Abstract
Recent studies demonstrated that long non-coding RNAs (lncRNAs) deregulated in many cancer tissues including nasopharyngeal carcinoma (NPC) and had critical roles in cancer progression and metastasis. In this study, we aimed to assess a lncRNA LINC01420 expression in NPC and explore its role in NPC pathogenesis. Our research revealed that the expression level of LINC01420 in NPC tissues were higher than nasopharyngeal epithelial (NPE) tissues. Moreover, NPC patients with high LINC01420 expression level showed poor overall survival. Knockdown LINC01420 inhibited NPC cell migration and invasion in vitro. In summary, LINC01420 may play a critical role in NPC progression and may serve as a potential prognostic biomarker in NPC patients.
Keywords: lncRNAs, nasopharyngeal carcinoma
Introduction
Head and neck cancer includes carcinomas arising from the mucosal epithelia of the head and neck region as well as various cell types of salivary glands and the thyroid 1. Nasopharyngeal carcinoma (NPC) is a kind of head and neck cancers, derived from the epithelial lining of the nasopharynx 2. NPC is rare (annual incidence, < 1/100,000) in most parts of the world. However, it is endemic in regions such as southern China and Southeast Asia, where the annual incidence ranges from 3 to 30 per 100,000 persons. Epstein-Barr virus, environmental influences and heredity each play important roles in NPC development 3-10. Undifferentiated carcinoma is the most common histological type of NPC and is associated with advanced local regional disease at diagnosis and a high incidence of distant metastasis. Distant relapse remains the major cause of treatment failure in NPC 11-17.
Long noncoding RNAs (lncRNAs) are non-protein-coding transcripts that are > 200 nucleotides in length and reside in the nucleus or cytoplasm 18. Although the function and mechanism of most lncRNAs remain unknown, accumulated evidence suggests that lncRNAs play important roles in the transcriptional, epigenetic, and post-transcriptional regulation of gene expression. More and more evidence has shown that lncRNAs are capable of influencing various cellular processes such as proliferation, cell cycle progression, cell growth, apoptosis and metastasis 19-22.
To find functional lncRNAs in NPC, we analyzed 2 previously published online datasets to find dysregulated lncRNAs in head and neck cancer. One novel lncRNA, LINC01420, was significantly overexpressed in the two head and neck cancer datasets. Then we examined the expression of LINC01420 in NPC tissues and demonstrated that LINC01420 might play a critical role in NPC progression and prognosis as a potential prognostic biomarker.
Materials and methods
Tissue samples
Two sets of tissue samples were collected for this study: Set 1 contained 26 NPC and 10 non-tumor NPE biopsies to verify LINC01420 expression with real-time PCR; Set 2 included 110 paraffin-embedded NPC tissue samples for in situ hybridization to confirm the expression of LINC01420. All tissue samples were collected from newly diagnosed NPC patients without any treatment at the Xiangya Hospital and the Affiliated Cancer Hospital of Central South University (Changsha, China). All the samples were handled according to the ethical and legal standards and approved by the Research Ethics Committee of the Xiangya Hospital and the Affiliated Cancer Hospital of Xiangya school of Medicine. All patients provided informed consent before surgery. After these specimens were confirmed by histopathological examination, these patients had received routine radiotherapy, and all of them were sensitive to radiotherapy 23.
RNA extraction and quantitative real-time PCR analyses (qRT-PCR)
Tissue RNA isolation and amplification were performed as our laboratory described previously 24-26. Cell RNA was extracted using TRIzol reagent (Invitrogen, Carlbad, CA, USA). For qRT-PCR, RNA was reverse transcribed to cDNA by using a PrimeScript RT reagent Kit (Takara, Dalian, China). qRT-PCR was performed using a SYBR_Premix ExTaqⅡ kit (Takara, Dalian, China) in the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) to determine the relative expression levels of target genes. The sequences of qRT-PCR primers: LINC01420: forward primer 5'-CACTCTACCCTCCGCACC-3' and reverse primer 5'-AGGAAGTGAAATCGTGCTGA-3'; β-actin: forward primer 5'-TCACCAACTGGGACGACATG-3' and reverse primer 5'-GTCACCGGAGTCCATCACGAT-3'; β-actin was used as reference and normalization control.
