ABSTRACT
The type III secretion system (T3SS) is a principal virulence determinant of the model bacterial plant pathogen Pseudomonas syringae. T3SS effector proteins inhibit plant defense signaling pathways in susceptible hosts and elicit evolved immunity in resistant plants. The extracytoplasmic function sigma factor HrpL coordinates the expression of most T3SS genes. Transcription of hrpL is dependent on sigma-54 and the codependent enhancer binding proteins HrpR and HrpS for hrpL promoter activation. hrpL is oriented adjacently to and divergently from the HrpL-dependent gene hrpJ, sharing an intergenic upstream regulatory region. We show that association of the RNA polymerase (RNAP)-HrpL complex with the hrpJ promoter element imposes negative autogenous control on hrpL transcription in P. syringae pv. tomato DC3000. The hrpL promoter was upregulated in a ΔhrpL mutant and was repressed by plasmid-borne hrpL. In a minimal Escherichia coli background, the activity of HrpL was sufficient to achieve repression of reconstituted hrpL transcription. This repression was relieved if both the HrpL DNA-binding function and the hrp-box sequence of the hrpJ promoter were compromised, implying dependence upon the hrpJ promoter. DNA-bound RNAP-HrpL entirely occluded the HrpRS and partially occluded the integration host factor (IHF) recognition elements of the hrpL promoter in vitro, implicating inhibition of DNA binding by these factors as a cause of negative autogenous control. A modest increase in the HrpL concentration caused hypersecretion of the HrpA1 pilus protein but intracellular accumulation of later T3SS substrates. We argue that negative feedback on HrpL activity fine-tunes expression of the T3SS regulon to minimize the elicitation of plant defenses.
IMPORTANCE
The United Nations Food and Agriculture Organization has warned that agriculture will need to satisfy a 50% to 70% increase in global food demand if the human population reaches 9 billion by 2050 as predicted. However, diseases caused by microbial pathogens represent a major threat to food security, accounting for over 10% of estimated yield losses in staple wheat, rice, and maize crops. Understanding the decision-making strategies employed by pathogens to coordinate virulence and to evade plant defenses is vital for informing crop resistance traits and management strategies. Many plant-pathogenic bacteria utilize the needle-like T3SS to inject virulence factors into host plant cells to suppress defense signaling. Pseudomonas syringae is an economically and environmentally devastating plant pathogen. We propose that the master regulator of its entire T3SS gene set, HrpL, downregulates its own expression to minimize elicitation of plant defenses. Revealing such conserved regulatory strategies will inform future antivirulence strategies targeting plant pathogens.
INTRODUCTION
Most agriculturally important bacterial plant pathogens utilize a type III secretion system (T3SS) as a channel for delivery of virulence proteins, known as effectors, into the plant cell cytoplasm (1). The T3SS consists of a conserved transmembrane base complex and a needle-like pilus appendage (2, 3). In plant pathogens, its early substrates, the harpins, function to form the translocon, a pore in the target cell membrane through which effectors are subsequently secreted. Effectors are structurally and functionally diverse (4), targeting key components of eukaryotic signaling pathways to suppress the two layers of plant immunity: (i) broadly acting innate defenses triggered by invariant pathogen-associated molecular patterns (PAMPs) and (ii) the rapid, localized, and pathogen-specific hypersensitive response (HR) triggered by evolved recognition of effectors (5).
Comprising over 50 disease-causing pathovars, many of which infect valuable crops such as tomato, bean, and rice, Pseudomonas syringae is the most highly developed model for T3SS-dependent plant pathogenesis and evolution of host specificity (6–8). P. syringae strains are found ubiquitously on leaf surfaces as well as in soil, freshwater, and precipitation. The P. syringae pv. tomato DC3000 pathovar (here DC3000) enters Arabidopsis thaliana leaves through wounds or stomata and replicates within the apoplast, causing chlorosis and necrotic lesions (8, 9).
The T3SS structural, helper, and regulatory proteins are encoded by a cluster of HR and conserved (hrc) and HR and pathogenicity (hrp) genes, flanked by variable effector loci within the Hrp pathogenicity island (10). In several plant pathogens, the extracytoplasmic function (ECF) sigma factor HrpL regulates the coordinated expression of the Hrp regulon via a conserved promoter motif, the hrp-box (11, 12). ECF sigma factors couple the expression of a functionally related gene set to perception of environmental cues (13). Transcription of hrpL is regulated by the alternative sigma-54 (σ54) factor, which requires activation by bacterial enhancer binding proteins (EBPs) bound at a distal promoter site called the upstream activation sequence (UAS) for transcription initiation (14). Integration host factor (IHF)-mediated DNA looping facilitates contact between the promoter-bound EBP complex, usually homohexameric, and the inactive RNA polymerase (RNAP)-σ54 complex. In P. syringae, transcription of hrpL is atypically activated by a heterohexamer comprising two codependent EBPs, HrpR and HrpS (15). Furthermore, HrpS is subjected to allosteric posttranslational inhibition by HrpV, which is in turn sequestered by HrpG (16).
Expression of the T3SS regulon is stimulated by minimal culture media mimicking the abiotic conditions of the leaf apoplast (17) and further enhanced by plant cells or soluble extracts (18). The importance of coordinated adjustments in gene expression for niche adaptation on the plant host is highlighted by the global changes in the P. syringae transcriptome evident upon transition from the leaf surface to the apoplast (19). However, how plant signals are perceived and transduced into the Hrp regulatory network remains poorly understood. Regulatory motifs such as feedback loops can influence the population-level behaviors of infecting pathogens. In particular, bistable expression of the T3SS drives heterogeneity and division of labor in both plant and animal pathogens (20, 21), including P. syringae (22). New insights into the regulatory networks underlying the T3SS and other bacterial virulence factors promise to inform strategies to manage plant disease. For example, by reducing the selection pressure for bacterial resistance, the use of antivirulence chemicals to “disarm” the T3SS by modulating its function or regulation represents an effective alternative to crop resistance breeding (23).
The hrpL gene is transcribed divergently with respect to the HrpL-dependent hrpJ operon, sharing an intergenic upstream regulatory region in which the respective UAS and hrp-box elements are directly adjacent (Fig. 1a). hrpJ encodes a putative regulator of T3SS substrate preference (24), while the downstream hrcV and hrcN genes encode conserved subunits of the base complex. We examined the control of the hrpL and hrpJ promoters to test the hypothesis that regulatory interplay might exist between them. We show that hrpL expression is subject to negative autogenous control (NAC), mediated via HrpL binding at the hrpJ promoter. The DNA footprint of the RNAP-HrpL complex suggests that HrpL achieves repression by occluding the UAS- and IHF-binding sites of the hrpL promoter. Quantitative proteomics suggests that T3SS function is highly sensitive to the HrpL concentration, allowing us to propose possible physiological advantages of negative-feedback mechanisms in the context of the host plant.
FIG 1 .
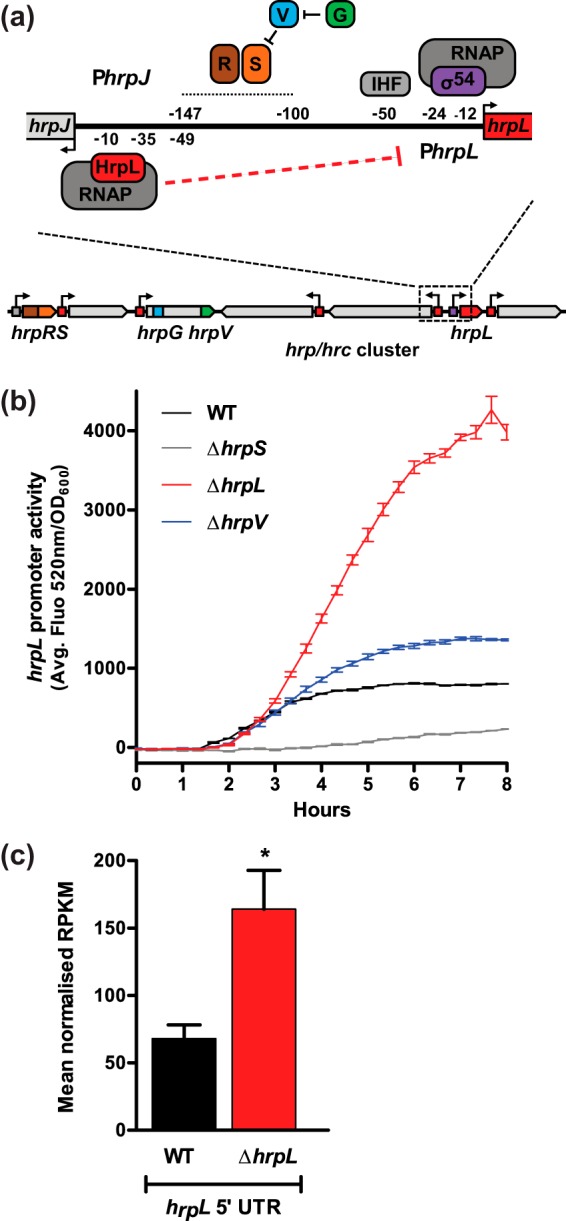
Negative feedback on hrpL transcription implied by the ΔhrpL mutant. (a) The regulation of hrpL transcription in P. syringae. The organization of the bidirectional promoter region between hrpL and hrpJ, including known binding sites for σ54, IHF, and HrpRS, is shown. HrpS activator function is regulated antagonistically by HrpV and HrpG. HrpL promotes transcription of hrpJ and other hrp-hrc operons via the hrp-box (red boxes). The study results propose a mechanism of negative autogenous control dependent on RNAP-HrpL binding at the hrpJ promoter (dashed red line). (b) hrpL promoter activity (fluorescence [Fluo]/OD600) in DC3000 wild-type (WT), ΔhrpS, ΔhrpL, and ΔhrpV strains carrying the pBBR1-PhrpL-gfp reporter plasmid under hrp-inducing conditions. Error bars represent standard errors of the means (SEM) of results of 3 biological replicates. (c) Transcription of chromosomal hrpL locus in wild-type and ΔhrpL strains inferred by RNA-seq. Data represent relative expression levels of a 5′ section of the hrpL transcript (−24 to +25 relative to ATG) under hrp-inducing conditions. The mean and quantile-normalized reads per kilobase per million (RPKM) values for two biological replicates per strain are shown with SEM. Differential expression is significant according to Baggerley’s test with false-discovery-rate (FDR) adjustment (P = 4.57 × 10−6).
