Abstract
Notch signaling plays a key role in the development of pancreatic cancer. Among the four identified Notch receptors, Notch1 and Notch2 share the highest homology. Notch1 has been reported to be an oncogene but some reports indicate that Notch2, not Notch1, plays a key role in pancreatic carcinogenesis. As both are transcription factors, examination of their genomic binding sites might reveal interesting functional differences between them. Notch proteins do not have DNA-binding domain. In the canonical Notch signaling pathway, ligand binding induces the release and nuclear translocation of Notch receptor intracellular domains (NICDs), which then interact with the transcription factor CSL, resulting in subsequent activation of the canonical Notch target genes. We investigated the binding site profiles of Notch1and Notch2 in the BxPC3 genome using CHIP-Seq and bioinfomatics. We found that Notch1, Notch2 and CSL generally bound to different target genes. We also found that only a small subset of Notch1 and Notch2 binding sites overlap with that of CSL, but about half of the CSL binding overlap with that of Notch1 or Notch2, indicating most Notch signaling activities are CSL-independent.
Keywords: Notch, Genome binding sites, Pancreatic Cancer.
Introduction
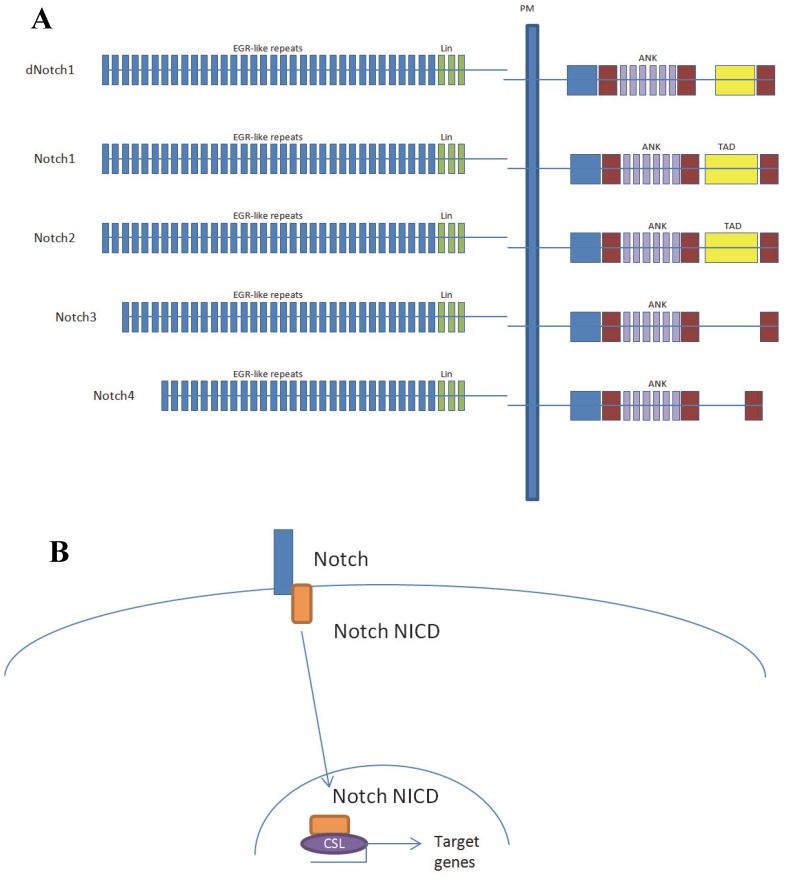
Aberrantly activated Notch signaling plays key roles in carcinogenesis and the progression of human malignancies 1. However, Notch signaling has been reported to play either oncogenic or tumor suppressor roles depending on the tissue context 2. The Notch signaling network is frequently deregulated in pancreatic cancer, with up-regulated expression of Notch receptors and their ligands 3. To date, four Notch receptors have been identified in mammals 4 and the presence of multiple Notch receptors and ligands raises the question whether different receptors play different roles in the same cell type. Of the four Notch receptors, Notch1 and Notch2 have the highest homology in both extracellular and intracellular domains (Fig. 1A). Nevertheless they have opposite effects on the growth of embryonic brain tumors growth, suggesting that they play opposing roles in a single tumor type 5.
Figure 1.
Notch protein domains and the canonical Notch signal transduction pathway. (A) Diagrammatic representation of the domains of the Drosophila Notch (dNotch) receptor and the four known mammalian receptors. Note that Notch3 and Notch4 contain no transcriptional activation domain TAD domain. PM, plasma membrane; EGFLR, epidermal growth-factor-like repeats; ANK, Ankyrin/CDC10 repeats; NLS, nuclear localization signals; TAD, transcriptional activation domain; PEST, Pro Glu Ser Thr (PEST) sequence. (B) Notch does not directly bind to DNA. In the canonical Notch signal transduction model, Notch does not directly bind to DNA, rather goes through CSL. In the absence of Notch intracellular domain (NICD), CSL binds to co-repressors. The interaction CSL with ANK repeats of the NICD displace these repressors to generate a transcriptional activator complex.