Data mining and analysis
To find functional lncRNAs in NPC, we downloaded two head and neck GEO expression data from GEO database: GSE6791 and GSE30784 (Affymetrix Human Genome U133 Plus 2.0 platform). We used Significant Analysis of Microarray (SAM) software to analyze the expression of lncRNAs between the non-tumor NPE biopsies and NPC tissue samples in the two datasets. The cut off value for differentially expressed lncRNA was set at ≥1.2-fold change and the false discovery ratio (FDR) was < 0.05.
In situ hybridization (ISH)
In situ hybridization was performed to detect LINC01420 expression in NPC specimens using three nucleotide probes from different LINC01420 regions.
The sequences of LINC01420 ISH probes: Probe-1: 5'-ATTTAAAGAGGGTGGGATTTGGTCAGAAACTCAC-3'; Probe-2: 5'-CAGGACTTGGACCTTCAACACGAAAAATTCAGAAT-3'; Probe-3: 5'-CACTTGAGAAAACCACTGTAGGACAAGAACAACAT-3'. The probes were synthesized and labeled with DIG-dUTP at the 3' and 5' end (Invitrogen, Shanghai, China). In situ hybridization was performed as previously described 12. All sections were independently scored by two pathologists who were blinded to the clinicopathological features and the clinical data.
Cell line and siRNAs
Human nasopharyngeal epithelium cell line NP69 and nasopharyngeal carcinoma HNE1, HK-1, HNE2 and 5-8F cell lines were maintained in our laboratory 27-33. Cell were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS, Invitrogen, Shanghai, China) in a humidified incubator with 5% CO2 at 37 ℃.
Three sequences of LINC01420 targeting siRNAs were: 5'-CAUCUCAGGUCUCUUGGCUUUGCCA-3'; 5'-GCGUUGGGAUUAUCCGGAAGGAACU-3'; and 5'-CCUCUGAGAUUUAAGGCCAUGCCCU-3'. Sequences of non-target scramble controls were provided by Invitrogen.
For gene knockdown, cells were seed overnight and transfected with either a mixture of three siRNAs that targeting LINC001420 or non-target scramble control siRNA (Invitrogen) using Lipofectamine RNAiMAX Reagent (Invitrogen) in OptiMEM medium (Invitrogen).
Cell migration and invasion assay
Cell migration was evaluated using a Transwell assay. A total of 2 × 104 cells in 200 µl of serum-free medium were added to the top chamber of the transwell (8 µm pore size, BD Biosciences, New Jersey, USA) at 24h after siRNA transfection. The bottom well contained growth medium with 20 % FBS. Cells were incubated at 37°C for 24 h. After 24 h, the cells that had migrated to the lower face of the filters were fixed with 100% methanol and stained with 0.5% crystal violet and counted 24, 25.
Matrigel invasion assays were performed as described previously 34. Briefly, filters coated with Matrigel in the upper compartment were loaded with 200µl serum-free medium containing 5 × 104 transfected cells, and the lower compartment filled with 20% FBS. After 24 h, migrated cells on the bottom surface were fixed with 100% methanol and counted after staining with 0.5% crystal violet.
Numbers of invaded cells were counted in six randomly selected fields under a microscope, and the average value was calculated. Each experiment was conducted in triplicate.
Statistical analysis
The expression levels of LINC01420 in NPC and NPE tissues were analyzed by unpaired t test. The chi-square and t tests were performed to assess the relationship between LINC01420 expression and clinicopathological features. Overall survival (OS) was calculated using the Kaplan-Meier method, and the results of the analysis were considered significant in a log-rank test if p < 0.05.
Results
LINC01420 is highly expressed in head and neck cancer
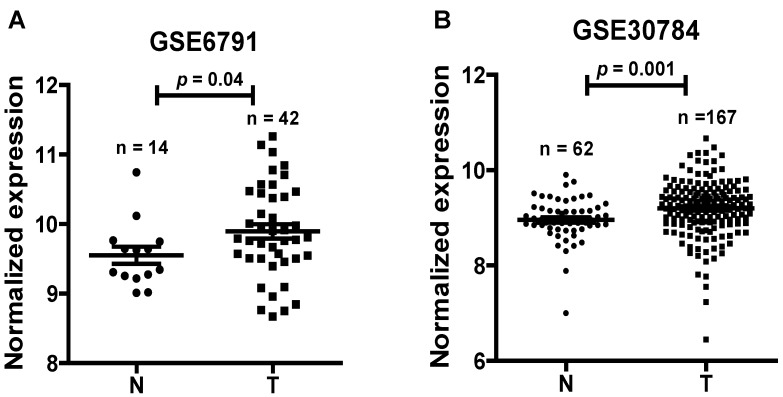
To find dysregulated lncRNAs in head and neck cancer, two online GEO datasets (GSE6791 and GSE30784) based on the Affymetrix HG_U133 Plus 2 arrays were reanalyzed. One novel lncRNA LINC01420 was significantly highly expressed in head and neck cancer compared with non-tumor tissues according to the GSE6791 (Normal, n = 14; Tumor, n = 42, p = 0.041, Fig. 1A) and GSE30784 (Normal, n = 62; Tumor, n = 167, p = 0.001, Fig. 1B) datasets.