RESULTS
HrpV-independent negative feedback on hrpL transcription.
The relative levels of activity of the hrpL promoter (PhrpL) across various regulatory mutant strains were compared using a transcriptional green fluorescent protein (GFP) fusion construct (pBBR1-PhrpL-gfp) encompassing the intergenic region shared between hrpL and hrpJ (Fig. 1a). Verifying the strict requirement of the HrpS coactivator for PhrpL activity under established hrp-inducing growth conditions, a basal level of GFP fluorescence, normalized for cell density, was observed in the ΔhrpS strain (Fig. 1b) (25). The striking 4-fold increase in fluorescence observed in the ΔhrpL strain in comparison to the wild-type (WT) strain after 8 h suggests that negative feedback acting on PhrpL in the wild-type strain had been relieved. Given that HrpV inhibits HrpS activity and that hrpV expression is directly dependent on the presence of HrpL, the downregulation of this regulator in the ΔhrpL strain partially accounts for the apparent negative feedback. Indeed, an increase in PhrpL activity in the ΔhrpV strain in comparison to wild type was observed. However, the fact that the upregulation of PhrpL activity apparent in the ΔhrpL strain was stronger than that in the ΔhrpV strain may suggest a novel HrpV-independent mechanism of negative feedback. The HrpL-mediated repression phenotype was verified at the level of the native transcript by transcriptome sequencing (RNA-seq), confirming that the differences in the levels of PhrpL activity observed were not an artifact of the reporter system. After 4 h under hrp-inducing conditions, while expression of the HrpL-dependent T3SS regulon was suppressed (see Data set S1 in the supplemental material), a 5′ section of the hrpL transcript (present in the ΔhrpL deletion construct) was upregulated in the ΔhrpL strain compared to the wild-type strain (Fig. 1c). The (+2.4-) fold change was approximately equivalent to the difference in the levels of reporter fluorescence observed at the same time point. Flow cytometry data confirmed that PhrpL activity conformed to approximately the normal distribution in both the wild-type and ΔhrpL strain populations (see Fig. S1a in the supplemental material). Moreover, broadly similar curves for optical density at 600 nm (OD600) confirmed that the differences in the levels of PhrpL activity across the strains tested were not artifacts of irregular cell growth (see Fig. S1b).
(Data 1) RNA-seq expression data for key T3SS genes in DC3000 wild-type and ΔhrpL. Statistically significant fold changes (FDR-adjusted P value < 0.05) are indicated in boldface characters. (Data 2) Peptide transitions analyzed by MRM-MS. Abbreviations: rep, replicate; ID, transition identifier; A, analyte; IS, internal standard; *, heavy amino acid residue. Download DATA SET S1, XLSX file, 0.02 MB (20.9KB, xlsx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of the ΔhrpL mutation on cell growth and the population distribution of hrpL promoter activity. (a) Histogram of GFP expression in populations of DC3000 cells after 8 h of growth in hrp-inducing medium. Triplicate populations of ΔhrpL (red shading) and wild-type (gray shading) strains were sampled for flow cytometry (10,000 events). Fluorescence signal is shown on a log10 scale. hrpL promoter activity followed a normal distribution across both the wild-type and ΔhrpL populations, with a higher average level of activity in the ΔhrpL strain. (b) Cell density (OD600) curves measured simultaneously with reporter fluorescence. ΔhrpS, ΔhrpL, and ΔhrpV gene deletions had a negligible effect on cell growth with respect to wild-type DC3000. Error bars represent SEM of results of 3 biological replicates. Download FIG S1, EPS file, 0.9 MB (966.2KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Negative feedback is dependent on HrpL concentration and DNA-binding function.
To examine whether HrpL-dependent negative feedback requires its canonical sigma factor function, the ΔhrpL strain was complemented with a variant of HrpL impaired in DNA binding. ECF sigma factors are strongly dependent on the C-terminal 4.2 region for interaction with promoter DNA at the −35 element (26) (Fig. 2a). The strong similarity between the predicted HrpL and known Escherichia coli σE protein structures (data not shown) was used to infer the location of the HrpL region 4.2. A truncated HrpL variant (positions 1 to 150; HrpLΔR4.2) was generated that was able to bind core RNA polymerase (see Fig. S2a and S2b) but unable to activate HrpL-dependent transcription (see Fig. S3a and S2c). In both the ΔhrpL strain and the wild-type strain, full-length HrpL hyperrepressed PhrpL activity when expressed from the pSEVA224 plasmid (Fig. 2b), suggesting that the intensity of negative feedback is dependent on the HrpL concentration. In contrast, the HrpLΔR4.2 DNA-binding mutant repressed PhrpL activity only weakly, confirming that negative feedback by HrpL is primarily dependent on promoter binding rather than an alternative function. This disqualifies DNA-independent competition between HrpL and σ54 for free RNAP molecules as a sole explanation for the observed differences in PhrpL activity.
FIG 2 .
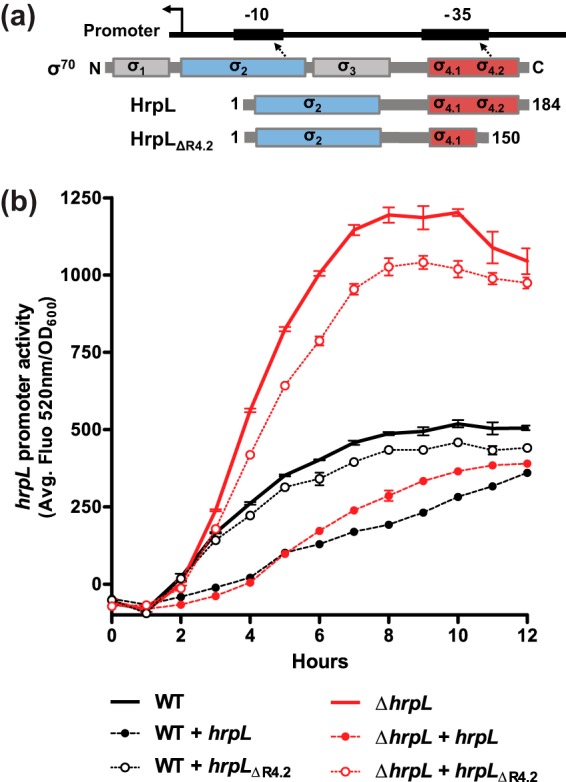
Negative feedback is dependent on the expression level and DNA-binding function of HrpL. (a) ECF σ factors retain the DNA-binding characteristics of the wider σ70-type family: region 2 (σ2) and region 4.2 (σ4.2) interact with the −10 and −35 promoter elements, respectively. The C-terminal truncation mutant, HrpLΔR4.2, lacks conserved −35 element binding determinants. (b) hrpL promoter activity in DC3000 wild-type and ΔhrpL strains carrying the pBBR1-PhrpL-gfp reporter and plasmid-encoded hrpL variants under hrp-inducing conditions. HrpL (filled circles) and HrpLΔR4.2 (hollow circles) were expressed from the pSEVA224-hrpL and pSEVA224-hrpLΔR4.2 plasmids, respectively, while empty pSEVA224 (bold line) was used to control for plasmid load. Error bars represent SEM of results of 3 biological replicates.
In vitro abortive transcription activity of copurified RNAP-HrpL and RNAP-HrpLΔR4.2 complexes on hrpJ promoter probes. (a) RNAp core (6His-β′) was coexpressed in E. coli with HrpL or HrpLΔR4.2. Total protein was sampled following cell disruption (Whole cell) and purification by Ni-affinity chromatography (Purified) and compared alongside size markers (L) by staining of SDS-PAGE gels with Coomassie dye. The following components are signified by arrows: RNAP α subunit (34.9 kDa); HrpL (22.6 kDa); HrpLΔR4.2 (18.9 kDa); RNAP ω subunit (10.5 kDa). (b) Detection of HrpL variants by Western blotting targeting a C-terminal Myc tag. Proteins were transferred directly from the gel shown in panel a to a polyvinylidene difluoride (PVDF) membrane and incubated with an anti-Myc antibody. (c) Abortive initiation on supercoiled PhrpJ or PhrpJ(Δ35e) promoter DNA was assayed in the presence of RNAP core enzyme or RNAP-HrpL or RNAP-HrpLΔR4.2 holoenzymes. An abortive small priming RNA (spRNA) product (predicted CpApUpApApU) was separated from free nucleotides by electrophoresis. The average level of initiation activity across 3 replicates is given as a percentage relative to RNAP-HrpL on wild-type promoter DNA. Initiation was inhibited >90% in the presence of RNAP-HrpLΔR4.2 and/or PhrpJ(Δ35e). Download FIG S2, EPS file, 7.9 MB (8.1MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid-borne expression of HrpL and HrpLΔR4.2 in DC3000. (a) HrpL dependence of hrpJ promoter fusion activity in DC3000. hrpJ promoter activity in wild-type (black) and ΔhrpL (red) strains carrying both pBBR1-rfp-PhrpL-gfp and plasmids for heterologous expression of hrpL under hrp-inducing conditions is reported. hrpL (full circles) or hrpLΔR4.2 (empty circles) genes were expressed from the pSEVA224 plasmid. Empty pSEVA224 vector (bold lines) was used as a plasmid load control. Autofluorescence by DC3000 in the absence of reporter was also measured (gray dashed line). Error bars represent SEM of results of 3 biological replicates. hrpJ promoter activity was negligible, with respect to autofluorescence, in the ΔhrpL strain, confirming the HrpL dependence of transcription. Complementation with plasmid-borne hrpL, but not inactive hrpLΔR4.2, resulted in strong upregulation of PhrpJ with respect to wild-type results. (b) Cell density (OD600) curves measured simultaneously with reporter fluorescence. hrpL and hrpLΔR4.2 overexpressed from pSEVA224 had a negligible effect on the cell growth of wild-type DC3000, with respect to an empty vector control for plasmid load. Error bars represent SEM of results of 3 biological replicates. Download FIG S3, EPS file, 1.1 MB (1.2MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
HrpL is sufficient for autogenous negative control.