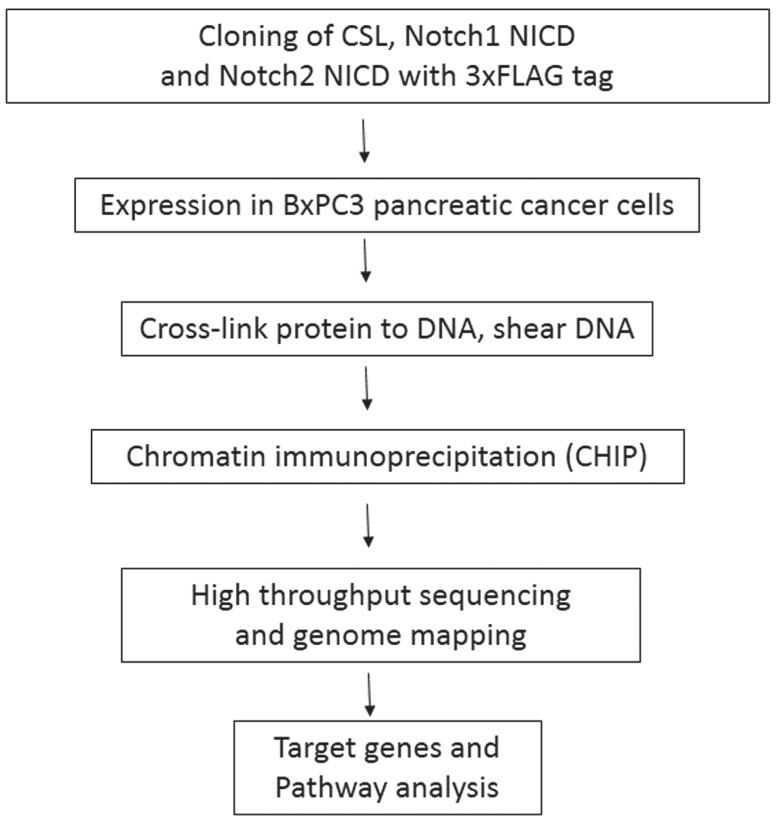
Although Notch1 has been reported to be oncogenic for pancreatic cancer, there is evidence that Notch2, rather than Notch1 plays a key role in pancreatic carcinogenesis 6, 7. Therefore, it is important to investigate the functional differences between Notch1 and Notch2 in pancreatic cancer cells. Notch receptors do not have DNA binding domains, so it is impossible to searching their targets based on genome sequences. In the canonical Notch signaling pathway, ligand binding induces a series of cleavages of the full-length receptors, and Notch receptor intracellular domains (NICDs) then translocate to the nucleus, interact with the transcription factor CSL (CBF1, Suppressor of Hairless, Lag1) which is then converted from a transcriptional repressor to a transcriptional activator 8 (Fig. 1B). As Notch1 and Notch2 are transcription factors, a closer examination of Notch1 and Notch2 genome binding data may reveal interesting functional differences in these Notch paralogues. CHIP-Seq was used for the identification of Notch1, Notch2 and CSL binding sites. Fig. 2 provides a schematic outline of the experimental procedure.
Figure 2.
A schematic diagram of the experimental procedures.
Materials and Methods
Cell culture
Human pancreatic BxPC3 cells were obtained from the ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Life Technologies, Gaithersburg, MD). Cells were incubated in 75cm2 culture flasks in a humidified atmosphere at 37°C with 5% carbon dioxide in air, and passaged on reaching 80% confluence.
Plasmid constructions
BxPC3 cells were grown on plates and total RNA was isolated using Trizol reagent (Life Technologies, Gaithersburg, MD), followed by clean up with an RNeasy® Mini kit (Qiagen, Valencia, CA). RNA was reverse-transcribed to cDNA. Notch1, Notch2 intracellular segments and CSL cDNA were amplified and cloned into pcDNA3.0-3×Flag. The ligation products were transformed into competent cells; transformants were cultured on solid LB medium overnight and suitable clones selected.
Western blotting
Cells were seeded at 1 × 106 per well in 5ml of culture medium in 75cm2 culture flasks, and allowed to grow overnight. SDS lysis buffer (0.05 mM Tris-HCl, 50mM BME, 2% SDS, 0.1% Bromophenol blue, 10% glycerol) was used to lyse the cells. Heat denatured proteins were then loaded, separated on an SDS-page gel, transferred onto a pure nitrocellulose membrane (Bio-Rad), and blocked with 5% milk. Membranes were incubated with anti-Flag antibody (1:500 dilution; M2, Sigma-Aldrich, St. Louis, MO) or mouse anti-human β-actin monoclonal antibody (1:100 dilution; Santa Cruz Biotechnology; Santa Cruz, CA) overnight at 4°C, then washed four times with TBST at room temperature for 5 min each wash, and incubated with horseradish peroxidase-conjugated rabbit antimouse immunoglobulin G (1:5000 dilution; Zymed, San Francisco, CA) for 60 min at room temperature. The membranes were washed again four times with TBST at room temperature (5 min each wash) and protein bands were visualized by enhanced chemiluminescence (ECL Detection System; Pierce, Rockford, IL).