Figure 1.
LINC01420 is highly expressed in head and neck cancer. LINC01420 significantly highly expressed in Gene Expression Profiling (GEP) datasets GSE6791 (Normal, n = 14; Tumor, n = 42, p = 0.041) and GSE30784 (Normal, n = 62; Tumor, n= 167, p = 0.001).
LINC01420 is overexpressed in NPC
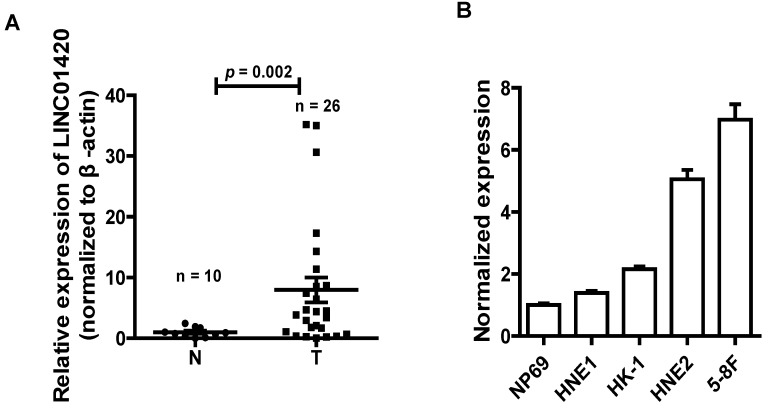
NPC is one kind of head and neck cancer that arise from cells in nasopharynx. To assess the role of LINC01420 in head and neck cancer, we examined the expression of LINC0142 in 26 NPC and 10 non-tumor nasopharyngeal epithelium samples using qRT-PCR. The results showed that LINC01420 was highly expressed in 26 NPC samples, compared with 10 non-tumor nasopharyngeal epithelium samples (p = 0.002, Fig. 2A). LINC01420 was also overexpressed in four NPC cell lines (HNE1, HK1, HNE2 and 5-8F) compared with NP69, a normal human nasopharyngeal epithelium cell line (Fig. 2B). These results suggested that LINC01420 was overexpressed in NPC.
Figure 2.
LINC01420 is highly expressed in NPC tissues and cell lines. (A) LINC01420 expression was higher in NPC samples (Tumor, n = 26) than that in non-cancerous tissues (Normal, n = 10). (B) LINC01420 expression was significantly increased in NPC cell lines (HNE1, HK1, HNE2, and 5-8F) compared with NP69, a normal human nasopharyngeal epithelium cell line.