E. coli has been utilized previously as a heterologous model system in which to study the initiation of hrpL transcription independently of the wider DC3000-specific regulatory network (15). In this study, a multiple plasmid-based system was engineered in the E. coli s17λpir strain with IPTG (isopropyl-β-d-thiogalactopyranoside)-induced heterologous expression of hrpRS from pAPT-hrpRS, driving activation of the pBBR1-PhrpL-gfp reporter. Negligible fluorescence was observed in the absence of IPTG (Fig. 3a). In this E. coli system, the additional effect of HrpL expression was studied by introducing the pSEVA614-hrpL plasmid, also induced via the use of IPTG. Compared to the results seen with the empty pSEVA614 vector control, HrpL expression strongly repressed PhrpL activity (by approximately 10-fold) after 8 h. Heterologous hrpL expression did not alter the levels of cell growth (see Fig. S3b), negating the possibility of pleiotropic effects on cell physiology. Furthermore, the results of both a transcriptional fusion assay and analysis of transcript levels indicated that HrpL does not significantly influence HrpRS expression (see Fig. S4 and Data set S1). These observations together suggest that HrpL alone, rather than a factor in the DC3000 HrpL-dependent regulon, is sufficient to autogenously repress PhrpL activity.
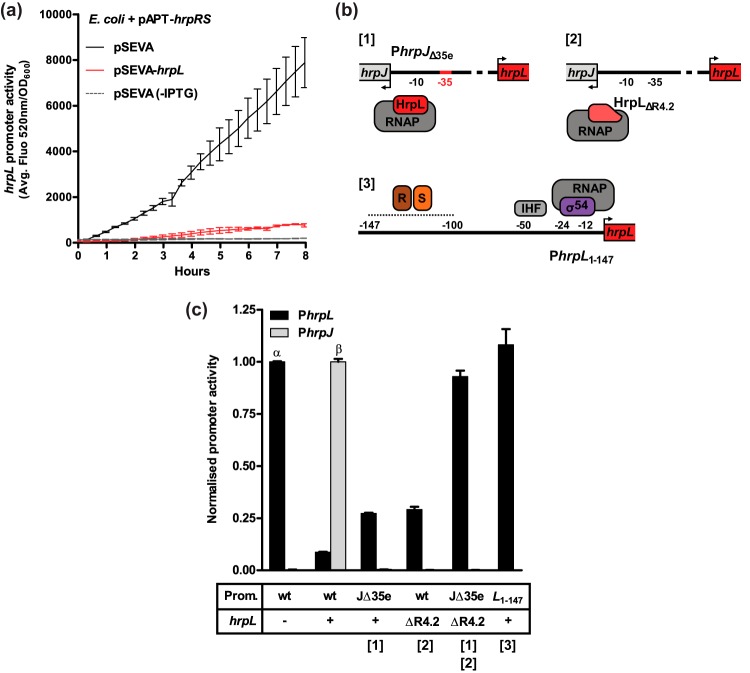
FIG 3 .
(a) hrpL promoter activity in an E. coli s17λpir strain carrying the pBBR1-PhrpL-gfp reporter in LB medium. Heterologous expression of hrpL from pSEVA614-hrpL (red) and hrpRS from pAPT-hrpRS was induced with 0.1 mM IPTG. The empty pSEVA614 vector (black) was used to control for plasmid load. HrpRS-independent PhrpL activity was measured in the absence of IPTG (dashed gray line). Error bars represent SEM of results of 3 biological replicates. (b) Mutations disrupting the association of the RNAP-HrpL with PhrpJ DNA. [1] Substitution of the hrp-box −35 element (GGAACT > AACACT). [2] Truncation of HrpL DNA-binding region 4.2. [3] Exclusion of PhrpJ using a minimal promoter sequence sufficient for PhrpL activity (−147 to +1). (c) The relative activity levels of PhrpL and PhrpJ seen when HrpL DNA-binding function was disrupted. The expression states of hrpL and the cis- and trans-acting mutations acting under each condition are tabulated below the graph. PhrpL (GFP) and PhrpJ (RFP) activities were measured simultaneously under all conditions using the reporters pBBR1-rfp-PhrpL-gfp (wt) or pBBR1-rfp-PhrpL(Δ35e)-gfp (JΔ35e), with the exception of the minimal pBBR1-PhrpL(147)-gfp reporter (L1-147; far-right column). hrpL (+) or hrpLΔR4.2 (ΔR4.2) were expressed from the pSEVA614 plasmid, the empty vector being used as a proxy for the ΔhrpL condition (−). HrpRS proteins were expressed from pAPT-hrpRS under all conditions. Promoter (Prom.) activities given are normalized relative to the maximum activity state for each condition after 8 h of growth in LB as follows: for PhrpL, empty pSEVA614/wild-type promoter (α); for PhrpJ, pSEVA614-hrpL/wild-type promoter (β). Error bars represent SEM of results of 3 biological replicates.
Activity of the hrpRS promoter in ΔhrpL. hrpRS promoter activity reported in WT (black) and ΔhrpL (red) strains carrying pBBR1-PhrpRS-gfp under hrp-inducing conditions. Error bars represent SEM of results of 3 biological replicates. No significant difference in hrpRS promoter activity is evident in the ΔhrpL strain with respect to wild-type DC3000 results during 8 h in hrp-inducing medium. Download FIG S4, EPS file, 0.5 MB (573KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Negative autogenous control is dependent on the adjacent hrpJ promoter.
The E. coli test system was developed further in order to test the hypothesis that HrpL exerts NAC as a result of σ factor function at the adjacent hrpJ promoter. pBBR1-PhrpL-gfp was modified to generate a bidirectional and dual-color reporter, pBBR1-rfp-PhrpL-gfp, in which the PhrpL and PhrpJ promoter elements are fused to GFP and red fluorescent protein (RFP), respectively, enabling their relative levels of activity to be measured simultaneously. A second reporter was derived in which the hrp-box −35 element at PhrpJ was disrupted by site-directed mutagenesis. Substitution of any nucleotide in the hrp-box −35 element (GGAAC) abolishes HrpL function (12). Therefore, a trinucleotide GGA>AAC substitution was introduced in the pBBR1-rfp-PhrpL(Δ35e)-gfp reporter to inhibit the association of HrpL at this site. Finally, a further reporter construct was generated consisting of the minimal sufficient PhrpL promoter sequence (−147 to +1) (15) but lacking the entire PhrpJ promoter, pBBR1-PhrpL(147)-gfp. Together with the pSEVA614-hrpL and pSEVA614-hrpLΔ4.2 expression plasmids, this series of reporters were used to investigate the specific role of HrpL DNA-binding function at PhrpJ for NAC (Fig. 3b). PhrpJ activity was observed in E. coli but only in the presence of full-length HrpL and an intact promoter sequence (here, the unmodified condition) (Fig. 3c). Both the hrp-box and HrpLΔR4.2 mutations abolished PhrpJ activity, inferred by negligible RFP fluorescence and inhibition of in vitro transcription from PhrpJ by >90% (see Fig. S2c). Compared to the results seen under the unmodified condition, in which NAC was apparent, both mutations induced a modest increase in PhrpL activity, corresponding to approximately 25% of that observed in the absence of HrpL. When the two mutations were combined in the same strain, maximum PhrpL activity was almost completely restored (93%). These data suggest that disrupting the ability of HrpL to associate with the PhrpJ hrp-box relieves NAC for PhrpL. The inability of either mutation alone to completely derepress PhrpL activity is suggestive of residual interactions between the RNAP-HrpL holoenzyme complex and promoter that are sufficient to impose partial NAC but unable to initiate transcription at PhrpJ. However, in a HrpLΔR4.2 background, the fact that the addition of the hrp-box mutation relieves NAC is sequence-specific evidence for a mechanism involving PhrpJ. Indeed, NAC was completely relieved in the absence of the entire PhrpJ element [pBBR1-PhrpL(147)-gfp].
The PhrpJ-bound RNAP-HrpL complex partially occludes the hrpL promoter.
The interaction between HrpL and the promoter DNA shared between hrpL and hrpJ was further characterized in vitro. HrpL readily formed insoluble inclusion bodies when overexpressed for protein purification, here (data not shown) and in previous studies (12, 27). Therefore, the solubility of HrpL and HrpLΔR4.2 was maintained via copurification in complex with an E. coli RNAP with a His tag at the β subunit (28). The purified RNAP-HrpL holoenzyme activated transcription from PhrpJ in vitro, confirming its ability to both bind promoter DNA and form an open promoter complex (see Fig. S2c). However, the RNAP-HrpLΔR4.2 mutant achieved approximately 4% of the wild-type activity. Exonuclease III (ExoIII) footprinting was performed on DNA probes comprising the entire PhrpJ-hrpL intergenic region, labeled at the PhrpJ terminus such that the 3′ to 5′ directionality of ExoIII might reveal the distal boundary of the RNAP-HrpL complex. The HrpL-RNAP complex blocked ExoIII digestion on both PhrpJ-hrpL and PhrpJ-hrpLΔ35e promoter probes and in the presence of nonspecific competitor DNA (Fig. 4a). However, both the RNAP-HrpLΔR4.2 holoenzyme and the RNAP core enzyme (no bound σ factor) failed to produce an equivalent footprint, suggesting that the ExoIII block is specific to the interaction between HrpL and promoter DNA. The fact that the footprint was observed on both promoter probes supports the conclusion that HrpL maintains some affinity for the mutated Δ35e hrp-box sequence, as suggested by the presence of residual NAC in vivo (Fig. 3c). In the context of DNA size markers and the IHF and RNAP-σ54 footprints, RNAP-HrpL blocked ExoIII digestion at a position within the IHF consensus recognition sequence of PhrpL (Fig. 4b). This implies that PhrpJ-bound RNAP-HrpL occludes a significant region of PhrpL, including the predicted UAS for HrpRS binding (15) and part of the IHF recognition sequence. NAC might therefore be manifest via interference with HrpRS and/or IHF DNA binding by RNAP-HrpL.
FIG 4 .