Chromatin immunoprecipitation (ChIP)
BxPC3 cells were cross-linked with 1% formaldehyde for 10 min at 37°C, then rinsed with cold PBS, harvested and lysed with 1% SDS, 10 mM EDTA, and 50 mM Tris-HCl (pH 8.1) containing a protease inhibitor cocktail (Roche, Indianapolis, IN). Chromatin was fragmented by two sequential sonications (2 min each) with 30 sec on/off cycles and a Bioruptor sonicator (Diagenode, Denville, NJ) at its highest intensity. The soluble chromatin was diluted in buffer containing 1% Triton, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl (pH 8.1) and added to Protein-G (Qiagen, Valencia, CA) pre-incubated with control IgG or anti-Flag antibody. After 24 h, precipitates were washed 5 times and immune complexes were eluted using 100 ml of 1% SDS and 0.1 M NaHCO3. The samples were incubated overnight at 65°C to reverse cross-linking, and the DNA was purified with a PCR purification kit (Qiagen, Valencia, CA).
DNA Library preparation and high-throughput sequencing
The purified DNA products were modified for Illumina Whole-Genome Chromatin IP sequencing using an Illumina Genomic DNA Sample Prep kit as follows: size-selected DNAs were end-repaired by T4 DNA polymerase and phosphorylated by T4 DNA polymerase and T4 polynucleotide kinase. The products were incubated with Klenow DNA Polymerase (Illumina) to generate 3′ adenine overhangs and then ligated to Illumina adapters, which contain 5′ thymine overhangs. The adapter-ligated products were purified on QIAquick spin columns (Qiagen), PCR-amplified with Phusion DNA Polymerase (Finnzymes) for 30 cycles using an Illumina genomic DNA primer set. The PCR products were purified on QIAquick and MinElute columns (Qiagen).The quality of the DNA was assessed and quantified using an Agilent DNA 1000 Series II assay and NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and the DNA was diluted to 10 nM. Cluster generation and sequencing were performed using a Standard Cluster Generation kit and a Cycle Solexa Sequencing kit on the Illumina Cluster Station and Illumina HiSeq2000 sequencer following the manufacturer's instructions. Sequencing was carried out by the Research & Cooperation Division, BGI-Shenzhen.
Bioinformatics
Primary sequencing data consisted of: (i) Basic read maps; (ii) Peak region scans, including peak region detection, peak counts, average peak length, median peak length; (iii) The associated genes with sample peaks; (iv) The depth of coverage distribution of the mapped reads of the samples in the gene region; (v) GO function notability enrichment analysis of peak-related genes. SOAPaligner/soap2 was used for mapping sequencing reads to the genome. The SOAPaligner/soap2 is a program for faster and efficient alignment for short oligonucleotide onto reference sequences Genome Reference Consortium GRCh37/hg19 was used as reference genome. After filtering, and aligning read tags to reference sequences http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/refGene.txt.gz), we calculated the average read coverage for all non-overlapping 50-bp slide windows of the genome. MACS 1.4.0 software was used for identifying potential peaks.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping is a procedure for mapping molecular datasets, especially large-scale datasets in genomics, transcriptomics, proteomics, and metabolomics, to the KEGG pathway maps for biological interpretation of higher-level systemic functions. Pathway annotations of the Notch1, Notch2 and CSL target genes were obtained from KEGG (http://www.genome.jp/kegg/). In the KEGG database, one gene may be involved in several pathways or interact with several other genes. Also, the KEGG Atlas website provides a mapping interface and allows mapping of genes as colored lines/circles in the global map. Pathway categories with a FDR <0.01 were marked.
Whole-genome expression profiling
The BxPC3 gene expression dataset was downloaded from GEO (accession number GSE15550). The dataset consists of microarray data, and the signal intensities were used as expression values.
Statistical analyses
All experiments were performed a minimum of three times. Data are means ± SDs and were compared using Student's t-tests. A P-value <0.05 was considered to be statistically significant. Statistical analyses were performed with Office 2013 software (Microsoft) for Windows.
Results
Notch1, Notch2 and CSL Binding Sites on the BxPC3 cell genome
As Notch 1 and Notch 2 have been implicated as important transcription factors in pancreatic cancer, and CSL is one of the downstream effectors for Notch family proteins, we mapped Notch1, Notch2 and CSL binding sites in the genome of BxPC3 pancreatic cancer cells. The quality of antibodies used for ChIP-Seq is a crucial factor that contributes to the quality of the data. Antibodies that offer high sensitivity and specificity are required for ChIP-Seq because they allow the detection of enrichment peaks without substantial background noise. It is also important to consider the potential cross-reactivity of antibodies with closely related protein family members. Because lack of specific CHIP-grade antibodies, FLAG-tagged CSL, Notch1 NICD and Notch2 NICD were expressed (Supplementary Fig S1), and anti-FLAG antibodies are used for the CHIP-Seq experiments.