Overexpression of LINC01420 predicts a poor prognosis and could be regarded as an independent predictor for overall survival in NPC
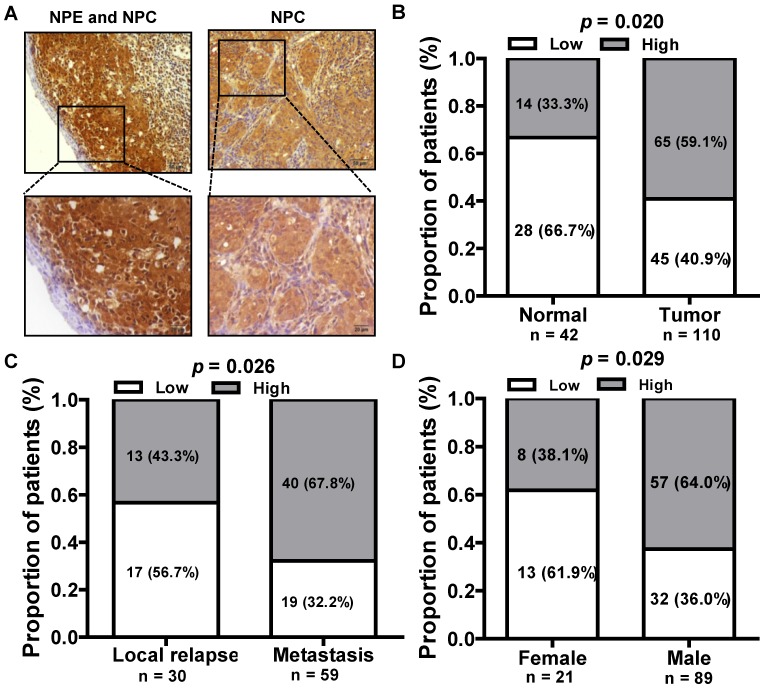
We next assessed the expression of LINC01420 in paraffin embedded NPC samples via in situ hybridization and found that LINC01420 expression was highly expressed in NPC cancer nests compared with surrounded nasopharyngeal epithelium (Fig. 3A and 3B). The correlation between LINC01420 expression and NPC clinicopathological features was also analyzed and the results demonstrated that the higher expression of LINC01420 was significantly correlated with NPC distant metastasis (p = 0.026, Fig. 3C) and the male NPC patients had a higher positive rate of LINC01420 than the female NPC patients (p = 0.029, Fig. 3D). However, LINC01420 expression did not correlate with other clinicopathological characteristics, such as age, clinical stage and relapse (data not show).
Figure 3.
The relationship between LINC01420 expression and clinicopathological features in NPC patients. (A) LINC01420 expression measured by in situ hybridization in paraffin embedded NPC biopsies. (B) LINC01420 expression was highly expressed in NPC cells (Tumor) compared to surrounded non-cancer NPE cells (Normal, p = 0.02). (C) High LINC01420 expression was associated with distant metastasis (p = 0.026). (D) More male NPC patients have high LINC01420 expression than female patients (p = 0.029).
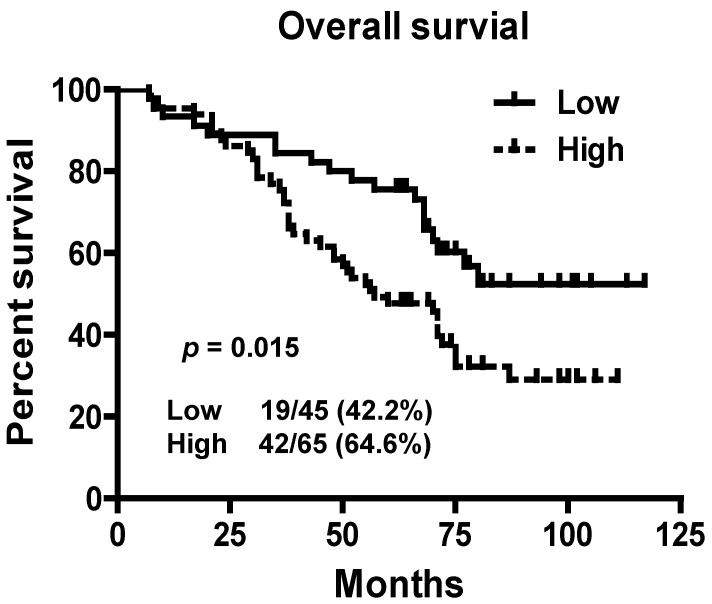
To assess prognostic value of LINC01420 expression in NPC patients, we examined the association between LINC01420 expression and overall survival using Kaplan-Meier analysis with the log-rank test. The expression of LINC01420 was negatively correlated with NPC patients' overall survival (p = 0.015, Fig. 4), NPC patients with high LINC01420 expression displaying lower overall survival. Taken together, these data indicated that high LINC01420 expression could be an independent risk factor for NPC patients.
Figure 4.
Kaplan-Meier survival curves of patients with NPC based on LINC01420 expression. Patients with high LINC001420 expression had a significantly unfavorable prognosis than those in low expression group (p = 0.015).