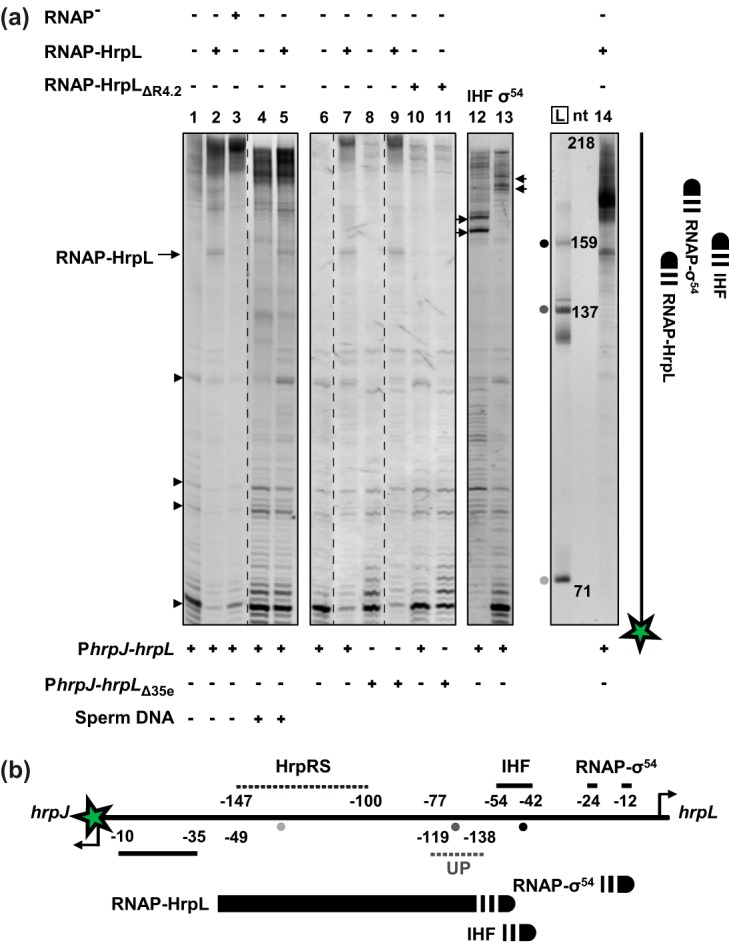
Exonuclease III footprint of RNAP-HrpL holoenzyme on hrpJ-hrpL promoter DNA. (a) PhrpJ-hrpL and PhrpJ-hrpLΔ35e promoter probes labeled with Cy3 dye at the PhrpJ terminus (green star) were incubated at 200 nM with protein complexes prior to incomplete 3′ to 5′ digestion by ExoIII and separation of the fragments in an 8% polyacrylamide gel. Proteins were analyzed at the following concentrations: RNAP-HrpL and RNAP-HrpLΔR4.2, 5 μM; RNAP core enzyme, 4 μM; IHF, 1 μM; RNAP-σ54, 400 nM and 2.4 μM. Salmon sperm DNA (1 μM) was used as nonspecific competitor DNA. Sites of 3′ digestion inhibition by DNA-bound complexes of RNAP-HrpL, IHF, and RNAP-σ54 are annotated (arrows, lanes 12 and 13) and summarized (far-right cartoon). Multiple independent gels (differentiated by boxes) are aligned with reference to consistent digestion fragments (arrowheads). Dashed lines signify exclusion of gel lanes for clarity. Size markers of lengths of 71, 137, and 159 nt (L) are shown alongside the RNAP-HrpL footprint (lane 14). (b) A schematic representation of the 3′ boundaries of DNA-bound RNAP-HrpL, IHF, and RNAP-σ54 complexes in the context of known regulatory elements of the PhrpJ-hrpL promoter region, annotated relative to the respective transcription start sites. The relative locations of the three size markers electrophoresed alongside footprint products, as annotated in panel a, are signified by shaded circles.
Misregulation of hrpL inhibits T3SS function in culture.
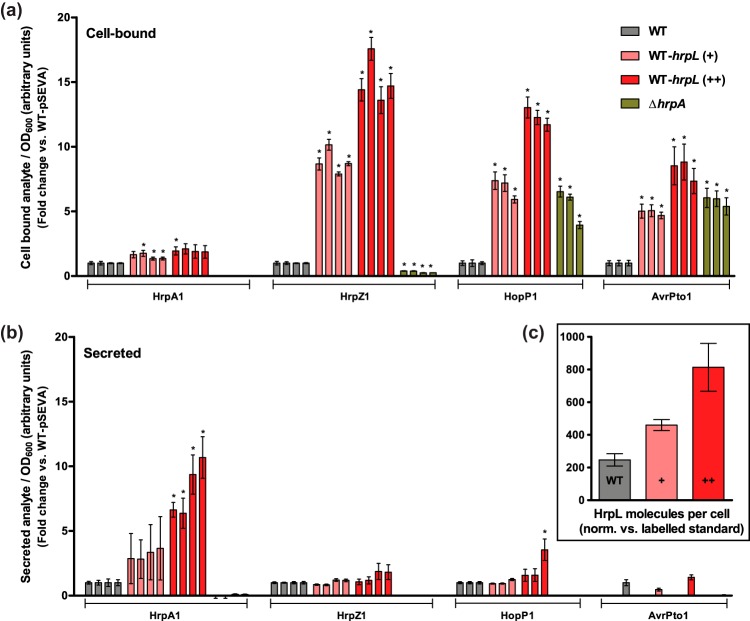
To infer the effect of NAC on T3SS function, the sensitivity of T3SS protein secretion to variations in HrpL copy numbers was investigated using targeted protein mass spectrometry (MS). We have previously developed a method for quantitative analysis of T3SS proteins secreted by DC3000 into hrp-inducing culture medium (HIM) (29). Briefly, shotgun MS was utilized to identify extracellular T3SS-associated proteins, prior to the most readily detectable signature peptide/fragment ion pairs (transitions) being selected for targeted and highly sensitive quantitation using multiple-reaction monitoring (MRM)-MS (30). Protein extract was “spiked” with a known concentration of heavy isotope-labeled standard such that absolute quantification of a specific target could be achieved upon ratiometric comparison of mass-distinguishable sample and standard transition peaks. In this study, the relative intracellular and extracellular abundances of four key T3SS-associated proteins (the pilus subunit HrpA1, the harpins HrpZ1 and HopP1, and the effector AvrPto1) were analyzed in a HrpL-concentration-dependent manner, using multiple peptide transitions for increased robustness. Given that HrpJ is a regulator of secretion (24), mutations that relieve NAC via disruption of PhrpJ function were considered unsuitable for assays of T3SS activity. Instead, HrpL was overexpressed from the pSEVA224-31-hrpL and pSEVA224-33-hrpL plasmids, using synthetic ribosome binding sites (strong and weak, respectively) to specify expression levels. Cell cultures were maintained for an extended 24 h under hrp-inducing conditions to enable accumulation of detectable extracellular protein before cell-bound and secreted protein fractions were extracted. The four T3SS-associated proteins were found in all supernatant samples with the exception of the ΔhrpA1 strain, confirming that their release was dependent on the presence of the T3SS pilus and was not an artifact of cell lysis. A heavy-isotope-labeled standard was used for the quantification of intracellular HrpL copy number (Fig. 5c). Approximately 200 to 250 copies of HrpL were detected in the wild-type cell. The addition of the pSEVA224-33-hrpL or pSEVA224-31-hrpL plasmid increased the HrpL copy number to approximately 450 or 800, respectively. The effect of the HrpL concentration on the abundance of T3SS-associated proteins was more significant in the intracellular fractions (Fig. 5a). A 3-fold increase in the HrpL concentration (pSEVA224-31-hrpL versus pSEVA224) resulted in 15-, 12-, and 8-fold increases in HrpZ1, HopP1, and AvrPto1 abundance, respectively. Similarly, a 2-fold increase in HrpL (pSEVA224-33-hrpL versus pSEVA224) resulted in a 5-fold or greater increase in the abundance of these proteins. The HrpL concentration had no effect on the intracellular abundance of the housekeeping protein σ70 (RpoD) or Lon protease (see Fig. S5), implying that its overexpression has negligible pleiotropic effects on protein synthesis and the wider proteome. In the secreted fraction, only the abundance of the HrpA1 pilus protein increased in correlation with the HrpL concentration (Fig. 5b). In contrast, with the exception of a single HopP1 transition (NSNS), there was no significant difference in the levels of extracellular abundance of HrpZ1, HopP1, or AvrPto1. These data suggest that an increase in the HrpL copy number results in an intracellular accumulation of harpins and effectors but not an increase in their secretion rate. Accumulation can be explained by increased T3SS protein expression or substrate saturation of the T3SS or both. HrpL-dependent overexpression of the T3SS regulon is implied both by the correlation between HrpA1 translocation rate and HrpL copy number and by the increase in PhrpJ activity observed upon addition of plasmid-borne hrpL (see Fig. S3a). However, given that a HrpL-dependent increase in secretion was observed only for HrpA1, the increase in intracellular abundance of the other T3SS substrates suggests that there was a limitation in translocation rate and that their wild-type abundance was nearly saturating. We propose that NAC by HrpL does not significantly downregulate T3SS activity under wild-type conditions but rather acts to prevent expression of surplus substrates. Supporting this hypothesis, an increased HrpL expression rate had a negligible effect on DC3000 fitness during infection of host Arabidopsis thaliana seedlings (see Fig. S6).
FIG 5 .
The effect of hrpL misregulation on expression and secretion of T3SS proteins. (a and b) Cell-bound (cell pellet) (a) and secreted (supernatant) (b) protein fractions were extracted from DC3000 cell cultures after 24 h in hrp-inducing medium for targeted protein quantification via LC-MRM-MS. The pSEVA224-33-hrpL (+) and pSEVA224-31-hrpL (++) plasmids were used to overexpress hrpL. An empty pSEVA224 vector was used as a plasmid load control. The ΔhrpA strain was used as a control for T3SS-independent release of protein. Multiple peptide transitions per protein were analyzed for calculation of relative target protein abundances between strains. From left to right, the columns shown represent the following transitions (see Data Set S1): for HrpA1, ISATa, ISATb, LTNLa, and LTNLb; for HrpZ1, AQFPa, AQFPb, SANSa, and SANSb; for HopP1, GQLNa, GQLNb, and NSNS; and for AvrPto1, HQLAa, HQLAb, and VSNN. The analyte transition peak intensities were normalized against sample cell density (OD600) and subsequently the wild-type (WT) pSEVA224 peak intensity to give data for fold change between strains. Absent column bars indicate that transitions were undetectable. (c) The absolute HrpL copy number in each intracellular sample was calculated using the ratio between the sample peptide (QPSS) and heavy-isotope standard peptide (QPSS-IS) transition peak intensities, normalizing for OD600 and standard concentration. Protein copy number data assume an OD600 of 1.0 = 109 CFU ml−1. Error bars represent SEM of results of 3 biological replicates.