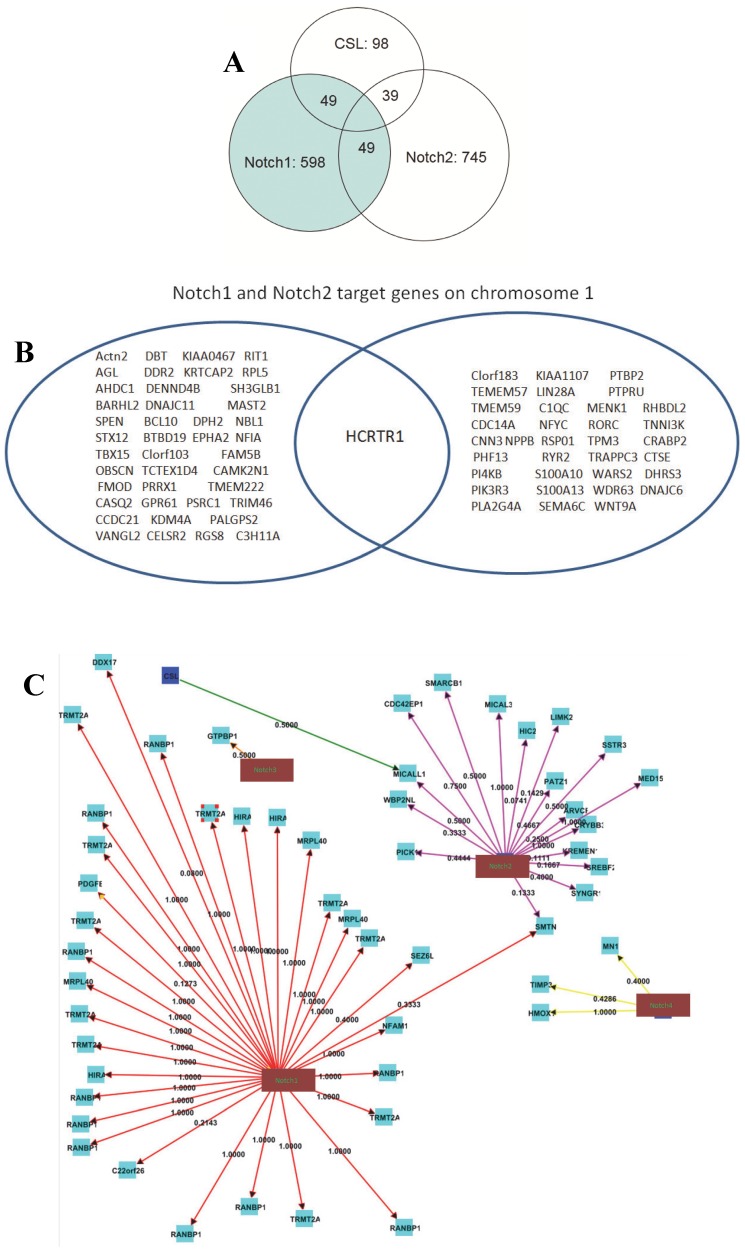
MACS (Model-based Analysis of ChIP-Seq) is a command line tool designed to analyze data generated by ChIP-Seq experiments in eukaryote. MACS is used to identify transcription factor binding sites and histone modification enriched regions based on the ChIP-Seq data with or without controls. We used MACS 1.4.0 software and peak-calling algorithms to identify potential binding sites referred to as enriched regions. Analysis of CHIP-seq data identified 598, 745 and 85 statistically significant peaks of reads for Notch1, Notch2 and CSL, respectively. Only 49 of 598 Notch1 peaks (8.19%) overlapped with CSL peaks, whereas 49 of the 98 CSL peaks (57.65%) overlapped with Notch1 peaks (Fig. 3A). Only 39 of the 745 Notch2 (5.23%) peaks overlapped with CSL peaks, whereas 45.88% of CSL peaks overlapped with Notch2 peaks (Fig. 3A). Thus, only small numbers of Notch1 and Notch2 binding sites overlap with those of CSL, indicating that most Notch1 and Notch2 functions are CSL-independent. Only 8.23% (49) of the Notch1 peaks overlapped with the Notch2 peaks, and 6.58% of the Notch2 peaks overlapped with the Notch1 peaks. We found that Notch1 and Notch2 have 48 and 43 target genes on chromosome 1, respectively, but they only share a common target, HCRTR1 (Fig. 3B; Supplementary Fig. S2). Other chromosomes have similar patterns, for example, on the chromosome 22, 32 and 17 target genes were found for Notch1 and Notch2, respectively, but only one gene (SMTN) was shared by Notch1 and Notch2 (Fig. 3C). Therefore Notch1 and Notch2 have different binding profiles in pancreatic cancer cells.
Figure 3.
Overlap of Notch1, Notch2 and CSL binding sites. Number of Notch1, Notch2 and CSL binding sites and overlapping binding sites are shown. (A) Analysis of CHIP-seq data identified 598, 745 and 85 statistically significant peaks of reads for Notch1, Notch2 and CSL. Number of Notch1, Notch2 and CSL binding sites and overlapping binding sites are shown. (B) Notch1 and Notch2 only share a few target genes. The figure shows Notch1 and Notch2 target genes on chromosome 1. Only one gene is targeted by both Notch1 and Notch2 on chromosome 1. (C) Target gene distribution of CSL and Notch family proteins in chromosome 22. Notch1 and Notch2 only share one gene (SMTN) on chromosome 22. The numbers are path coefficients.