Knockdown of LINC01420 inhibited cell migration and invasion in NPC
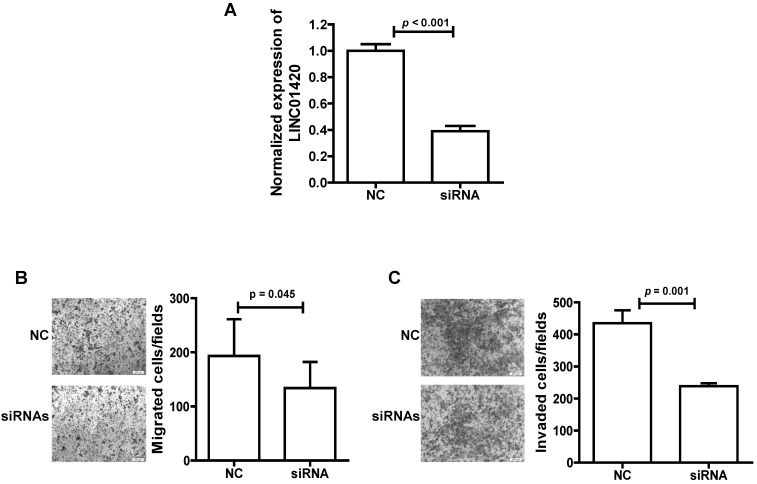
We also explored the effect of LINC01420 knockdown on NPC cell migration/invasion. First, the expression of LINC01420 was examined and successfully decreased after knockdown by a mixture of three siRNAs in 5-8F cells (p < 0.001, Fig. 5A). Then, cell migration capacity was evaluated using a Transwell assay without Matrigel. The results showed that knockdown of LINC01420 inhibited 5-8F cells migration capacity compared to the control group (p = 0.045, Fig. 5B). Matrigel invasion assays also demonstrated that 5-8F cells transfected with LINC01420 siRNAs had lower invasive capability (p = 0.001, Fig. 5C).
Figure 5.
LINC01420 Knockdown suppressed tumor cell migration and invasion in vitro. (A) siRNAs dramatically suppressed LINC01420 expression in 5-8F cells (p < 0.001). (B and C) 5-8F cells were grown and transfected with LINC01420 siRNAs, or scramble siRNA. Twenty-four hours after transfection, cells were subjected to a Transwell assay without or with Matrigel to measure migration or invasion capacity. LINC01420 knockdown significantly inhibited 5-8F cell migration (p = 0.045) and invasion (p = 0.01).
Discussion
Cancer is still the major cause of morbidity and mortality in most areas in the world 35. The incidence of NPC is high in endemic regions, with a 5-year overall survival rate of approximately 70% 36; thus, it is important to identify new molecular targets for the diagnosis, prognosis, and treatment of NPC.
LncRNAs participate in many biological processes and many studies have implicated that abnormal expression of lncRNAs is closely related to the occurrence and development of malignant tumors 37-40. Several lncRNAs were shown to be associated with NPC and indicate poor prognosis, including LINC00312 23, H19 41, NEAT1 42, HNF1A-AS1 43, HOTAIR 44, AFAP1-AS1 34 and lncRNA-LET 45. LINC01420 is a new lncRNA which is firstly reported in this paper.
In this study, we reported a novel functional lncRNA LINC01420 which was significantly high expressed in NPC samples and correlated with a poor prognosis of NPC patients. LINC01420 was poly (A)-positive and locates on chr X (p11.21). As we all know, there is a famous lncRNA, Xist (X inactive specific transcript), locating in the X inactivation center and its product is transcribed from the inactive X chromosome 46. It is typically expressed by all female somatic cells but lost in female breast, ovarian, and cervical cancer cell lines 47-49. In mammals, X chromosome inactivation (XCI) is triggered by Xist to equalize gene expression between the sexes 50. Studies demonstrated that Xist directly interacts with SHARP to silence transcription through HDAC3 51 and the histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression 52. While another research reported that Xist reduction in breast cancer upregulated AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression 53. Xist also play important role in cervical cancer 54, non-small cell lung cancer 55 glioblastoma 56 and so on. In our study, we found that LINC01420 is significantly high expressed in male NPC patients. The role of LINC01420 as an lncRNA in gender is worth of our further study.
In summary, the findings presented in this study suggested that LINC01420 expression was commonly high expressed in NPC and significantly correlated with the distant metastasis in NPC patients. Furthermore, high expression of LINC01420 was an independent poor prognostic for NPC patients. We also found that the LINC01420 knockdown significantly suppressed the invasive abilities of tumor cells, indicating that further investigation of LINC01420 might lead to the development of novel tumor therapies.