(a) RpoD and (b) Lon protease abundance in intracellular protein fractions. Total cell-bound (cell pellet) protein was extracted from DC3000 cell cultures after 24 h in hrp-inducing medium for targeted protein quantification via LC-MRM-MS. The pSEVA224-33-hrpL (+) and pSEVA224-31-hrpL (++) plasmids were used to overexpress hrpL. Empty pSEVA224 was used as a plasmid load control in the wild-type strain. From left to right, the column bars shown represent the transitions analyzed for relative protein abundance (see Data Set S1) as follows: for RpoD, FGAVa, FGAVb, IPVHa, and IPVHb; for Lon, TSLAa and TSLAb. The analyte transition signal intensities were normalized against sample cell density (OD600). Error bars represent SEM of results of 3 biological replicates. Download FIG S5, EPS file, 0.7 MB (724.9KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Arabidopsis thaliana seedling infection assays. (a) Col-0 seedlings were flood inoculated with bacterial suspensions of DC3000 wild-type and ΔhrpL strains at a cell density of 5 × 105 CFU ml−1. Three replicate groups of 16 seedlings were flooded with each strain. Mock inoculations were performed with 10 mM MgCl2 solution as a control. Bacterial populations were quantified at 0, 1, 2, and 3 days postinfection (dpi). Error bars represent SEM for 3 samples, each consisting of 3 seedlings. Asterisks denote statistical significance in population size with respect to wild-type control results, based on a two-tailed t test. (b) Disease phenotypes of groups of three representative seedlings photographed 3 days postinfection. Symptoms, including chlorosis and lesion formation, are visible after infection with the wild-type strain but not after infection with the ΔhrpL strain or mock infection. (c) The effect of graded HrpL overexpression on in planta fitness. Seedlings were flood inoculated with DC3000 bacterial suspensions as described for panel a. The growth of wild-type populations carrying a series of pBBR1MCS4 plasmids for hrpL (plain bars) overexpression was monitored over 3 dpi. hrpL expression was regulated by synthetic ribosome binding sites (RBSs) (activity of pSEVA224-31-hrpL [31] > activity of pSEVA224-33-hrpL [33]). The pBBR1MCS4 vector was used as a plasmid control (black). hrpLΔR4.2 (hashed bars) was similarly overexpressed as a control for sigma-independent effects of HrpL. Neither HrpL overexpression nor HrpLΔR4.2 overexpression had any significant effect on in planta fitness with respect to plasmid control results (two-tailed t test). Download FIG S6, EPS file, 16.4 MB (16.8MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Feedback control mechanisms are utilized widely in mechanical and electrical systems to allow optimal performance to be maintained autogenously. Analogous mechanisms have evolved in a variety of biological systems, from gene networks to predator-prey cycles, and impart robustness to molecular or environmental fluctuations. Feedback loops represent common motifs in bacterial genetic circuits, which they endow with complex regulatory dynamics such as switching and oscillation. Around 40% of transcription factors in E. coli are subject to negative autogenous control (NAC) (31). Side-by-side comparisons of synthetic gene circuits have shown that the advantages of this particular mechanism can include robustness against gene expression noise, rapid response time, and population-wide homogeneity (32, 33). We now show that HrpL, the master regulator of T3SS expression in DC3000, exerts NAC of hrpL transcription as a direct result of its canonical σ-factor function at the adjacent hrpJ promoter, thus validating the hypothesis that the close synteny of these two genes imposes novel regulatory coupling between them. Independent lines of evidence, from both a plasmid-borne reporter fusion and the transcriptome, suggest that PhrpL is subject to negative feedback, relieved by the ΔhrpL deletion. Furthermore, heterologous reconstitution of hrpL transcription in E. coli, independently of the wider DC3000-specific regulatory network, reveals that a significant component of negative feedback by HrpL is both autogenous and dependent on the hrpJ promoter element.
Negative autogenous control of a σ54-depdendent promoter.
The hrp-box at PhrpJ is situated only 15 nucleotides (nt) upstream of the putative UAS site proposed by Jovanovic et al. (15). Extrinsic cis-regulatory elements can repress σ54-dependent transcription initiation by altering local DNA geometry (34, 35) or interfering with DNA binding of either the activator complex (36) or σ54 itself (37). The example most analogous to the control of hrpL described here is the regulation of the divergent atrZ-atzDEF promoter region in Pseudomonas sp. strain ADP. Upon binding at a single recognition site, the LysR-type transcription factor AtrZ both activates atzDEF expression and represses σ54-dependent transcription of atrZ (37, 38). However, a notable contrast with hrpL is that activation of atrZ transcription by NtrC occurs independently of a UAS.
The in vivo analyses presented suggest that the mechanism of NAC by HrpL is dependent on DNA binding at PhrpJ but not subsequent transcription initiation. The torque generated by transcription leads to negative supercoiling in upstream DNA which can be sufficient to alter neighboring gene expression (39). However, the fact that NAC is maintained despite the elimination of PhrpJ activity due to either the hrp-box (Δ35e) or HrpLΔR4.2 mutation alone negates torsional stress as its primary mechanism. The requirement for the two mutations to be acting in parallel for complete relief of repression implies that residual and potentially low-affinity interactions between the RNAP-HrpL holoenzyme and promoter DNA are sufficient to impose autogenous control. In support of this idea, the Δ35e mutation does not destabilize the RNAP-HrpL footprint on promoter DNA, corroborating previous evidence that HrpL maintains binding affinity for the hrp-box in spite of substitutions that inactivate transcription (12). Furthermore, the necessity of the Δ35e mutation for complete relief of NAC confirms that its mechanism is DNA sequence specific rather than acting from solution, for example, via competition between σ factors for RNAP.
As a classical ECF σ factor, the RNAP-σE holoenzyme occupies heat shock promoter DNA up to position −60 (40). If this DNA-binding property is common to the members of the ECF family, then it might be predicted a priori that PhrpJ-bound RNAP-HrpL overlaps the distal region of the PhrpL UAS. Surprisingly, the ExoIII footprinting data presented here suggest that this holoenzyme occupies promoter DNA further than 130 nt upstream of the transcription site, occluding completely the UAS and partially the IHF recognition sequence of PhrpL. One possible explanation relates to the function of the C-terminal domain of the RNAP α subunit (α-CTD) which, separated from the RNAP core enzyme by a flexible linker, can associate with distal promoter elements upstream of the σ recognition sequence (41). The regulation of the E. coli lac operon by the cyclic AMP receptor protein (CRP) requires an interaction with the α-CTD which can occur at the −92 position (42). The α-CTD also interacts with AT-rich upstream (UP) elements independently of protein interaction partners (43). Interestingly, a sequence resembling the UP element consensus sequence (43) is present upstream of PhrpJ, close to the location of the RNAP-HrpL footprint (Fig. 4b and Fig. S7 in the supplemental material). It is therefore plausible that the α-CTD is mediating NAC via an interaction with this putative UP element, although further work is required to verify this hypothesis. Given the location of the footprint, the most likely mechanisms of repression include inhibition of (i) HrpRS binding, (ii) IHF binding, or (iii) IHF-dependent DNA looping.
Putative UP element sequence upstream of the hrpJ promoter. (a) Position of putative UP element (UP J) with respect to the IHF recognition sequence and σ54 promoter of the PhrpL. (a) Alignment of UP J and known functional UP elements (rrnB1 P1, rrnD1 P1) with the consensus UP element (Estrem et al. [43]). Conserved positions are indicated in boldface and underlined characters. W, A or T nucleotide; N, any nucleotide. Download FIG S7, EPS file, 0.7 MB (686.1KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Examples demonstrating direct repression of σ54-dependent transcription are rare because strict activator-dependent initiation usually negates the need for additional control. Therefore, the fact that multiple feedback mechanisms converge during hrpL transcription highlights the importance of fine-tuning T3SS expression in DC3000. In addition to NAC by HrpL, described here, HrpS activity is negatively regulated by HrpV binding (16) and hrpRS transcription is believed to be positively regulated via HrpA1, albeit by an unknown mechanism (44, 45). The fact that negative feedback on hrpL expression is partially autogenous represents an interesting contrast with the elegant negative-feedback system that couples T3SS expression to injectisome assembly in Pseudomonas aeruginosa (46). The AraC-type master regulator of T3SS gene expression, ExsA, is posttranslationally regulated by a series of antiactivators, which sequester one another in turn. Once the T3SS secretion channel is open, the export of ExsE triggers a signaling cascade, which ultimately liberates ExsA for upregulation of T3SS gene expression. There is no evidence to suggest that HrpL is regulated by an anti-σ factor, as is common among the ECF family, but it is plausible that the reported positive-feedback mechanism mediated by HrpA1 is by definition coupled to pilus function.
The physiological significance of negative feedback by HrpL.
Given that the T3SS is a key determinant of P. syringae pathogenicity, it is assumed that any mechanism inhibiting its expression must impart a net positive fitness effect or be subject to negative selection. Singh and Hespanha propose that the potential fitness cost of decreased gene expression due to NAC can be outweighed if the gene in question is environmentally induced, if it functions at a low protein copy number, and if stochastic transitions between threshold expression states are unfavorable (47). Our current understanding of HrpL expression suggests that it fits these criteria closely. This study has shown that HrpL functions at relatively low abundance, as is generally the case for ECF family σ factors (48). Moreover, given that it responds to both the metabolic state (49) and plant-derived signals (18), the probability is high that considerable extrinsic noise is associated with hrpL expression. Finally, it can be assumed that stochastic inactivation of T3SS expression during an established interaction with the plant cell is deleterious.
Limitation of T3SS expression may be advantageous in the context of P. syringae ecology. A side effect of strong host specificity, defined by the existence of highly evolved effector protein repertoires, is susceptibility to the adaptive immune responses of non-host-plant species. Many P. syringae pathovars can survive asymptomatically on species outside their host range (9, 50). Thus, given that T3SS expression is broadly induced by cell-free exudates of both host and nonhost plants (18), it is plausible that there is a fitness trade-off between virulence on susceptible hosts and elicitation of non-host-plant defenses. Indeed, the need for tight regulation of effector expression has been noted previously in light of evidence suggesting that some effectors stimulate the hypersensitive response, or otherwise decrease bacterial fitness in planta, in a dose-dependent manner (51, 52). Given that nonspecific, abiotic factors introduce an element of randomness in the dispersal of P. syringae cells (53), those genotypes that impose negative feedback on HrpL expression may experience a positive fitness effect compared to the otherwise more virulent strains when populations are spread across a variety of plant hosts.
