Abstract
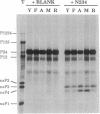
Upon infection of animal cells by Sindbis virus, four nonstructural (ns) proteins, termed nsP1-4 in order from 5' to 3' in the genome, are produced by posttranslational cleavage of a polyprotein. nsP4 is believed to function as the viral RNA polymerase and is short-lived in infected cells. We show here that nsP4 produced in reticulocyte lysates is degraded by the N-end rule pathway, one ubiquitin-dependent proteolytic pathway. When the N-terminal residue of nsP4 is changed by mutagenesis, the metabolic stabilities of the mutant nsP4s follow the N-end rule, in that the half-life of nsP4 bearing different N-terminal residues decreases in the order Met greater than Ala greater than Tyr greater than or equal to Phe greater than Agr. Addition of dipeptides Tyr-Ala, Trp-Ala, or Phe-Ala to the translation mixture inhibits degradation of Tyr-nsP4 and Phe-nsP4, but not of Arg-nsP4. Conversely, dipeptides His-Ala, Arg-Ala, and Lys-Ala inhibit the degradation of Arg-nsP4 but not of Tyr-nsP4 or Phe-nsP4. We found that there is no lysine in the first 43 residues of nsP4 that is required for its degradation, indicating that a more distal lysine functions as the ubiquitin acceptor. Strict control of nsP4 concentration appears to be an important aspect of the virus life cycle, since the concentration of nsP4 in infected cells is regulated at three levels: translation of nsP4 requires read-through of an opal termination codon such that it is underproduced; differential processing by the virus-encoded proteinase results in temporal regulation of nsP4; and nsP4 itself is a short-lived protein degraded by the ubiquitin-dependent N-end rule pathway.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D. J., Sawicki S. G., Sawicki D. L. Solubilization and immunoprecipitation of alphavirus replication complexes. J Virol. 1991 Mar;65(3):1496–1506. doi: 10.1128/jvi.65.3.1496-1506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum E. Z., Hyman L. E., Wormington W. M. Post-translational control of ribosomal protein L1 accumulation in Xenopus oocytes. Dev Biol. 1988 Mar;126(1):141–149. doi: 10.1016/0012-1606(88)90247-3. [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., DiGiuseppe J. A., Bercovich B., Orian A., Richter J. D., Schwartz A. L., Brodeur G. M. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L. How are substrates recognized by the ubiquitin-mediated proteolytic system? Trends Biochem Sci. 1989 Dec;14(12):483–488. doi: 10.1016/0968-0004(89)90180-1. [DOI] [PubMed] [Google Scholar]
- Ding M. X., Schlesinger M. J. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology. 1989 Jul;171(1):280–284. doi: 10.1016/0042-6822(89)90539-4. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989 Oct 5;264(28):16700–16712. [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Hahn Y. S., de Groot R. J., Strauss E. G., Strauss J. H. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology. 1990 Jul;177(1):199–208. doi: 10.1016/0042-6822(90)90473-5. [DOI] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J Virol. 1988 Mar;62(3):998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989 Nov;63(11):4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Ubiquitin-phytochrome conjugates. Pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989 Mar 25;264(9):4998–5005. [PubMed] [Google Scholar]
- Johnson E. S., Gonda D. K., Varshavsky A. cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990 Jul 19;346(6281):287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S., Ruohonen L. Nonstructural proteins of Semliki Forest virus: synthesis, processing, and stability in infected cells. J Virol. 1983 Sep;47(3):505–515. doi: 10.1128/jvi.47.3.505-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. J., Niesters H. G., Hong Z., Strauss J. H. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991 Jun;182(2):430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- Kulesh D. A., Ceceña G., Darmon Y. M., Vasseur M., Oshima R. G. Posttranslational regulation of keratins: degradation of mouse and human keratins 18 and 8. Mol Cell Biol. 1989 Apr;9(4):1553–1565. doi: 10.1128/mcb.9.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. P., Rice C. M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989 Mar;63(3):1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y., Kaim D., Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988 Feb 25;263(6):2693–2698. [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology. 1985 Jul 15;144(1):20–34. doi: 10.1016/0042-6822(85)90301-0. [DOI] [PubMed] [Google Scholar]
- Shaeffer J. R. ATP-dependent proteolysis of hemoglobin alpha chains in beta-thalassemic hemolysates is ubiquitin-dependent. J Biol Chem. 1988 Sep 25;263(27):13663–13669. [PubMed] [Google Scholar]
- Shirako Y., Strauss J. H. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990 Jul;177(1):54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takkinen K., Peränen J., Keränen S., Söderlund H., Käriäinen L. The Semliki-Forest-virus-specific nonstructural protein nsP4 is an autoproteinase. Eur J Biochem. 1990 Apr 20;189(1):33–38. doi: 10.1111/j.1432-1033.1990.tb15456.x. [DOI] [PubMed] [Google Scholar]
- Wellink J., van Kammen A. Proteases involved in the processing of viral polyproteins. Brief review. Arch Virol. 1988;98(1-2):1–26. doi: 10.1007/BF01321002. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Hardy W. R., Shirako Y., Strauss J. H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990 Aug;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]