Acknowledgments
This study was supported in part by grants from The National Natural Science Foundation of China (81372907, 81301757, 81472531, 81572787, 81672683 and 81672993), the 111 Project (111-2-12), the Natural Science Foundation of Hunan Province (14JJ1010, 2015JJ1022 and 2016JC2035), and the Fundamental Research Funds for the Central Universities of Central South University (502210003).
References
- 1.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Z, Huang H, Zhang W. et al. Nasopharyngeal carcinoma: advances in genomics and molecular genetics. Sci China Life Sci. 2011;54:966–75. doi: 10.1007/s11427-011-4223-5. [DOI] [PubMed] [Google Scholar]
- 3.Xiong W, Zeng ZY, Xia JH. et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–4. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 4.Zeng Z, Zhou Y, Zhang W. et al. Family-based association analysis validates chromosome 3p21 as a putative nasopharyngeal carcinoma susceptibility locus. Genet Med. 2006;8:156–60. doi: 10.1097/01.gim.0000196821.87655.d0. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Z, Huang H, Huang L. et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315–26. doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 6.Yan Q, Zeng Z, Gong Z. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–82. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y, Li X, Zeng Z. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–36799. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Z, Fan S, Zhang X. et al. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2016;18:206–11. doi: 10.1007/s12094-015-1354-3. [DOI] [PubMed] [Google Scholar]
- 9.Xiao K, Yu Z, Li X. et al. Genome-wide Analysis of Epstein-Barr Virus (EBV) Integration and Strain in C666-1 and Raji Cells. J Cancer. 2016;7:214–24. doi: 10.7150/jca.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He B, Li W, Wu Y. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Z, Zhou Y, Xiong W. et al. Analysis of gene expression identifies candidate molecular markers in nasopharyngeal carcinoma using microdissection and cDNA microarray. J Cancer Res Clin Oncol. 2007;133:71–81. doi: 10.1007/s00432-006-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng ZY, Zhou YH, Zhang WL. et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol. 2007;38:120–33. doi: 10.1016/j.humpath.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Liao Q, Wei F. et al. LPLUNC1 inhibits nasopharyngeal carcinoma cell growth via down-regulation of the MAP kinase and cyclin D1/E2F pathways. PLoS One. 2013;8:e62869. doi: 10.1371/journal.pone.0062869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao Q, Zeng Z, Guo X. et al. LPLUNC1 suppresses IL-6-induced nasopharyngeal carcinoma cell proliferation via inhibiting the Stat3 activation. Oncogene. 2014;33:2098–109. doi: 10.1038/onc.2013.161. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Liao Q, Li X. et al. HYOU1, Regulated by LPLUNC1, Is Up-Regulated in Nasopharyngeal Carcinoma and Associated with Poor Prognosis. J Cancer. 2016;7:367–76. doi: 10.7150/jca.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Zeng Z, Fan S. et al. Evaluation of the prognostic value of TGF-beta superfamily type I receptor and TGF-beta type II receptor expression in nasopharyngeal carcinoma using high-throughput tissue microarrays. J Mol Histol. 2012;43:297–306. doi: 10.1007/s10735-012-9392-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Fan S, Zou G. et al. Lactotransferrin could be a novel independent molecular prognosticator of nasopharyngeal carcinoma. Tumour Biol. 2015;36:675–83. doi: 10.1007/s13277-014-2650-1. [DOI] [PubMed] [Google Scholar]
- 18.Gong Z, Zhang S, Zhang W. et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120–4. doi: 10.1007/s11427-012-4413-9. [DOI] [PubMed] [Google Scholar]
- 19.Gong Z, Yang Q, Zeng Z. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683–95. doi: 10.1007/s13277-015-4156-x. [DOI] [PubMed] [Google Scholar]
- 20.Gong Z, Zhang S, Zeng Z. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674. doi: 10.1371/journal.pone.0110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Xue D, Li Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991–1001. doi: 10.7150/jca.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Z, Bo H, Gong Z. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–37. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Huang C, Gong Z. et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545–54. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Chen P, Zeng Z, Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol; 2016. [DOI] [PubMed] [Google Scholar]
- 25.Liang F, Li Q, Li X. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046–56. doi: 10.3892/ijo.2016.3599. [DOI] [PubMed] [Google Scholar]