In support of this model, we present evidence to suggest that the function of the DC3000 T3SS is highly sensitive to an only modest increase in the concentration of HrpL. Graded constitutive expression of HrpL in excess of its native concentration was performed as a proxy for relief of negative feedback. In the absence of a target host plant cell, only a small subset of T3SS-associated proteins are secreted into culture medium (54), including the harpins HrpZ1 and HopP1 and the effector AvrPto1 (29). Comparing the relative abundances of these T3SS substrates between intracellular and secreted protein fractions, it is apparent that a 2-fold increase in HrpL copy number is sufficient to saturate T3SS activity. Whereas the HrpA1 pilus subunit is more rapidly exported in response to a HrpL-dependent increase in expression, the HrpZ1, HopP1, and AvrPto1 substrates accumulate inside the cell. A restriction on T3SS activity experienced specifically by substrates translocated through the pilus but not the pilus subunit itself might arise due to (i) a pilus-dependent rate-limiting translocation step or (ii) an inability to switch from pilus formation to substrate secretion. Given that the Salmonella enterica serovar Typhimurium T3SS needle channel is less than 3 nm in diameter (2), necessitating that effectors transverse it in a fully unfolded state, substrate saturation of the pilus is very plausible. Alternatively, an imbalance in the normal stoichiometry of T3SS proteins caused by HrpL overexpression may affect the dynamics of substrate switching. A conserved cytoplasmic sorting platform governs the hierarchical, chaperone-dependent loading of substrates at the base of the S. Typhimurium T3SS (55), and a conformational change in this region accompanies the switch from needle formation to effector secretion (2). How substrate switching is regulated by the T3SSs of plant pathogens has yet to be fully explored, although it has been hypothesized that the switch from pilus assembly to effector secretion is coupled to penetration of the host cell membrane (54). It is plausible that substrate overexpression, in particular, that of HrpA1, inhibits a concentration-dependent regulatory event required for harpin translocation. In support of this hypothesis, it has been noted that plasmid-mediated overexpression of hrpA1 can inhibit the ability of P. syringae to elicit the hypersensitive response (45).
In either scenario, tight control of HrpL expression is theoretically advantageous. In the first case, the accumulation of surplus, nonsecreted substrates represents a futile metabolic cost. Given that a DC3000 cell can secrete on the order of 105 HrpA1 and 104 AvrPto1 molecules per hour (29), the burden of T3SS expression is sizeable. Indeed, ΔT3SS mutants have a growth advantage over wild-type cells (56). In the second scenario, the hierarchical dynamics of T3SS function depends on coordinated expression of the T3SS regulon and therefore on HrpL abundance. This is exemplified by the increase in the level of HrpA1 released into the cell supernatant when HrpL is modestly overexpressed. The presence of HrpA1 in this fraction can be attributed to mechanical shearing or depolymerization of the pilus or to complete secretion into the extracellular space. Although the relative levels of significance of these processes in planta are uncertain, the pilus protein is thought to be a general elicitor of plant immune defenses. The hrpA sequence displays signatures of positive selection for substitutions that enable escape from immune recognition (57). Clearly, negative feedback by HrpL is advantageous in the context of immune evasion.
A notable limitation of our study was that the significance of NAC by HrpL was explored predominantly ex planta. This requires several assumptions to be made, the principal being that fine control of HrpL is as relevant in the complex plant environment as is apparent here in culture medium, albeit one mimicking the apoplast. No fitness effect of HrpL misregulation was observed in our simplified model system for host infection. More elaborate plant assays, also performed on nonhosts, are required to validate this. Similarly, our interpretation of the T3SS activity data assumes that the rates of substrate expression and secretion observed are intrinsic rather than regulated. Instead, it is plausible that T3SS function is regulated differently in planta from in culture. However, several technical challenges currently limit the applicability of quantitative proteomics to complex in planta samples.
Concluding remarks.
Recognizing negative autogenous control by HrpL advances our knowledge of the regulatory system controlling T3SS gene expression in DC3000. Not only is this mechanism of fundamental interest with regard to σ54-regulated transcription, but it also highlights the importance of exploring the complexity that underlies otherwise well-defined genetic networks. We also argue that negative feedback on HrpL expression has important implications for the ecology of the DC3000 pathovar. It will be of future interest to determine the extent to which NAC is conserved among P. syringae strains and other Hrp group 1 plant pathogens and whether there exists a correlation between the expression levels of T3SS components and different pathogenic strategies.
MATERIALS AND METHODS
General microbiology and molecular biology.
The bacterial strains and plasmids used in this study are described in Table S1 in the supplemental material. E. coli and DC3000 were grown in lysogeny broth (LB) at 37°C and 28°C, respectively. hrp gene expression in DC3000 was stimulated with hrp-inducing medium (HIM) (25) at 25°C. Plasmid cloning procedures, protein purifications, and markerless gene deletions in DC3000 are described in Text S1.
Strains and plasmids used in this work. Download TABLE S1, DOCX file, 0.04 MB (44KB, docx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplementary methods. Download TEXT S1, DOCX file, 0.04 MB (41.4KB, docx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assay of in vivo promoter activity.
Cell fluorescence derived from transcriptional fusion reporter plasmids was measured simultaneously with OD600 at the population level under microwell conditions using a FLUOstar fluorometer (BMG). GFP fluorescence (485-nm excitation [ex.]/520-nm ± 10-nm emission [em.]) and RFP fluorescence (584-nm ex./620-nm ± 10-nm em.) were detected using standard settings. Fluorescence per unit cell growth, blank corrected against growth medium autofluorescence, was measured across three biological replicate cultures at 20-min intervals over 20 h. Starting cell culture densities were normalized to an OD600 of 0.25 for DC3000 and of 0.05 for E. coli.
Analysis of T3SS transcript expression by RNA sequencing.
DC3000 and ΔhrpL strains were grown in duplicate in LB medium for 16 h at 28°C before being washed in 10 mM MgCl2 and resuspended to an OD600 of 0.25 in 500 ml HIM. After 4 h of growth at 25°C, the cells were fixed with a 1/10 vol of 5% phenol–95% ethanol (vol/vol) and harvested by centrifugation. Adopting a 5′ end-selective methodology for analysis of the primary transcriptome (58), whole-cell RNA preparation, library quality control, next-generation sequencing, and read alignment experiments were performed commercially (Vertis Biotechnologie). The protocol used is detailed further in Text S1. Briefly, total RNA was treated to enrich for primary transcripts and fragmented (50 to 100 nt) enzymatically and the derivative cDNA libraries were sequenced via the use of an Illumina HiSeq 2000 platform, with parameters optimized for high sequencing depth values (stranded, 100 million reads, 50-nt read length). Reads were aligned to the DC3000 genome (GenBank accession no. AE016853) prior to sample library normalization and statistical analysis (CLC genomics workbench; CLC Bio). The alignment template was modified to include two additional features in the hrpL locus: a 50-nt 5′ section present in both strains [“hrpL(5′)”; nt 1542813 to 1542862] and a 100-nt section of the open reading frame (ORF) absent in the ΔhrpL strain due to markerless deletion [“hrpL(ORF)”; nt 1542987 to 1543086]. Gene expression values accounting for variation in sample libraries and gene length were inferred from the counts of uniquely mapped reads subject to quantile (59) and reads per kilobase per million (RPKM) (60) normalizations. Differential expression analysis was performed on normalized gene expression values (mean of two replicates) using Baggerley’s beta-binomial test (61) with a false discovery rate (FDR) threshold of 0.05 (62).
Analysis of intracellular and secreted protein fractions.
Differential analysis of DC3000 protein fractions by multiple reaction monitoring–mass spectrometry (MRM-MS) was performed as described previously (29). HrpL and AvrPto1 proteins were doubly labeled at arginine and lysine residues in vivo for protein standard absolute quantification (PSAQ). Briefly, the coding sequences were PCR amplified and cloned into pET28b+ (Novagen) in frame with an N-terminal histidine tag. Gutnick minimal medium supplemented with 0.4% glucose, 10 mM NH4Cl, 1 mM heavy-labeled l-(13C6,15N2)-arginine, and l-(13C6,15N2)-lysine (Sigma-Aldrich) and 18 unlabeled amino acids (each at a 1 mM concentration) was used for protein expression in a modified ΔargA ΔlysA BL21 strain (63). Proteins were purified from inclusion bodies by nickel (Ni)-affinity chromatography in the presence of 7 M urea. Specific protein standard concentrations were calculated using a Bradford-based assay, correcting for impurities estimated by SDS-PAGE and fluorescent Sypro Ruby staining (Bio-Rad). DC3000 strains were grown in duplicate in LB medium for 16 h at 28°C before being washed in 10 mM MgCl2 and resuspended to an OD600 of 0.25 in triplicate 75-ml cultures in HIM (pH 6). After a further 24 h of growth at 25°C, the extracellular supernatant (“secreted”) and cell-associated (“intracellular”) protein fractions were subsequently separated by centrifugation of 30-ml samples. A mix of labeled HrpL and AvrPto1 standards was added to the supernatant fractions prior to concentration to 200 µl using an Ultra-15 centrifugal filter unit (Millipore) (molecular weight cutoff [MWCO], 15). The concentrated sample was incubated overnight at 37°C with 2 µg modified trypsin (Promega) and buffer T {100 mM Tris-HCl [pH 8], 50 mM NH4HCO3, 1 mM TCEP [tris(2-carboxyethyl)phosphine]}. Complete tryptic digestion was confirmed by SDS-PAGE prior to the addition of 2% formic acid. The corresponding cell pellet fractions were resuspended in 1 ml HIM supplemented with 7 M urea prior to disruption by sonication. A 20-µl sample of intracellular protein was subjected to tryptic digestion as described above. Tryptic peptides were analyzed using a QTrap 6500 mass spectrometer coupled to an ekspert nanoLC 400 liquid chromatography (LC) system (AB Sciex). Details of the working settings of the liquid chromatography-mass spectrometry (LC-MS) analysis are provided in Text S1. The peptide transitions analyzed by MRM are listed in Data set S1, adapting the previously optimized method (29). Data analysis was performed using Analyst software (AB Sciex). Relative levels of sample protein abundance were estimated using analyte peak intensities normalized for cell density at sampling.
Exonuclease III footprinting.
