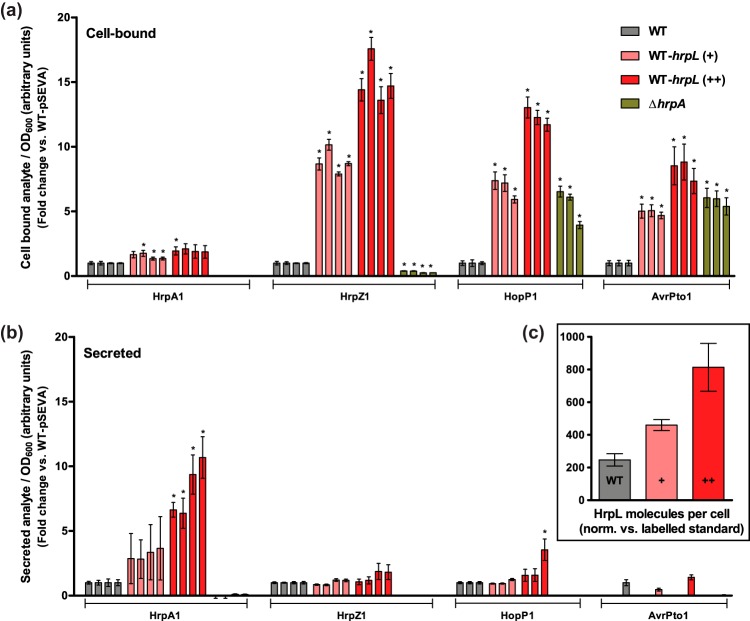
FIG 5 .
The effect of hrpL misregulation on expression and secretion of T3SS proteins. (a and b) Cell-bound (cell pellet) (a) and secreted (supernatant) (b) protein fractions were extracted from DC3000 cell cultures after 24 h in hrp-inducing medium for targeted protein quantification via LC-MRM-MS. The pSEVA224-33-hrpL (+) and pSEVA224-31-hrpL (++) plasmids were used to overexpress hrpL. An empty pSEVA224 vector was used as a plasmid load control. The ΔhrpA strain was used as a control for T3SS-independent release of protein. Multiple peptide transitions per protein were analyzed for calculation of relative target protein abundances between strains. From left to right, the columns shown represent the following transitions (see Data Set S1): for HrpA1, ISATa, ISATb, LTNLa, and LTNLb; for HrpZ1, AQFPa, AQFPb, SANSa, and SANSb; for HopP1, GQLNa, GQLNb, and NSNS; and for AvrPto1, HQLAa, HQLAb, and VSNN. The analyte transition peak intensities were normalized against sample cell density (OD600) and subsequently the wild-type (WT) pSEVA224 peak intensity to give data for fold change between strains. Absent column bars indicate that transitions were undetectable. (c) The absolute HrpL copy number in each intracellular sample was calculated using the ratio between the sample peptide (QPSS) and heavy-isotope standard peptide (QPSS-IS) transition peak intensities, normalizing for OD600 and standard concentration. Protein copy number data assume an OD600 of 1.0 = 109 CFU ml−1. Error bars represent SEM of results of 3 biological replicates.