Pathways relevant to Notch1, Notch2 and CSL target genes
We used the KEGG pathway mapping tool to identify the pathways biologically related to the ChIP-Seq-based Notch1, Notch2 and CSL target genes, consisting of the genes significantly enriched in each set. Table 1 shows the top 10 pathways associated with the Notch1, Notch2 and CSL target genes. The PI3K-AKT pathway is an intracellular signaling pathway important in regulating the cell cycle. Therefore, it is directly related to cellular quiescence and proliferation; if this pathway is overactive, it will reduce apoptosis and allow proliferation. We identified PI3K-AKT pathway as the most significant canonical pathway associated with the target genes of all three proteins, Notch1, Notch2 and CSL, thus supporting their roles in cell cycle control and cell growth.
Table 1.
Top 10 KEGG pathways relevant to CHIP-Seq-based Notch1, Notch2 and CSL target genes.
| Pathways (gene numbers) | |||
|---|---|---|---|
| Rank | Notch1 target genes | Notch2 target genes | CSL target genes |
| 1 | PI3K-Akt signaling pathway (15) | PI3K-Akt signaling pathway (16) | Regulation of actin cytoskeleton (4) |
| 2 | Transcriptional misregulation in cancer (10) | Pathways in cancer (13) | PI3K-Akt signaling pathway (4) |
| 3 | Metabolic pathways (10) | Transcriptional misregulation in cancer (10) | cAMP signaling pathway (3) |
| 4 | Pathways in cancer (9) | Viral carcinogenesis (9) | Tight junction (2) |
| 5 | Ras signaling pathway (9) | cAMP signaling pathway (9) | Arrhythmogenic right ventricular cardiomyopathy (ARVC) (2) |
| 6 | MAPK signaling pathway (9) | Metabolic pathways (9) | Calcium signaling pathway (2) |
| 7 | Proteoglycans in cancer (9) | HTLV-I infection (9) | Jak-STAT signaling pathway (2) |
| 8 | Regulation of actin cytoskeleton (8) | Neuroactive ligand-receptor interaction (9) | Hypertrophic cardiomyopathy (HCM) (2) |
| 9 | HTLV-I infection (8) | Ras signaling pathway (8) | Herpes simplex infection (2) |
| 10 | MicroRNAs in cancer (7) | Epstein-Barr virus infection (7) | MicroRNAs in cancer (2) |
Notes: By importing Entrez Gene IDs of ChIP-Seq-based Notch1, Notch2 and CSL target genes into the Functional Annotation tool of KEGG, KEGG pathways showing significant relevance to the set of imported genes were identified.
Besides the PI3K-AKT pathway, we identified transcriptional misregulation in cancer, metabolic pathways, the Ras pathway and the MAP kinase pathway as the most significant pathways linked to the Notch1 target genes. Pathways in cancer, transcriptional misregulation in cancer and viral carcinogenesis were associated with Notch2 target genes. Some Notch1, Notch2 and CSL target genes are associated with the PI3K-AKT pathway, the actin cytoskeleton and cancer microRNAs in cancer, but most of these target genes do not overlap. In the PI3K-AKT pathway, CSL only shares the target genes IL-4 and TSC1 with both Notch1 and Notch2, ITGB4 with Notch1, and CHRM1 with Notch2, but most of the target genes implicated in the same pathway are different. For example, Notch1 and Notch2 have 15 and 16 target genes, respectively, in the PI3K-AKT pathway, but only 3 are shared: CDKN1A, IL-4 and TSC1 (data not shown).
Notch1 target genes and cancer pathways
The Notch1 target genes associated with cancer pathways are CCDC6, CDKN1A, FGFR2, GNB3, MET, MYC, PDGFB, PTCH1, RUNX1T1 (Table 2; Supplementary Fig. S3). In the gene expression dataset for BxPC3 in the NCBI GEO database (Dataset GSE15550), the average expression level is 433.66 for the all probes used. In that dataset, the Notch1 target genes associated with cancer pathways, namely genes MET, MYC, CCDC6 and CDKN1A, have expression levels exceeding this average level (i.e. >433.6). The gene CDKN1A codes for p21, which is a potent cyclin-dependent kinase inhibitor (CKI) 9. CCDC6 encodes a coiled-coil domain-containing protein and may function as a tumor suppressor 10.
Table 2.
List and expression level of Notch1 target genes in cancer pathway.
| Notch1 target genes | Full name of the gene | Illumina ID | Gene expression level |
|---|---|---|---|
| MET | met proto-oncogene (hepatocyte growth factor receptor) | ILMN_12011 | 4048.37 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | ILMN_28130 | 3006.24 |
| CCDC6 | coiled-coil domain containing 6 | ILMN_19206 | 2311.41 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | ILMN_5895 | 1260.56 |
| GNB3 | guanine nucleotide binding protein (G protein), beta polypeptide 3 | ILMN_4600 | 87.09 |
| PDGFB | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | ILMN_18749 | 80.97 |
| PTCH1 | patched homolog 1 (Drosophila) | ILMN_18640 | 76.33 |
| FGFR2 | fibroblast growth factor receptor 2 | ILMN_20933 | 71.53 |
| RUNX1T1 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | ILMN_13370 | 67.37 |
The KEGG Atlas website provides a mapping interface to allow mapping of genes as colored lines/circles in the global map. MET codes for the hepatocyte growth factor (HGF) receptor c-Met and has been implicated in driving proliferation and invasion, and is associated with a poor prognosis in pancreatic cancer 11. c-Myc is a member of the MYC family of transcription factors 12. MET and MYCs are thought to be oncogenes, and CCDC6 and CDKN1A tumor suppressor genes (TSGs). These results indicate that Notch1 targets both oncogenes and TSGs, and not all Notch1 target genes are highly expressed.