Exonuclease III (ExoIII) footprinting was performed on variant PhrpJ-hrpL double-stranded DNA (dsDNA) promoter probes, labeled at the PhrpJ terminus with a 5′ cyanine (Cy3) dye molecule during PCR amplification using pTE103-PhrpJ and pTE103-PhrpJ(Δ35e) as the templates. Footprinting reactions (using 12-μl reaction mixtures) were performed in STA buffer (2.5 mM Tris-acetate [pH 8], 8 mM Mg-acetate, 10 mM KCl, 1 mM dithiothreitol, 3.5% [wt/vol] polyethylene glycol [PEG] 8000). The copurification of RNAP-HrpL and RNAP-HrpLΔR4.2 complexes is described in Text S1. IHF and σ54 protein samples were sourced from the laboratory collection and purified as previously described (15). Protein-DNA complexes were preincubated at 22°C for 10 min before the addition of 50 units ExoIII and proprietary buffer (Promega). Digestion was performed for 2 min before ExoIII was inactivated with 20 mM EDTA for 10 min at 70°C. Partially digested single-stranded DNA (ssDNA) products were run on an 8% urea footprinting gel. Cy3 fluorescence was detected using a FLA-5000 phosphorimager (Fujifilm) with a 488-nm excitation laser and a 532-nm emission filter.
ACKNOWLEDGMENTS
We acknowledge B. Wang (University of Edinburgh) for plasmids pSB4A3-PhrpL-gfp and pSB4A3-PhrpL-rfp, C. R. Beuzon (University of Malaga) for plasmid pGEM-T-KanFRT, D. W. Holden (Imperial College London) for the E. coli s17 λpir strain, and R. T. Hay (University of Dundee) for the BL21(DE3) Δlys Δarg strain.
This work was supported by a doctoral training grant (BBSRC, BB/F017324/1) awarded to C.W. and in part by BBSRC (BB/G020434/1) and Leverhulme Trust (F/07 058/BM) project grants. The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
We have no conflict of interest to declare in relation to the work presented.
Footnotes
Citation Waite C, Schumacher J, Jovanovic M, Bennett M, Buck M. 2017. Negative autogenous control of the master type III secretion system regulator HrpL in Pseudomonas syringae. mBio 8:e02273-16. https://doi.org/10.1128/mBio.02273-16.
REFERENCES
- 1.Büttner D, Bonas U. 2006. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol 9:193–200. doi: 10.1016/j.mib.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Q, He SY. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294:2556–2558. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- 4.Block A, Alfano JR. 2011. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14:39–46. doi: 10.1016/j.mib.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/Nature05286. [DOI] [PubMed] [Google Scholar]
- 6.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindeberg M, Cunnac S, Collmer A. 2012. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol 20:199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Xin XF, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 9.Hirano SS, Upper CD. 1990. Population biology and epidemiology of Pseudomonas syringae. Annu Rev Phytopathol 28:155–177. doi: 10.1146/annurev.py.28.090190.001103. [DOI] [Google Scholar]
- 10.Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci U S A 97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan L, Deng X, Zhou J, Tang X. 2006. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS. Mol Plant Microbe Interact 19:976–987. doi: 10.1094/MPMI-19-0976. [DOI] [PubMed] [Google Scholar]
- 12.Nissan G, Manulis S, Weinthal DM, Sessa G, Barash I. 2005. Analysis of promoters recognized by HrpL, an alternative sigma-factor protein from Pantoea agglomerans pv. gypsophilae. Mol Plant Microbe Interact 18:634–643. doi: 10.1094/MPMI-18-0634. [DOI] [PubMed] [Google Scholar]
- 13.Mascher T. 2013. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr Opin Microbiol 16:148–155. doi: 10.1016/j.mib.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M. 2008. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol Microbiol 68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 15.Jovanovic M, James EH, Burrows PC, Rego FGM, Buck M, Schumacher J. 2011. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat Commun 2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jovanovic M, Lawton E, Schumacher J, Buck M. 2014. Interplay among Pseudomonas syringae HrpR, HrpS and HrpV proteins for regulation of the type III secretion system. FEMS Microbiol Lett 356:201–211. doi: 10.1111/1574-6968.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahme LG, Mindrinos MN, Panopoulos NJ. 1992. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol 174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haapalainen M, van Gestel K, Pirhonen M, Taira S. 2009. Soluble plant cell signals induce the expression of the type III secretion system of Pseudomonas syringae and upregulate the production of pilus protein HrpA. Mol Plant Microbe Interact 22:282–290. doi: 10.1094/MPMI-22-3-0282. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2013. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci U S A 110:E425–E434. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czechowska K, McKeithen-Mead S, Al Moussawi K, Kazmierczak BI. 2014. Cheating by type 3 secretion system-negative Pseudomonas aeruginosa during pulmonary infection. Proc Natl Acad Sci U S A 111:7801–7806. doi: 10.1073/pnas.1400782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Q, Laiosa MD, Steeber DA, Biddle EM, Peng Q, Yang CH. 2012. Cell individuality: the bistable gene expression of the type III secretion system in Dickeya dadantii 3937. Mol Plant Microbe Interact 25:37–47. doi: 10.1094/MPMI-05-11-0105. [DOI] [PubMed] [Google Scholar]
- 22.Rufián JS, Sánchez-Romero MA, López-Márquez D, Macho AP, Mansfield JW, Arnold DL, Ruiz-Albert J, Casadesús J, Beuzón CR. 2016. Pseudomonas syringae differentiates into phenotypically distinct subpopulations during colonization of a plant host. Environ Microbiol 18:3593–3605. doi: 10.1111/1462-2920.13497. [DOI] [PubMed] [Google Scholar]
- 23.Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect Immun 73:3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crabill E, Karpisek A, Alfano JR. 2012. The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol Microbiol 85:225–238 doi: 10.1111/j.1365-2958.2012.08097.x. [DOI] [PubMed] [Google Scholar]
- 25.Huynh TV, Dahlbeck D, Staskawicz BJ. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 26.Lane WJ, Darst SA. 2006. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol 4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Peng Q, Zhang Q, Zou L, Li Y, Robert C, Pritchard L, Liu H, Hovey R, Wang Q, Birch P, Toth IK, Yang CH. 2010. Genome-wide identification of HrpL-regulated genes in the necrotrophic phytopathogen Dickeya dadantii 3937. PLoS One 5:e13472. doi: 10.1371/journal.pone.0013472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. 2007. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell 26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher J, Waite CJ, Bennett MH, Perez MF, Shethi K, Buck M. 2014. Differential secretome analysis of Pseudomonas syringae pv. tomato using gel-free MS proteomics. Front Plant Sci 5:242. doi: 10.3389/fpls.2014.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picotti P, Aebersold R. 2012. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 31.Thieffry D, Huerta AM, Pérez-Rueda E, Collado-Vides J. 1998. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20:433–440. doi:. [DOI] [PubMed] [Google Scholar]
- 32.Camas FM, Blázquez J, Poyatos JF. 2006. Autogenous and nonautogenous control of response in a genetic network. Proc Natl Acad Sci U S A 103:12718–12723. doi: 10.1073/pnas.0602119103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee KY, Opel M, Ito E, Hung S-p, Arfin SM, Hatfield GW. 1999. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc Natl Acad Sci U S A 96:14294–14299. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao XJ, Huo YX, Buck M, Kolb A, Wang YP. 2007. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res 35:1432–1440. doi: 10.1093/nar/gkl1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J, Goss TJ, Bender RA, Ninfa AJ. 1995. Repression of the Klebsiella aerogenes nac promoter. J Bacteriol 177:5535–5538. doi: 10.1128/jb.177.19.5535-5538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitale E, Milani A, Renzi F, Galli E, Rescalli E, de Lorenzo V, Bertoni G. 2008. Transcriptional wiring of the TOL plasmid regulatory network to its host involves the submission of the sigma54-promoter Pu to the response regulator PprA. Mol Microbiol 69:698–713. doi: 10.1111/j.1365-2958.2008.06321.x. [DOI] [PubMed] [Google Scholar]
- 37.Porrúa O, García-González V, Santero E, Shingler V, Govantes F. 2009. Activation and repression of a σN-dependent promoter naturally lacking upstream activation sequences. Mol Microbiol 73:419–433. doi: 10.1111/j.1365-2958.2009.06779.x. [DOI] [PubMed] [Google Scholar]
- 38.García-González V, Govantes F, Porrúa O, Santero E. 2005. Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon. J Bacteriol 187:155–167. doi: 10.1128/JB.187.1.155-167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee KY, Opel M, Ito E, Hung Sp, Arfin SM, Hatfield GW. 1999. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc Natl Acad Sci U S A 96:14294–14299. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowing DW, Gross CA. 1989. Interaction of Escherichia coli RNA polymerase holoenzyme containing σ 32 with heat shock promoters. J Mol Biol 210:513–520. doi: 10.1016/0022-2836(89)90127-7. [DOI] [PubMed] [Google Scholar]
- 41.Lee DJ, Busby SJW. 2012. Repression by cyclic AMP receptor protein at a distance. mBio 3:e00289-12. doi: 10.1128/mBio.00289-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ushida C, Aiba H. 1990. Helical phase dependent action of CRP: effect of the distance between the CRP site and the −35 region on promoter activity. Nucleic Acids Res 18:6325–6330. doi: 10.1093/nar/18.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estrem ST, Gaal T, Ross W, Gourse RL. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A 95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei W, Plovanich-Jones A, Deng WL, Jin QL, Collmer A, Huang HC, He SY. 2000. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc Natl Acad Sci U S A 97:2247–2252. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortiz-Martín I, Thwaites R, Macho AP, Mansfield JW, Beuzón CR. 2010. Positive regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 23:665–681. doi: 10.1094/MPMI-23-5-0665. [DOI] [PubMed] [Google Scholar]
- 46.Cisz M, Lee PC, Rietsch A. 2008. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J Bacteriol 190:2726–2738. doi: 10.1128/JB.01553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh A, Hespanha JP. 2009. Evolution of gene auto-regulation in the presence of noise. IET Syst Biol 3:368–378. doi: 10.1049/iet-syb.2009.0002. [DOI] [PubMed] [Google Scholar]