- 26.Xu K, Xiong W, Zhou M. et al. Integrating ChIP-sequencing and digital gene expression profiling to identify BRD7 downstream genes and construct their regulating network. Mol Cell Biochem. 2016;411:57–71. doi: 10.1007/s11010-015-2568-y. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Zhou H, Yang Y. et al. Lipopolysaccharide (LPS) regulates TLR4 signal transduction in nasopharynx epithelial cell line 5-8F via NFkappaB and MAPKs signaling pathways. Mol Immunol. 2007;44:984–92. doi: 10.1016/j.molimm.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Zheng H, Li M, Ren W. et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol Immunol. 2007;44:2221–7. doi: 10.1016/j.molimm.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Duan Z, Zheng H, Xu S. et al. Activation of the Ig Ialpha1 promoter by the transcription factor Ets-1 triggers Ig Ialpha1-Calpha1 germline transcription in epithelial cancer cells. Cell Mol Immunol. 2014;11:197–205. doi: 10.1038/cmi.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu D, Duan Z, Li M. et al. Heterogeneity of aberrant immunoglobulin expression in cancer cells. Cell Mol Immunol. 2011;8:479–85. doi: 10.1038/cmi.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu D, Zheng H, Liu H. et al. Immunoglobulin expression and its biological significance in cancer cells. Cell Mol Immunol. 2008;5:319–24. doi: 10.1038/cmi.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Zheng H, Duan Z. et al. Promotion of cell proliferation and inhibition of ADCC by cancerous immunoglobulin expressed in cancer cell lines. Cell Mol Immunol. 2012;9:54–61. doi: 10.1038/cmi.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao R, Liu Y, Wang H, BRD7 plays an anti-inflammatory role during early acute inflammation by inhibiting activation of the NF-small ka, CyrillicB signaling pathway. Cell Mol Immunol; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bo H, Gong Z, Zhang W. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–18. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bray F, Ren JS, Masuyer E. et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 36.Yau TK, Lee AW, Wong DH. et al. Treatment of Stage IV(A-B) nasopharyngeal carcinoma by induction-concurrent chemoradiotherapy and accelerated fractionation: impact of chemotherapy schemes. Int J Radiat Oncol Biol Phys. 2006;66:1004–10. doi: 10.1016/j.ijrobp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Riquelme I, Ili C, Roa JC, Long non-coding RNAs in gastric cancer: mechanisms and potential applications. Oncotarget; 2016. [Google Scholar]
- 38.Nobili L, Lionetti M, Neri A. Long non-coding RNAs in normal and malignant hematopoiesis. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Lin Y, Yang X. et al. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473:913–9. doi: 10.1016/j.bbrc.2016.03.150. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Li T, Wei G, The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol; 2016. [DOI] [PubMed] [Google Scholar]
- 43.Zhuang K, Wu Q, Jin CS. et al. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomark. 2016;16:291–300. doi: 10.3233/CBM-150567. [DOI] [PubMed] [Google Scholar]
- 44.Fu WM, Lu YF, Hu BG. et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7:4712–23. doi: 10.18632/oncotarget.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Q, Liu H, Li L. et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol. 2015;32:226. doi: 10.1007/s12032-015-0673-0. [DOI] [PubMed] [Google Scholar]
- 46.Brown CJ, Ballabio A, Rupert JL. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 47.Weakley SM, Wang H, Yao Q. et al. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–6. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawakami T, Zhang C, Taniguchi T. et al. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004;23:6163–9. doi: 10.1038/sj.onc.1207808. [DOI] [PubMed] [Google Scholar]
- 49.Benoit MH, Hudson TJ, Maire G. et al. Global analysis of chromosome X gene expression in primary cultures of normal ovarian surface epithelial cells and epithelial ovarian cancer cell lines. Int J Oncol. 2007;30:5–17. [PubMed] [Google Scholar]
- 50.Yildirim E, Kirby JE, Brown DE. et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–42. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McHugh CA, Chen CK, Chow A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–6. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvador MA, Wicinski J, Cabaud O. et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res. 2013;19:6520–31. doi: 10.1158/1078-0432.CCR-13-0877. [DOI] [PubMed] [Google Scholar]
- 53.Huang YS, Chang CC, Lee SS, Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren C, Li X, Wang T. et al. Functions and Mechanisms of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2015;25:566–9. doi: 10.1097/IGC.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 55.Tantai J, Hu D, Yang Y. et al. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–95. [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Y, Ma J, Xue Y. et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]