Notch2 target genes and cancer pathways
Notch2 target genes associated with cancer pathway are TPM3, NF-κB IA, NF-κB 1, CDKN1A, GSK-3β, GNAQ, BAX, MDM2, PGF, FOXO1, RASGRP2 (Table 3; Supplementary Fig. S4). TPM3 has the highest expression level among the genes in the gene expression dataset for BxPC3 in the NCBI GEO database, with a value of 11010.90. The expression level of TPM3, NF-κB IA, NF-κB 1, CDKN1A, GSK-3β, and GNAQ are also above the average, and the expression levels of BAX, MDM2, PGF, FOXO1 and RASGRP2 is lower than the average value. TPM3, NF-κB 1, GSK-3β and GNAQ are potential oncogenes and NF-κB IA and BAX are thought to be TSGs. These findings suggest that Notch2 targets both oncogenes and TSGs. Fig. 3C shows the Notch1 and Notch2 target different cancer-related pathways in the BxPC3 pancreatic cancer cells.
Table 3.
List and expression level of Notch2 target genes in cancer pathway.
| Notch2 target genes | Full name of the gene | Ilumina ID | Gene expression |
|---|---|---|---|
| TPM3 | tropomyosin 3 | ILMN_17262 | 11010.9 |
| NF-κB IA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | ILMN_6745 | 10786.49 |
| NF-κB 1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | ILMN_26921 | 6345.35 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | ILMN_5895 | 1260.56 |
| GSK3B | glycogen synthase kinase 3 beta | ILMN_7421 | 932.78 |
| GNAQ | guanine nucleotide binding protein (G protein), q polypeptide | ILMN_18320 | 621.61 |
| BAX | BCL2-associated X protein | ILMN_10832 | 140 |
| MDM2 | Mdm2 p53 binding protein homolog (mouse) | ILMN_21265 | 114.98 |
| PGF | placental growth factor | ILMN_27436 | 90.5 |
| FOXO1 | forkhead box O1 | ILMN_4600 | 87.09 |
| RASGRP2 | RAS guanyl releasing protein 2 (calcium and DAG-regulated) | ILMN_10397 | 82.93 |
| FN1 | fibronectin 1 | ILMN_4516 | 72.35 |
| GNG3 | guanine nucleotide binding protein (G protein), gamma 3 | ILMN_7558 | 71.26 |
Discussion
The domains of Notch1 and Notch2 are highly conserved, and although both genes are expressed in pancreatic cancer, their functions (and differences in function) are not clear. Identification of their target genes may help to understand their roles in pancreatic cancer. CSL is a DNA binding transcription factor. In the canonical Notch signaling pathway, when ligands on neighboring cells interact with Notch receptors, Notch is proteolytically cleaved, and the notch receptor intracellular domain (NICD) translocates from the cell membrane to the nucleus where it forms a transcriptionally active ternary complex with CSL and a member of the Mastermind (MAM) family of coactivators. Assembly of the CSL-NICD-MAM ternary complex at a target gene is the switch for up-regulating transcription of that locus 13.
Our results indicate that most Notch1 and Notch2 target genes are CSL-independent. We found only a small subset of Notch1 and Notch2 peaks that overlapped with CSL peaks, indicating that most Notch1 and Notch2 binding sites in the genome are not dependent on CSL. Factors other than CSL may be involved in Notch1 and Notch2 binding to the genome. Since about half of the CSL peaks overlap with Notch1 or Notch2 peaks, the functions of CSL appear to be more closely related to those of Notch1 and Notch2, than those of Notch1 and Notch2 are related to CSL. In addition, a few of some other co-factors may also be involved in CSL's activities. For example, the bHLH transcription factor Ptf1a was found to be a co-factor of CSL and forms transcription complexes that can activate CSL target gene transcription 14.
Our results indicate that the profiles of the major pathways in which Notch1, Notch2 and CSL target genes are different. Moreover different Notch1, Notch2 and CSL target genes are associated with a given common pathway. For example, Notch1, Notch2 and CSL only share a few target genes in the PI3K-AKT pathway, namely IL-4, TSC1 and CDKN1A, while most of the other target genes do not overlap. This results support the notion that Notch1, Notch2 and CSL tend to play different roles even in the same pathway. Further studies are needed to identify the roles of Notch1, Notch2 and CSL in regulating the activity of a pathway that they have in common: do they play parallel, opposing or synergistic roles?
We found that Notch1 and Notch2 both bind to the same target gene CDKN1A in the cancer pathway. Gene CDKN1A codes for p21(CIP1/WAF1), which binds to and inhibits the activity of cyclin-CDK2, -CDK1, and -CDK4/6 complexes, and thus functions as a regulator of cell cycle progression in G1 and S phases. In addition to growth arrest, p21 can mediate cellular senescence. The expression of this gene is tightly controlled by tumor suppressor protein p53, through which p21 mediates p53-dependent cell cycle arrest in G1 in response to a variety of stress stimuli 9. When DNA damage or inhibitory extracellular signals are present, p53 increases and stimulates transcription of the p21 gene. As CDKN1A is a target of both Notch1 and Notch2, further study is required to establish the different roles of Notch1 and Notch2 in regulation of p21 expression.