- 48.Maeda H, Jishage M, Nomura T, Fujita N, Ishihama A. 2000. Two extracytoplasmic function sigma subunits, sigma(E) and sigma(FecI), of Escherichia coli: promoter selectivity and intracellular levels. J Bacteriol 182:1181–1184. doi: 10.1128/JB.182.4.1181-1184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stauber JL, Loginicheva E, Schechter LM. 2012. Carbon source and cell density-dependent regulation of type III secretion system gene expression in Pseudomonas syringae pathovar tomato DC3000. Res Microbiol 163:531–539. doi: 10.1016/j.resmic.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Mishina TE, Zeier J. 2007. Bacterial non-host resistance: interactions of Arabidopsis with non-adapted Pseudomonas syringae strains. Physiol Plant 131:448–461. doi: 10.1111/j.1399-3054.2007.00977.x. [DOI] [PubMed] [Google Scholar]
- 51.Macho AP, Ruiz-Albert J, Tornero P, Beuzón CR. 2009. Identification of new type III effectors and analysis of the plant response by competitive index. Mol Plant Pathol 10:69–80. doi: 10.1111/j.1364-3703.2008.00511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macho AP, Zumaquero A, Gonzalez-Plaza JJ, Ortiz-Martín I, Rufián JS, Beuzón CR. 2012. Genetic analysis of the individual contribution to virulence of the type III effector inventory of Pseudomonas syringae pv. phaseolicola. PLoS One 7:e35871. doi: 10.1371/journal.pone.0035871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris CE, Monteil CL, Berge O. 2013. The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu Rev Phytopathol 51:85–104. doi: 10.1146/annurev-phyto-082712-102402. [DOI] [PubMed] [Google Scholar]
- 54.Wei HL, Collmer A. 2012. Multiple lessons from the multiple functions of a regulator of type III secretion system assembly in the plant pathogen Pseudomonas syringae. Mol Microbiol 85:195–200. doi: 10.1111/j.1365-2958.2012.08119.x. [DOI] [PubMed] [Google Scholar]
- 55.Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diard M, Garcia V, Maier L, Remus-Emsermann MNP, Regoes RR, Ackermann M, Hardt WD. 2013. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494:353–356. doi: 10.1038/nature11913. [DOI] [PubMed] [Google Scholar]
- 57.Guttman DS, Gropp SJ, Morgan RL, Wang PW. 2006. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae. Mol Biol Evol 23:2342–2354. doi: 10.1093/molbev/msl103. [DOI] [PubMed] [Google Scholar]
- 58.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 59.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 60.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 61.Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
- 63.Matic I, Jaffray EG, Oxenham SK, Groves MJ, Barratt CL, Tauro S, Stanley-Wall NR, Hay RT. 2011. Absolute SILAC-compatible expression strain allows sumo-2 copy number determination in clinical samples. J Proteome Res 10:4869–4875. doi: 10.1021/pr2004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Data 1) RNA-seq expression data for key T3SS genes in DC3000 wild-type and ΔhrpL. Statistically significant fold changes (FDR-adjusted P value < 0.05) are indicated in boldface characters. (Data 2) Peptide transitions analyzed by MRM-MS. Abbreviations: rep, replicate; ID, transition identifier; A, analyte; IS, internal standard; *, heavy amino acid residue. Download DATA SET S1, XLSX file, 0.02 MB (20.9KB, xlsx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of the ΔhrpL mutation on cell growth and the population distribution of hrpL promoter activity. (a) Histogram of GFP expression in populations of DC3000 cells after 8 h of growth in hrp-inducing medium. Triplicate populations of ΔhrpL (red shading) and wild-type (gray shading) strains were sampled for flow cytometry (10,000 events). Fluorescence signal is shown on a log10 scale. hrpL promoter activity followed a normal distribution across both the wild-type and ΔhrpL populations, with a higher average level of activity in the ΔhrpL strain. (b) Cell density (OD600) curves measured simultaneously with reporter fluorescence. ΔhrpS, ΔhrpL, and ΔhrpV gene deletions had a negligible effect on cell growth with respect to wild-type DC3000. Error bars represent SEM of results of 3 biological replicates. Download FIG S1, EPS file, 0.9 MB (966.2KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro abortive transcription activity of copurified RNAP-HrpL and RNAP-HrpLΔR4.2 complexes on hrpJ promoter probes. (a) RNAp core (6His-β′) was coexpressed in E. coli with HrpL or HrpLΔR4.2. Total protein was sampled following cell disruption (Whole cell) and purification by Ni-affinity chromatography (Purified) and compared alongside size markers (L) by staining of SDS-PAGE gels with Coomassie dye. The following components are signified by arrows: RNAP α subunit (34.9 kDa); HrpL (22.6 kDa); HrpLΔR4.2 (18.9 kDa); RNAP ω subunit (10.5 kDa). (b) Detection of HrpL variants by Western blotting targeting a C-terminal Myc tag. Proteins were transferred directly from the gel shown in panel a to a polyvinylidene difluoride (PVDF) membrane and incubated with an anti-Myc antibody. (c) Abortive initiation on supercoiled PhrpJ or PhrpJ(Δ35e) promoter DNA was assayed in the presence of RNAP core enzyme or RNAP-HrpL or RNAP-HrpLΔR4.2 holoenzymes. An abortive small priming RNA (spRNA) product (predicted CpApUpApApU) was separated from free nucleotides by electrophoresis. The average level of initiation activity across 3 replicates is given as a percentage relative to RNAP-HrpL on wild-type promoter DNA. Initiation was inhibited >90% in the presence of RNAP-HrpLΔR4.2 and/or PhrpJ(Δ35e). Download FIG S2, EPS file, 7.9 MB (8.1MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid-borne expression of HrpL and HrpLΔR4.2 in DC3000. (a) HrpL dependence of hrpJ promoter fusion activity in DC3000. hrpJ promoter activity in wild-type (black) and ΔhrpL (red) strains carrying both pBBR1-rfp-PhrpL-gfp and plasmids for heterologous expression of hrpL under hrp-inducing conditions is reported. hrpL (full circles) or hrpLΔR4.2 (empty circles) genes were expressed from the pSEVA224 plasmid. Empty pSEVA224 vector (bold lines) was used as a plasmid load control. Autofluorescence by DC3000 in the absence of reporter was also measured (gray dashed line). Error bars represent SEM of results of 3 biological replicates. hrpJ promoter activity was negligible, with respect to autofluorescence, in the ΔhrpL strain, confirming the HrpL dependence of transcription. Complementation with plasmid-borne hrpL, but not inactive hrpLΔR4.2, resulted in strong upregulation of PhrpJ with respect to wild-type results. (b) Cell density (OD600) curves measured simultaneously with reporter fluorescence. hrpL and hrpLΔR4.2 overexpressed from pSEVA224 had a negligible effect on the cell growth of wild-type DC3000, with respect to an empty vector control for plasmid load. Error bars represent SEM of results of 3 biological replicates. Download FIG S3, EPS file, 1.1 MB (1.2MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Activity of the hrpRS promoter in ΔhrpL. hrpRS promoter activity reported in WT (black) and ΔhrpL (red) strains carrying pBBR1-PhrpRS-gfp under hrp-inducing conditions. Error bars represent SEM of results of 3 biological replicates. No significant difference in hrpRS promoter activity is evident in the ΔhrpL strain with respect to wild-type DC3000 results during 8 h in hrp-inducing medium. Download FIG S4, EPS file, 0.5 MB (573KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) RpoD and (b) Lon protease abundance in intracellular protein fractions. Total cell-bound (cell pellet) protein was extracted from DC3000 cell cultures after 24 h in hrp-inducing medium for targeted protein quantification via LC-MRM-MS. The pSEVA224-33-hrpL (+) and pSEVA224-31-hrpL (++) plasmids were used to overexpress hrpL. Empty pSEVA224 was used as a plasmid load control in the wild-type strain. From left to right, the column bars shown represent the transitions analyzed for relative protein abundance (see Data Set S1) as follows: for RpoD, FGAVa, FGAVb, IPVHa, and IPVHb; for Lon, TSLAa and TSLAb. The analyte transition signal intensities were normalized against sample cell density (OD600). Error bars represent SEM of results of 3 biological replicates. Download FIG S5, EPS file, 0.7 MB (724.9KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Arabidopsis thaliana seedling infection assays. (a) Col-0 seedlings were flood inoculated with bacterial suspensions of DC3000 wild-type and ΔhrpL strains at a cell density of 5 × 105 CFU ml−1. Three replicate groups of 16 seedlings were flooded with each strain. Mock inoculations were performed with 10 mM MgCl2 solution as a control. Bacterial populations were quantified at 0, 1, 2, and 3 days postinfection (dpi). Error bars represent SEM for 3 samples, each consisting of 3 seedlings. Asterisks denote statistical significance in population size with respect to wild-type control results, based on a two-tailed t test. (b) Disease phenotypes of groups of three representative seedlings photographed 3 days postinfection. Symptoms, including chlorosis and lesion formation, are visible after infection with the wild-type strain but not after infection with the ΔhrpL strain or mock infection. (c) The effect of graded HrpL overexpression on in planta fitness. Seedlings were flood inoculated with DC3000 bacterial suspensions as described for panel a. The growth of wild-type populations carrying a series of pBBR1MCS4 plasmids for hrpL (plain bars) overexpression was monitored over 3 dpi. hrpL expression was regulated by synthetic ribosome binding sites (RBSs) (activity of pSEVA224-31-hrpL [31] > activity of pSEVA224-33-hrpL [33]). The pBBR1MCS4 vector was used as a plasmid control (black). hrpLΔR4.2 (hashed bars) was similarly overexpressed as a control for sigma-independent effects of HrpL. Neither HrpL overexpression nor HrpLΔR4.2 overexpression had any significant effect on in planta fitness with respect to plasmid control results (two-tailed t test). Download FIG S6, EPS file, 16.4 MB (16.8MB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative UP element sequence upstream of the hrpJ promoter. (a) Position of putative UP element (UP J) with respect to the IHF recognition sequence and σ54 promoter of the PhrpL. (a) Alignment of UP J and known functional UP elements (rrnB1 P1, rrnD1 P1) with the consensus UP element (Estrem et al. [43]). Conserved positions are indicated in boldface and underlined characters. W, A or T nucleotide; N, any nucleotide. Download FIG S7, EPS file, 0.7 MB (686.1KB, eps) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this work. Download TABLE S1, DOCX file, 0.04 MB (44KB, docx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplementary methods. Download TEXT S1, DOCX file, 0.04 MB (41.4KB, docx) .
Copyright © 2017 Waite et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.