007/s15010-005-4012-9 [DOI] [PubMed] [Google Scholar]
- Kluytmans J., Vanbelkum A., Verbrugh H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Weidenmaier C., Peschel A. (2009). Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 191 4482–4484. 10.1128/JB.00221-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H., Oh J., Deming C., Conlan S., Grice E., Beatson M. (2012). Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22 850–859. 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B., Lofdahl S., Betley M., Oreilly M., Schlievert P., Bergdoll M., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305 709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- Lampe M., Burlingame A., Whitney J., Williams M., Brown B., Roitman E., et al. (1983). Human stratum corneum lipids- characterization and regional variations. J. Lipid Res. 24 120–130. [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Diep B., Villaruz A., Braughton K., Jiang X., Deleo F., et al. (2009). Evolution of Virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. PNAS 106 5883–5888. 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libberton B. (2011). The Ecology of Staphylococcus aureus. Ph.D. thesis, University of Liverpool, Liverpool. [Google Scholar]
- Lindgren J., Thomas V., Olson M., Chaudhari S., Nuxoll A., Schaeffer C., et al. (2014). Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J. Bacteriol. 196 2277–2289. 10.1128/JB.00051-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D., Siemssen N., Laufs R. (1992). Parallel induction by glucose of adherence and a polysacharide antigen specific for plastic-adherent-Staphylococcus-epidermidis - evidence for functional relation to intercellular-adhesion. Infect. Immun. 60 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran G., Krishnadasan A., Gorwitz R., Fosheim G., Linda M., Mcdougal L., et al. (2006). Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355 666–674. 10.1056/NEJMoa055356 [DOI] [PubMed] [Google Scholar]
- Moran J., Horsburgh M. (2016). Whole-genome sequence of Staphylococcus epidermidis Tü3298. Genome Announc. 4 e112–e116. 10.1128/genomeA.00112-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Squire J. (1948). Nose and skin carriage of Staphylococcus aureus in patients receiving penicillin. Lancet 1 320–325. 10.1016/S0140-6736(48)92088-1 [DOI] [PubMed] [Google Scholar]
- Nagarajan V., Elasri M. (2007). SAMMD: Staphylococcus aureus microarray meta-database. BMC Genomics 8:351 10.1186/1471-2164-8-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann Y., Ohlsen K., Donat S., Engelmann S., Kusch H., Albrecht D., et al. (2015). The effect of skin fatty acids on Staphylococcus aureus. Arch. Microbiol. 197 245–267. 10.1007/s00203-014-1048-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Hanzelmann D., Härtner T., Peschel A., Götz F. (2015). Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect. Immun. 84 205–215. 10.1128/IAI.00822-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilkens S., Koch-Singenstreu M., Niemann V., Gotz F., Stehle T., Unden G. (2014). Nitrate/oxygen co-sensing by an NreA/NreB sensor complex of Staphylococcus carnosus. Mol. Microbiol. 91 381–393. 10.1111/mmi.12464 [DOI] [PubMed] [Google Scholar]
- Nolan T., Hands R., Bustin S. (2006). Quantification of mRNA using real-time RT-PCR. Nature Protocols 1 1559–1582. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- Parsons J., Broussard T., Bose J., Rosch J., Jackson P., Subramanian C., et al. (2014). Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111 10532–10537. 10.1073/pnas.1408797111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J., Yao J., Frank M., Jackson P., Rock C. (2012). Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J. Bacteriol. 194 5294–5304. 10.1128/JB.00743-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietiainen M., Francois P., Hyyrylainen H., Tangomo M., Sass V., Sahl H., et al. (2009). Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429 10.1186/1471-2164-10-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Mccarthy D., Smyth G. (2010). EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11 R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosberg-Cody E., Liavonchanka A., Gobel C., Ross R., O’sullivan O., Fitzgerald G., et al. (2011). Myosin-cross-reactive antigen (MCRA) protein from Bifidobacterium breve is a FAD-dependent fatty acid hydratase which has a function in stress protection. BMC Biochem 12:9 10.1186/1471-2091-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannasiddappa T., Hood G., Hanson K., Costabile A., Gibson G., Clarke S. (2015). Staphylococcus aureus MnhF mediates cholate efflux and facilitates survival under human colonic conditions. Infect. Immun. 83 2350–2357. 10.1128/IAI.00238-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer L., Kragballe K. (1991). Abnormalities in epidermal lipid-metabolism in patients with Atopic dermatitis. J. Investig. Dermatol. 96 10–15. 10.1111/1523-1747.ep12514648 [DOI] [PubMed] [Google Scholar]
- Schlag S., Fuchs S., Nerz C., Gaupp R., Engelmann S., Liebeke M., et al. (2008). Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 190 7847–7858. 10.1128/JB.00905-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J., Lopes C., Tavaria F., Delgado L., Pintado M. (2013). A diversity profile from the staphylococcal community on atopic dermatitis skin: a molecular approach. J. Appl. Microbiol. 115 1411–1419. 10.1111/jam.12296 [DOI] [PubMed] [Google Scholar]
- Swartz T., Ito M., Ohira T., Natsui S., Hicks D., Krulwich T. (2007). Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 189 3081–3090. 10.1128/JB.00021-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigawa H., Nakagawa H., Kuzukawa M., Mori H., Imokawa G. (2005). Deficient production of hexadecenoic acid in the skin is associated in part with the vulnerability of atopic dermatitis patients to colonization by Staphylococcus aureus. Dermatology 211 240–248. 10.1159/000087018 [DOI] [PubMed] [Google Scholar]
- Volkov A., Liavonchanka A., Kamneva O., Fiedler T., Goebel C., Kreikemeyer B., et al. (2010). Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285 10353–10361. 10.1074/jbc.M109.081851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. (2012). Primer-BLAST: a tool to design target-specific primers for a polymerase chain reaction. BMC Bioinformatics 13:134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.