The main Notch1 target genes are MET, MYC and CCDC6. HGF and its receptor, c-Met, have been implicated in driving proliferation, invasion, and a poor prognosis in pancreatic cancer 11. In one report, HGF and MET mRNAs were measured in 59 primary pancreatic cancers and 51 normal samples, and the results showed that both factors are highly expressed in pancreatic cancer. This suggests that the HGF/c-Met axis plays an important role in the progression of pancreatic cancer and that targeting c-Met may be of therapeutic value 11. Mounting evidence suggests that c-Myc participates in most aspects of cellular function, including metabolism, growth, differentiation, apoptosis, adhesion, and migration 12. Amplification and/or over-expression of c-Myc are frequently detected in cancers including pancreatic cancer 12.
Besides CDKN1A, Notch1 also targets the potential tumor suppressor gene CCDC6, which encodes a coiled-coil domain-containing protein that is involved in DNA damage repair. When thyroid cancer cells were treated with etoposide or ionizing radiation (IR), CCDC6 underwent ATM-mediated phosphorylation at Thr 434, stabilizing nuclear CCDC6, indicating that this protein is involved in the response to ATM-mediated DNA damage. Impairment of CCDC6 gene function may have a role in thyroid carcinogenesis 13.
The Notch2 target genes in the cancer pathway are TPM3, NF-κBIA, NF-κB1, GSK-3β and GNAQ. TPM3 gene encodes a member of the tropomyosin family of actin-binding proteins and is a potential oncogene. TPM3 was overexpressed in human hepatocellular carcinoma and there was a potential link to epithelial-mesenchymal transition 14. Also, TPM3 overexpression is associated with high-grade gliomas and increased mortality. The role of TPM3 in pancreatic cancer deserves further investigation.
It is interesting that Notch2 targets both NF-κB and the NF-κB inhibitor, NF-κBIA, and both genes are highly expressed in BxPC3 pancreatic cancer cells. NF-κB plays a key role in proliferation, cell survival, and invasion by pancreatic cancer cells. It is constitutively activated in pancreatic cancer where its inhibition enhances the sensitivity of cancer cells to chemotherapeutic agents and death receptor-mediated apoptosis. NF-κBIA, also known as IκBα, is a member of the family of cellular proteins that inhibit the activity of NF-κB by masking its nuclear localization signal (NLS) and keeping it sequestered in an inactive state in the cytoplasm 15. It may be a tumor suppressor 16. Notch2 is potentially a key regulator of NF-κB and NF-κBIA expression. Further experiments are needed to identify additional factors involved in the regulation of NF-κB and NF-κBIA expression by Notch2. Obviously the NF-κB /NF-κBIA ratio is important for the activity of NF-κB signaling.
GSK-3β is also a potential Notch2 target gene. GSK-3β is a serine/threonine protein kinase that regulates multiple signaling pathways. Although its role in cancer is still debated, the overall results so far indicate that aberrant expression and activity of GSK-3β is a fundamental characteristic of a broad spectrum of cancers 17. The potential involvement of GSK-3β in the invasiveness of pancreatic cancers and their resistance to gemcitabine and ionizing radiation, the two major obstacles to more effective treatment, have also been reported 18.
The role of GNAQ in pancreatic cancer has not been examined. GNAQ encodes the alpha subunit of the q class of heterotrimeric GTP binding protein (Gq) that transduces signals from G-protein-coupled receptors (GPCRs), and stimulates β phospholipase C (PLCβ), which catalyzes the hydrolysis of phosphatidylinositol biphosphate (PIP2). Frequent somatic mutations were found in GNAQ in a subset of melanocytic neoplasms that did not have alterations in RAS or BRAF 19. Genetic, biochemical and biological analysis has indicated that GNAQ is a bona fide human oncogene. The role of GNAQ in pancreatic cancer remains to be defined.
Notch2 may be involved in the regulation of the p53 pathway. Mdm2 is an important negative regulator of p53 and is a Notch2 target gene. Mdm2 functions both as an E3 ubiquitin ligase that recognizes the N-terminal trans-activation domain (TAD) of p53 and an inhibitor of p53 transcriptional activation 20. It has been reported that single-nucleotide polymorphisms are associated with the risk of cancer 21-23. Detection of mutations and genomic polymorphisms in the Notch target sequences may provide valuable information about differences among individuals in terms of the Notch activity and help us to understand how the Notch pathways are involved in the risk of pancreatic cancer. Chen et al. reported that 8 genetic variants are predictive of early onset pancreatic cancer, and all of the 8 SNPs were predicted to play functional roles in the disruption of transcription factor and/or enhancer binding sites and most of them were expression quantitative trait loci (eQTL) of the target genes 24.
The Notch family protein-related gene regulation network is complex. As Notch1 and Notch2 target both oncogenes and TSGs, it is hard to define them as oncogenes or TSGs. Nevertheless, as they have different target gene profiles they may play distinct roles in pancreatic cancer.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by The National Nature Science Foundation of China (grant no. 81372156) and a grant from the Major State Basic Research Development Program (2010CB529400).
References
- 1.Bray SJ. Notch signaling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006 Sep;7(9):678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 2.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med. 2011 Sep 26;208(10):1931–5. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullendore ME, Koorstra JB, Li YM. et al. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009 Apr 1;15(7):2291–301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011 Sep;138(17):3593–612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 5.Fan X, Mikolaenko I, Elhassan I. et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004 Nov 1;64(21):7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto Y, Maitra A, Ghosh B. et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003 Jun;3(6):565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 7.Mazur PK, Einwächter H, Lee M. et al. Notch2 is required forprogression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2010 Jul 27;107(30):13438–43. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009 Apr 17;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabliauskaite K, Hehl AB, Seleznik GM. et al. p21(WAF1) (/Cip1) limits senescence and acinar-to-ductal metaplasia formation during pancreatitis. J Pathol. 2015 Feb;235(3):502–14. doi: 10.1002/path.4440. [DOI] [PubMed] [Google Scholar]
- 10.Inuzuka H, Guggino G, Monaco R. et al. FBXW7 and USP7 regulate CCDC6 turnover during the cell cycle and affect cancer drugs susceptibility in NSCLC. Oncotarget. 2015 May 20;6(14):12697–709. doi: 10.18632/oncotarget.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H, Yang R, Zheng Z. et al. MetMAb,the one-armed 5D5 anti-c-Met antibody, inhibits orthotopic pancreatic tumor growth and improves survival. Cancer Res. 2008 Jun 1;68(11):4360–8. doi: 10.1158/0008-5472.CAN-07-5960. [DOI] [PubMed] [Google Scholar]
- 12.Morra F, Luise C, Merolla F, Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J Natl Cancer Inst; 2014. Oct 11;106(12) [DOI] [PubMed] [Google Scholar]
- 13.Merolla F, Pentimalli F, Pacelli R. et al. Involvement of H4(D10S170) protein in ATM-dependent response to DNA damage. Oncogene. 2007 Sep 13;26(42):6167–75. doi: 10.1038/sj.onc.1210446. [DOI] [PubMed] [Google Scholar]
- 14.Choi HS, Yim SH, Xu HD. et al. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymaltransition in human hepatocellular carcinoma. BMC Cancer. 2010 Apr 1;10:122. doi: 10.1186/1471-2407-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbone C, Melisi D. NF-κB as a target for pancreatic cancer therapy. Expert Opin Ther Targets. 2012 Apr;16(Suppl 2):S1–10. doi: 10.1517/14728222.2011.645806. [DOI] [PubMed] [Google Scholar]
- 16.Prigent M, Barlat I, Langen H. et al. IkappaBalpha and IkappaBalpha/NF-kappa B complex are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000 Nov 17;275(46):36441–9. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 17.Wilson W 3rd, Baldwin AS. Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res. 2008 Oct 1;68(19):8156–63. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimasaki T, Ishigaki Y, Nakamura Y. et al. Glycogen synthase kinase 3β inhibition sensitizes pancreatic cancer cells to gemcitabine. J Gastroenterol. 2012 Mar;47(3):321–33. doi: 10.1007/s00535-011-0484-9. [DOI] [PubMed] [Google Scholar]
- 19.Lamba S, Felicioni L, Buttitta F. et al. Mutational profile of GNAQQ209 in human tumors. PLoS One. 2009 Aug 31;4(8):e6833. doi: 10.1371/journal.pone.0006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffy MJ, Synnott NC, McGowan PM. et al. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014 Dec;40(10):1153–60. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Kurosaki T, Suzuki M, Enomoto Y, Nishimatsu H, Arai T, Sawabe M, Hosoi T, Homma Y, Kitamura T. Significance of common variants on human chromosome 8q24 in relation to the risk of prostate cancer in native Japanese men. BMC Genet. 2009 Jul 14;10:37. doi: 10.1186/1471-2156-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Suzuki M, Arai T, Sawabe M, Enomoto Y, Nishimatsu H, Kume H, Homma Y, Kitamura T. A replication study examining three common single-nucleotide polymorphisms and the risk of prostate cancer in a Japanese population. Prostate. 2011 Jul;71(10):1023–32. doi: 10.1002/pros.21317. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M, Liu M, Kurosaki T, Suzuki M, Arai T, Sawabe M, Kasuya Y, Kato M, Fujimura T, Fukuhara H, Enomoto Y, Nishimatsu H, Ishikawa A, Kume H, Homma Y, Kitamura T. Association of rs6983561 polymorphism at 8q24 with prostate cancer mortality in a Japanese population. Clin Genitourin Cancer. 2011 Sep;9(1):46–52. doi: 10.1016/j.clgc.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Wu X, Huang Y, Chen W, Brand RE, Killary AM, Sen S, Frazier ML. Identification of genetic variants predictive of early onset pancreatic cancer through a population science analysis of functional genomic datasets. Oncotarget. 2016 Jul 29. doi: 10.18632/oncotarget.10924. [Epub ahead of print] PubMed. doi: 10.18632/oncotarget.10924. PMID: 27486767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.